Optimizing Ribonucleoprotein (RNP) Complex Microinjection in Zebrafish Embryos: A Guide for Precision Genome Editing
The direct delivery of pre-assembled Cas protein-gRNA ribonucleoprotein (RNP) complexes into zebrafish embryos via microinjection represents a transformative approach for precision genome editing.
Optimizing Ribonucleoprotein (RNP) Complex Microinjection in Zebrafish Embryos: A Guide for Precision Genome Editing
Abstract
The direct delivery of pre-assembled Cas protein-gRNA ribonucleoprotein (RNP) complexes into zebrafish embryos via microinjection represents a transformative approach for precision genome editing. This method offers immediate activity, reduced off-target effects, and high editing efficiency, making it indispensable for creating accurate disease models and for functional genomics. This article provides a comprehensive resource for researchers, covering the foundational principles of RNP complexes, detailed microinjection protocols for techniques like prime editing and knock-in, advanced strategies to overcome efficiency challenges, and a comparative analysis with other delivery methods. By synthesizing the latest advancements, we aim to equip scientists with the knowledge to fully leverage RNP technology in zebrafish for biomedical and therapeutic discovery.
Understanding RNP Complexes: The Foundation of Precision Editing in Zebrafish
What are RNP Complexes? Defining the Cas Protein and Guide RNA Cargo
Ribonucleoprotein (RNP) complexes, fundamental assemblies of RNA and RNA-binding proteins, have emerged as a powerful cargo format for delivering CRISPR-based genome editing tools. This Application Note defines RNP complexes within the context of zebrafish embryo research, detailing their composition, advantages over alternative cargo formats, and providing a standardized protocol for RNP microinjection. We present quantitative data demonstrating the superior efficiency and reduced cytotoxicity of RNP delivery, along with essential reagent solutions and workflow visualizations to facilitate adoption in functional genomics and drug discovery research.
Definition and Basic Composition
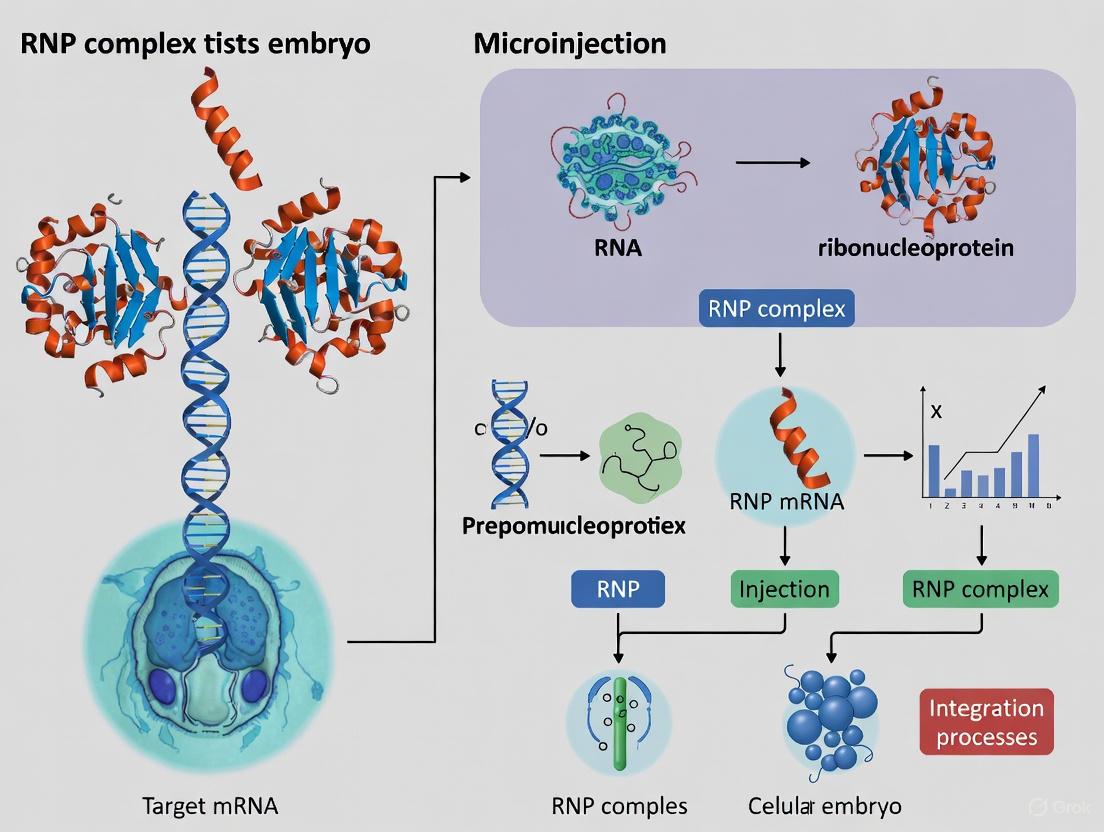
A ribonucleoprotein (RNP) complex is a fundamental biological structure formed through the association of RNA molecules with RNA-binding proteins (RBPs) [1]. These complexes play integral roles in numerous cellular processes, including transcription, translation, and gene expression regulation [2]. In the context of CRISPR genome editing, the term "RNP complex" specifically refers to the pre-assembled complex comprising the Cas nuclease protein (such as Cas9 or Cas12) bound to its corresponding guide RNA (gRNA) [3]. This active enzyme complex is capable of recognizing and cleaving specific DNA sequences complementary to the gRNA [4].
The formation of CRISPR RNP complexes is driven by molecular interactions between aromatic amino acid residues in the Cas protein and the RNA nucleobases, creating stacking interactions that stabilize the complex. Additionally, positively charged lysine residues in the helical regions of the Cas protein interact with the negatively charged phosphate backbone of the gRNA through electrostatic attraction [1] [2]. These interactions result in a stable RNP complex that functions as a programmable DNA-cutting machine.
RNP Complexes in Cellular Contexts
Beyond CRISPR applications, RNP complexes exist naturally as diverse intracellular compartments. These include stress granules, processing bodies (P-bodies), and other RNP granules that function in the storage, processing, degradation, and transportation of RNA transcripts [1]. In somatic cells, many RNP granules are highly specialized; for example, chromatoid bodies are found exclusively in male germ cells, while transport granules have so far been identified only in neurons and oocytes [1]. These natural RNP complexes are particularly important in cell types where post-transcriptional regulation is critical, such as in neurons where transcripts must be transported and stored in dendrites for synaptic formation and strengthening [1].
RNP Complexes as CRISPR Cargo: Advantages and Applications
Comparison of CRISPR Delivery Cargo Formats
CRISPR genome editing components can be delivered to cells in three primary formats: DNA plasmids, mRNA-gRNA combinations, and pre-assembled RNP complexes [5]. Each format presents distinct advantages and limitations for experimental and therapeutic applications.
Table 1: Comparison of CRISPR Delivery Cargo Formats
| Cargo Format | Composition | Key Advantages | Key Limitations |
|---|---|---|---|
| DNA Plasmid | Plasmid encoding both Cas9 and gRNA sequences | Inexpensive, easy to work with, can include selectable markers [4] | High off-target effects, random integration risk, cytotoxic, variable editing efficiency [4] |
| mRNA + gRNA | mRNA for Cas9 translation + separate gRNA | Reduced integration risk compared to DNA | Requires cellular translation, intermediate off-target risk, immune response potential |
| RNP Complex | Pre-assembled Cas9 protein + gRNA | Immediate activity, highest specificity, minimal off-target effects, no integration risk, reduced cytotoxicity [4] [6] | More expensive, limited shelf life, challenging delivery for some cell types |
Key Advantages of RNP Delivery
The pre-assembled RNP format offers several significant advantages for genome editing applications:
Reduced Off-Target Effects: RNP complexes have a shorter intracellular half-life (approximately 24 hours) compared to plasmid-based systems, which can persist for weeks. This limited activity window significantly decreases opportunities for erroneous editing, with studies demonstrating a 28-fold lower off-target to on-target mutation ratio for RNPs compared to plasmid DNA [4].
Elimination of Integration Risk: Unlike plasmid DNA, which may randomly integrate into the host genome at on- or off-target sites, RNP delivery completely avoids the risk of foreign DNA integration, enhancing safety for therapeutic applications [4].
Reduced Cellular Toxicity: RNP transfection demonstrates significantly higher cell viability compared to plasmid transfection. In various studies, RNP delivery resulted in at least 2x more viable colonies in embryonic stem cells relative to plasmid transfection [4].
Immediate Activity and High Efficiency: Since RNPs are pre-assembled and active immediately upon delivery, they bypass the need for transcription and translation steps required by plasmid-based systems. This results in faster editing onset and higher efficiency, particularly for homology-directed repair [6] [4].
Adaptability to Advanced Editing Systems: The RNP format has been successfully adapted for advanced CRISPR applications, including prime editing. Recent research demonstrates that PE7 protein complexed with La-accessible pegRNA forms efficient RNP complexes for precise editing in zebrafish embryos [7].
Quantitative Performance Data
Recent studies provide compelling quantitative evidence supporting the superiority of RNP delivery across multiple performance metrics.
Table 2: Quantitative Performance Comparison of RNP vs. Alternative Delivery Methods
| Performance Metric | RNP Complexes | DNA Plasmids | Experimental Context |
|---|---|---|---|
| Off-target/On-target Ratio | 28-fold lower [4] | Baseline | OT3-18 gene editing in human cells |
| Cell Viability | >80% [6] | Significant reduction, dose-dependent [4] | Immortalized cell lines |
| Editing Efficiency | Up to 50% integration efficiency [6] | Variable, typically lower [4] | CHO-K1 cells with cyclodextrin-based polymer delivery |
| Experimental Timeline | 50% reduction [4] | Baseline | Workflow comparison including cell sorting |
| Prime Editing Efficiency | 15.99% (6.81-11.46x improvement over PE2) [7] | Baseline | Zebrafish embryos with PE7 RNP |
RNP Microinjection in Zebrafish Embryos: Detailed Protocol
Zebrafish embryos represent an ideal model system for RNP-based genome editing due to their external development, optical clarity, and high fecundity. The one-cell stage microinjection protocol ensures that genetic edits are incorporated throughout the developing organism.
Reagent Preparation
RNP Complex Assembly:
- Combine purified Cas9 protein (or alternative editors like PE7) with synthetic sgRNA in a 2:1 molar ratio (protein:RNA) [8] [7].
- For prime editing applications: Use PE7 protein complexed with La-accessible pegRNA containing 3' polyU modifications to enhance stability and editing efficiency [7].
- Incubate the mixture at room temperature for 5-10 minutes to allow complete RNP complex formation [8].
- Add phenol red to a final concentration of 0.25% to visualize injection success [8].
- Adjust final concentration to 750 ng/μL Cas protein and 240 ng/μL gRNA/pegRNA using nuclease-free water [7].
Embryo Preparation:
- Place zebrafish breeding pairs in divided tanks the night before injection.
- Remove dividers in the morning and collect embryos immediately after spawning.
- Wash embryos with E3 medium containing methylene blue (0.0001%) to prevent fungal growth [8].
- Under a microscope, identify and select fertilized eggs (distinguished by dark yolk membrane) [8].
Microinjection Setup
Needle Preparation:
- Use a micropipette puller to create fine-tipped injection needles from 1.0 mm glass capillaries.
- Cut the needle tip with a razor blade at an angle to create an opening of approximately 10-15 μm.
- Load the prepared RNP solution into the needle using microloader tips.
Injection System Calibration:
Microinjection Procedure
- Align 20-30 dechorionated embryos along the trough of an injection plate filled with E3 medium.
- Position the injection needle at a 30-45° angle relative to the embryos.
- Gently penetrate the chorion and yolk membrane of the first embryo.
- Deliver 1-2 nL of RNP solution directly into the yolk cytoplasm at the one-cell stage [7].
- Retract the needle carefully and proceed to the next embryo.
- After injecting the entire batch, transfer embryos to fresh E3 medium with methylene blue.
- Maintain injected embryos and uninjected controls at 28.5°C in a humidified incubator [8] [7].
- Monitor embryonic development daily, removing any deceased or abnormally developing embryos.
- Change E3 medium daily until analysis or hatching.
Post-Injection Analysis
Genomic DNA Extraction:
- At 2 days post-fertilization (dpf), collect 6-8 normally developed embryos.
- Extract genomic DNA using commercial kits (e.g., QIAamp DNA Mini Kit) [7].
Editing Efficiency Assessment:
- Amplify target regions using site-specific primers in a first-round PCR.
- Add barcodes in a second-round PCR for multiplexed sequencing.
- Sequence using next-generation sequencing platforms (e.g., Illumina Novaseq X Plus) [7].
- Analyze sequencing data for precise editing rates and indel spectra.
Workflow for Zebrafish RNP Microinjection
The Scientist's Toolkit: Essential Research Reagents
Successful implementation of RNP-based genome editing in zebrafish requires specific reagents and equipment optimized for this model system.
Table 3: Essential Reagents for RNP Microinjection in Zebrafish Research
| Reagent Category | Specific Products/Components | Function and Application Notes |
|---|---|---|
| Core Editing Components | Cas9 protein (wild-type or high-fidelity variants), Cas12 protein, PE7 protein [7] | Engineered versions (e.g., high-fidelity Cas9) can reduce off-target effects; PE7 enhances prime editing efficiency |
| Guide RNA | Synthetic sgRNA, chemically modified sgRNA, La-accessible pegRNA with 3' polyU [7] [4] | Chemical modifications (methylated or phosphorothioate linkages) enhance stability and reduce degradation |
| Delivery Materials | Cationic cyclodextrin-based polymers (Ppoly), lipid nanoparticles [6] | Alternative delivery vehicles for challenging cell types; Ppoly shows >90% encapsulation efficiency and >80% cell viability |
| Embryo Handling | E3 embryo medium, methylene blue, low-melt agarose for mounting | E3 medium: 5mM NaCl, 0.17mM KCl, 0.33mM CaClâ‚‚, 0.33mM MgSOâ‚„; methylene blue prevents fungal growth |
| Microinjection Supplies | 1.0 mm glass capillaries, microloader tips, injection molds | Capillaries with internal filaments improve sample loading consistency |
| Analysis Reagents | DNA extraction kits (QIAamp), PCR reagents, barcoded sequencing primers, restriction enzymes | Barcoded primers enable multiplexed sequencing of multiple samples |
| Croverin | Croverin, MF:C21H22O6, MW:370.4 g/mol | Chemical Reagent |
| HG-12-6 | HG-12-6, MF:C29H27F3N6O2S, MW:580.6 g/mol | Chemical Reagent |
Advanced Delivery Systems for RNP Complexes
While direct microinjection remains the gold standard for zebrafish embryo delivery, several advanced delivery systems have been developed for RNP complexes that may have applications in other model systems or for specific zebrafish research needs.
Nanoparticle-Based Delivery
Cationic hyperbranched cyclodextrin-based polymers (Ppoly) have demonstrated remarkable efficiency in RNP delivery, achieving over 90% encapsulation efficiency while maintaining cell viability above 80% [6]. These nanosponges facilitate effective transport of RNP complexes to target cells, with one study reporting 50% integration efficiency in CHO-K1 cells, significantly outperforming commercial reagents [6].
Extracellular Vesicles
Engineered extracellular vesicles (EVs) represent a promising platform for RNP delivery, offering high biocompatibility, reduced immunogenicity, and inherent biological barrier crossing capabilities [9]. EV-mediated RNP delivery demonstrates particular promise for therapeutic applications where viral vectors pose safety concerns.
Virus-Like Particles (VLPs)
VLPs provide an empty viral capsid without viral genetic material, offering the cell entry advantages of viral vectors without associated safety concerns such as integration [5]. Although manufacturing challenges remain, VLPs enable transient delivery of CRISPR components while reducing the possibility of long-term expression and off-target editing [5].
RNP Delivery Methods and Applications
Troubleshooting and Optimization Guidelines
Successful implementation of RNP-based genome editing requires attention to potential challenges and optimization opportunities.
Low Editing Efficiency:
- Verify RNP complex assembly through electrophoretic mobility shift assays or other quality control measures.
- Optimize protein:RNA ratio (typically 2:1 molar ratio) for specific Cas variants.
- Ensure gRNA quality through HPLC or capillary electrophoresis purification.
- Increase injection concentration up to 1000 ng/μL Cas9 protein if needed, balancing with potential toxicity.
Poor Embryo Survival:
- Reduce injection volume to 1 nL or less for sensitive strains.
- Optimize injection timing strictly to the one-cell stage.
- Use sharper injection needles to minimize mechanical damage.
- Include proper controls to distinguish injection-related toxicity from RNP toxicity.
Variable Editing Outcomes:
- Standardize injection technique across all experimental groups.
- Use the same RNP preparation batch for comparative experiments.
- Include positive control gRNAs with known efficiency.
- Implement barcoded sequencing to track individual embryo outcomes.
RNP complexes represent the optimal cargo format for precise genome editing in zebrafish embryos, combining high efficiency with minimal off-target effects and cellular toxicity. The pre-assembled nature of RNP complexes enables immediate activity upon delivery, bypassing transcription and translation steps required by alternative formats. As CRISPR technologies continue to evolve, including the development of prime editors, base editors, and CRISPR-associated transposases, the RNP delivery format provides a versatile platform for implementing these advanced tools in zebrafish research. The protocols and guidelines presented in this Application Note provide researchers with a comprehensive framework for implementing RNP-based genome editing in zebrafish models, supporting advancements in functional genomics, disease modeling, and drug discovery.
Ribonucleoprotein (RNP) complex delivery represents a transformative approach for CRISPR-based genome editing in zebrafish embryos. An RNP complex is a pre-assembled unit composed of a Cas nuclease (such as Cas9) bound to its guide RNA (sgRNA), forming a fully functional editing machinery ready for direct cellular delivery [10] [11]. Unlike DNA-based methods (plasmids) or RNA (mRNA) that require cellular transcription and/or translation, RNPs are immediately active upon delivery and are rapidly degraded, minimizing prolonged exposure in cells [5] [11]. Within the context of zebrafish research, microinjection of RNPs into single-cell embryos has become the gold standard for achieving highly efficient and precise genetic modifications, enabling advanced functional genomics and disease modeling [12] [13] [14].
Core Advantages of RNP Delivery
The superiority of the RNP delivery method in zebrafish embryos is anchored in two principal advantages that address critical challenges in genome editing: precision and kinetics.
Reduced Off-Target Effects
The transient presence of the RNP complex in the cell is a key factor in enhancing editing precision. Because the Cas9 protein and sgRNA are pre-complexed and degrade quickly after delivery, the window for unintended genomic interactions is significantly shortened [11]. This reduction in off-target risk is a major reason for the high safety profile of RNP-based therapies, including the first FDA-approved CRISPR therapy, Casgevy [11].
Evidence from zebrafish models substantiates this advantage. When compared to Cas9-mediated homology-directed repair (HDR), prime editing delivered as RNP consistently induced fewer unwanted edits at target sites, demonstrating its higher relative precision [12]. Furthermore, a comprehensive study investigating structural variants in zebrafish found that microinjection of RNP complexes resulted in efficient on-target editing with a defined spectrum of off-target activity, allowing for careful experimental planning and validation [13].
Immediate Activity and High Editing Efficiency
RNP complexes bypass the need for intracellular transcription and translation, leading to rapid and efficient genome editing. The editing machinery is active immediately upon delivery, with maximum editing efficiency typically achieved within 24 hours [11]. This immediate activity is crucial in fast-developing systems like zebrafish embryos.
Quantitative data from zebrafish studies confirm high efficiency across various editing platforms. The table below summarizes somatic editing efficiencies achieved via RNP microinjection in zebrafish embryos.
Table 1: Editing Efficiencies of CRISPR Systems Delivered as RNP in Zebrafish
| Editing System | Type of Edit | Target Gene | Reported Somatic Efficiency | Citation |
|---|---|---|---|---|
| Prime Editor (PE2) RNP | Point Mutation (G→C/T) | Multiple zebrafish genes | PPE*: 0.28% - 4.01% (PE2) | [12] |
| Prime Editor (PE2) RNP | 5-bp Deletion | Three target sites | PPE: 4.13% - 33.61% | [12] |
| Prime Editor (PE2) RNP | 18-bp Insertion | Two target sites | PPE: Up to 18.00% | [12] |
| Prime Editor (PE2) RNP | Pathogenic Variants (tyr P302L, kras G12V) | tyr, kras | PPE: Up to 6.53% | [12] |
| Cytosine Base Editor (BE3) RNP | C:G to T:A conversion | Multiple targets | 9.25% - 28.57% | [14] [15] |
| AncBE4max RNP | C:G to T:A conversion | Multiple targets | ~3x higher than BE3 | [14] [15] |
| CBE4max-SpRY RNP | C:G to T:A conversion | Multiple targets | Up to 87% at some loci | [14] [15] |
*PPE: "Pure Prime Edits" - alleles with only the intended edit.
Application Notes and Protocols
Standardized Microinjection Protocol for RNP Complexes in Zebrafish
The following detailed protocol ensures consistent and high-efficiency genome editing in zebrafish embryos using RNP complexes.
Part 1: Preparation of Zebrafish Embryos
- Egg Collection: The night before injection, set up adult zebrafish in breeding tanks with dividers. Remove dividers the following morning to allow for mating. Collect eggs immediately after being laid using a strainer [16].
- Embryo Selection: Rinse eggs with egg water into a Petri dish. Remove unfertilized eggs and debris with a transfer pipette. It is critical that embryos are injected before the four-cell stage, ideally at the one-cell stage, to ensure distribution of the edit throughout the organism [16] [17].
- Alignment for Injection: Place a microscope slide in the inverted lid of a 100mm Petri dish. Use a transfer pipette to line up the eggs against the slide, forming a single column. Remove excess water with a Kimwipe to secure the embryos in place [16]. Alternatively, use an agarose injection mold to create troughs for holding embryos [17].
Part 2: RNP Complex Assembly and Needle Preparation
- RNP Formation: Pre-assemble the RNP complex by incubating purified Cas9 (or base editor/prime editor) protein with a molar excess of synthesized sgRNA (or pegRNA for prime editing) in a suitable buffer for 10-20 minutes at room temperature [12] [10]. A dye such as phenol red can be added to the injection mixture to visualize delivery [17].
- Needle Pulling: Using a micropipette puller, pull a 1.0mm OD glass capillary into two fine-tipped needles. Needles can be prepared in advance and stored in a 150mm Petri dish [16].
- Needle Loading: Backload the needle with 3 µL of the prepared RNP mixture using a microloader pipette. Gently shake the bolus toward the needle tip to minimize air bubbles [16].
Part 3: Microinjection System Calibration
- System Setup: Turn on the air source and microinjector. Insert the loaded needle into the microinjector, ensuring a tight seal. Position the micromanipulator for a wide range of movement [16] [17].
- Needle Trimming: Bring the needle tip into the microscope's view. Use sharp forceps to break the tip at a point that allows it to pierce the chorion and yolk but still deliver a consistent volume [16].
- Volume Calibration: Adjust the injection pressure and time on the microinjector. To calibrate the volume, inject into a drop of mineral oil on a micrometer slide. A bead with a diameter of 0.1 mm corresponds to approximately 500 pL. Adjust settings until the desired bead size is consistently produced. Ideal injection volumes should fill about 10% of the egg volume [16] [17].
Part 4: Embryo Microinjection
- Injection Procedure: Lower the needle toward the column of aligned eggs. In one smooth stroke, pierce the chorion and enter the yolk. Gently press the foot pedal to expel the RNP solution, visible as a punctuate spot in the yolk. Avoid injecting air bubbles or stretching the yolk, as this can be lethal [16] [17].
- Post-Injection Care: After injecting a column, use a gentle stream of egg water to transfer the embryos to a fresh Petri dish. Keep uninjected siblings as controls. Incubate embryos at 28.5°C, periodically replacing the egg water to prevent infection. Remove dead embryos over the following days [16].
The workflow below summarizes the RNP microinjection process.
The Scientist's Toolkit: Essential Research Reagents
Successful RNP microinjection requires a suite of specialized reagents and equipment. The following table details the key materials and their functions.
Table 2: Essential Reagents for RNP Microinjection in Zebrafish
| Item | Function/Description | Key Considerations |
|---|---|---|
| Cas Nuclease | Engineered protein (e.g., SpCas9, HiFi-Cas9, Base Editor) that performs the DNA cut or chemical conversion. | Select for high fidelity (e.g., HiFi-Cas9) to minimize off-targets. Base editors (BE) enable single-nucleotide changes without double-strand breaks [10] [14]. |
| sgRNA/pegRNA | Synthetic guide RNA that directs the Cas protein to the specific genomic target sequence. | Chemically modified sgRNAs (e.g., 2'-O-methyl) enhance stability and efficiency. For prime editing, a specialized pegRNA is required [12] [10]. |
| Microinjector | Apparatus that delivers precise, pressurized pulses to expel the RNP solution from the needle. | Allows calibration of injection volume (pressure and time) for consistency [16] [17]. |
| Micromanipulator | Device that holds and allows fine, three-dimensional movement of the injection needle. | Essential for precise control when targeting the tiny yolk of a zebrafish embryo [17]. |
| Glass Capillaries | Thin glass tubes that are heated and pulled to create fine, sharp injection needles. | Needle tip quality is crucial for piercing the chorion without damaging the embryo [16]. |
| Agarose Plates | Plates with molded grooves used to hold embryos stationary during the injection process. | Critical for aligning and stabilizing dozens of embryos for rapid, sequential injection [17]. |
| AM-5308 | AM-5308, MF:C26H35N5O5S, MW:529.7 g/mol | Chemical Reagent |
| BMS-986144 | BMS-986144, MF:C40H51F4N5O9S, MW:856.9 g/mol | Chemical Reagent |
Discussion and Future Perspectives
The implementation of RNP delivery in zebrafish research has set a new benchmark for precision and efficiency in genome editing. The combined advantages of reduced off-target effects and immediate activity make it an indispensable tool for generating robust and reliable functional genomic data and disease models [12] [11]. The protocol outlined here provides a reliable foundation, though the field continues to advance with the development of more sophisticated editors like near PAM-less cytidine base editors, which have achieved efficiencies of up to 87% in zebrafish when delivered as RNP [14] [15].
Future directions will focus on optimizing delivery methods further, including the use of lipid nanoparticles (LNPs) and engineered virus-like particles (eVLP) for in vivo RNP delivery, which could expand applications beyond microinjection [5] [11]. Furthermore, as new CRISPR systems and editors are discovered, their rapid testing and application in zebrafish via the RNP route will continue to accelerate translational research, bridging the gap between basic science and therapeutic development. The ongoing refinement of RNP-based protocols ensures that zebrafish will remain at the forefront of modeling human disease and validating genetic discoveries.
The Zebrafish Embryo as an Ideal Model System for RNP Microinjection
The zebrafish (Danio rerio) has emerged as a preeminent model organism in developmental biology and functional genomics, offering unique advantages for ribonucleoprotein (RNP) complex delivery. Its external fertilization, rapid embryonic development, and optical clarity during early stages provide an unparalleled system for microinjection-based genome editing techniques [15]. The high genetic similarity to humans, with approximately 70% gene homology, further positions zebrafish as a critical translational bridge between basic research and therapeutic development [18]. The application of RNP complexes—preassembled complexes of Cas protein and guide RNA—represents a transformative approach in zebrafish genome engineering, enabling precise genetic modifications with reduced off-target effects and minimal cytotoxicity compared to DNA-based delivery methods [19].
RNP microinjection into single-cell zebrafish embryos has revolutionized genetic engineering approaches by delivering the fully functional editing machinery directly to the site of action. This technique leverages the immediate availability of the nuclease complex, which is rapidly degraded after editing, creating a transient editing window that significantly minimizes off-target effects [19] [20]. The zebrafish embryo's large size and robust nature facilitate high survival rates post-injection, making it an ideal model for high-efficiency genetic screens and the generation of stable mutant lines. This application note details standardized protocols and quantitative outcomes for implementing RNP microinjection in zebrafish embryos, providing researchers with a comprehensive framework for advancing functional genomics and disease modeling.
Quantitative Analysis of Editing Efficiencies Across RNP Platforms
The efficacy of RNP microinjection in zebrafish embryos has been quantitatively demonstrated across multiple genome-editing platforms. Table 1 summarizes the performance metrics of various editing systems delivered as RNP complexes, highlighting the significant advancements in editing efficiency and specificity.
Table 1: Editing Efficiencies of RNP Complexes in Zebrafish Embryos
| Editing System | Target Loci | Editing Efficiency | Key Improvement | Reference |
|---|---|---|---|---|
| PE7 RNP + La-pegRNA | tyr, adgrf3b | Up to 15.99% | 6.81- to 11.46-fold over PE2 | [7] |
| CRISPR-RfxCas13d RNP | nanog, smad5 | High efficiency (maternal mRNAs) | Effective cytosolic mRNA knockdown | [21] |
| AncBE4max (CBE) | Various oncogenic mutations | ~90% efficiency with AncBE4max | ~3-fold increase over BE3 system | [15] |
| CBE4max-SpRY | Multiple loci | Up to 87% | Near PAM-less targeting capability | [15] |
The data demonstrate that contemporary RNP systems achieve remarkable efficiencies. Prime editing with the PE7 system and specialized pegRNAs shows substantial improvement over earlier generations, enabling precise base substitutions, insertions, and deletions without double-strand breaks [7]. Similarly, cytosine base editors like AncBE4max and CBE4max-SpRY achieve efficiencies previously thought impossible with earlier editing platforms, with the latter system bypassing traditional PAM sequence constraints to dramatically expand the targetable genome space [15].
Experimental Protocols for RNP Microinjection
RNP Complex Preparation and Quality Control
The preparation of functional RNP complexes requires precise assembly conditions. For prime editing applications, incubate PE7 protein at a concentration of 750 ng/μL with La-accessible pegRNA (240 ng/μL) to form stable RNP complexes [7]. For standard CRISPR-Cas9 editing, pre-complex purified Cas9 protein with chemically modified single-guide RNAs (sgRNAs) featuring 2'-O-methyl analogs and 3'-phosphorothioate linkages at the terminal nucleotides to enhance nuclease stability and editing efficiency [21]. Following complex assembly, incubate the mixture at 25-37°C for 10-15 minutes to ensure proper ribonucleoprotein formation before microinjection.
Quality control measures are essential for successful editing outcomes. Verify RNP complex integrity using native gel electrophoresis, which should show a mobility shift compared to free protein or RNA components. For functional validation, perform in vitro cleavage assays with target DNA fragments to confirm enzymatic activity before proceeding to embryo injections.
Zebrafish Embryo Collection and Microinjection
Embryo Collection and Preparation: Collect naturally spawned embryos within 15 minutes post-fertilization. Maintain embryos at 28.5°C in a humidified incubator and align them on an agarose injection mold (1.5-2.0%) in a Petri dish filled with embryo medium [7] [18]. Remove excess medium to prevent embryo floating during injection.
Microinjection Setup: Prepare injection needles from borosilicate glass capillaries using a pipette puller. Load 2-3 μL of RNP complex solution into the needle using a microloader tip. Calibrate injection volume to 2 nL per embryo using a microinjector and stereomicroscope; this typically corresponds to a droplet diameter of approximately 0.2-0.3 mm [7].
Injection Technique and Post-Injection Care: Position the injection needle at a 30-45° angle relative to the embryo surface. For single-cell injections, target the yolk cytoplasm near the blastomere at the one-cell stage. Following injection, transfer embryos to fresh embryo medium and incubate at 28.5°C. Monitor development daily, removing unviable embryos to maintain water quality.
Validation and Genotyping Protocols
At 2 days post-fertilization (dpf), extract genomic DNA from 6-8 normally developed embryos using commercial kits (e.g., QIAamp DNA Mini Kit) following manufacturer protocols [7]. For phenotypic screening of successful editing at the tyr locus, anesthetize larvae at 2 dpf with 0.03% Tricaine and image using standardized microscopy systems [7]. Reduced melanin pigmentation provides visual confirmation of successful editing.
For molecular validation, perform deep amplicon sequencing through a two-step PCR process. In the initial amplification, use locus-specific primers to amplify the target region from genomic DNA. In the second PCR, add barcodes and sequencing adapters to the amplicons. Pool equal amounts of PCR products and sequence using high-throughput platforms (e.g., Illumina Novaseq X plus). Analyze sequencing data to quantify editing efficiencies and identify specific sequence modifications at target loci [7].
Diagram Title: RNP Microinjection Workflow for Zebrafish Embryos
The Scientist's Toolkit: Essential Research Reagents
Successful implementation of RNP microinjection requires carefully selected reagents and equipment. Table 2 catalogs the essential components of the zebrafish RNP microinjection workflow, providing researchers with a comprehensive resource for experimental setup.
Table 2: Essential Research Reagents for Zebrafish RNP Microinjection
| Category | Specific Product/Component | Function & Application Notes |
|---|---|---|
| Editor Proteins | PE7, Cas9 nuclease, RfxCas13d, AncBE4max | Engineered nucleases for specific editing applications; PE7 enhances prime editing efficiency through La fusion [7] [21]. |
| Guide RNAs | La-accessible pegRNA, chemically modified sgRNAs | La-pegRNAs contain 3' polyU extensions for improved PE7 interaction [7]; chemically modified guides (2'-O-methyl, phosphorothioate) enhance stability [21]. |
| Microinjection Equipment | Microinjector, micromanipulator, borosilicate capillaries | Precision delivery systems for consistent 2 nL injection volumes into single-cell embryos [7]. |
| Zebrafish Strains | Wild-type AB, golden (slc24a5), nacre (mitfa) | Specific strains facilitate phenotypic screening; golden mutation reduces pigmentation for visual editing confirmation [22]. |
| Imaging & Analysis | Mueller matrix OCT, silver staining micro-CT | Advanced imaging for 3D quantitative phenotyping; silver staining specifically highlights melanin for pigment quantification [18] [22]. |
| BI-1950 | BI-1950, MF:C32H26Cl2FN7O3, MW:646.5 g/mol | Chemical Reagent |
| Saucerneol | Saucerneol, MF:C31H38O8, MW:538.6 g/mol | Chemical Reagent |
Additional critical reagents include embryo medium (E3 or Danieau solution), Tricaine (MS-222) for anesthesia, agarose for injection molds, and DNA extraction kits for downstream genotyping. The selection of appropriate zebrafish strains is particularly important, with pigmentation mutants like golden (slc24a5) providing visually screenable phenotypes for rapid assessment of editing efficiency [22].
Advanced Imaging and Phenotypic Characterization
Contemporary imaging technologies enable comprehensive phenotypic characterization following RNP-mediated genome editing. Mueller matrix optical coherence tomography (OCT) provides non-invasive, three-dimensional imaging of zebrafish development from 1 to 19 days post-fertilization, allowing quantitative analysis of organ volume and morphology without harmful radiation [18]. When combined with deep learning-based segmentation algorithms, this approach can automatically identify and quantify structures including eyes, spine, yolk sac, and swim bladder throughout development.
For specific quantification of melanin patterns resulting from editing of pigmentation genes like tyr, silver deposition micro-CT offers exceptional resolution and specificity. This technique adapts the histological Fontana-Masson staining principle for whole-organism imaging, enabling three-dimensional computational analysis of regional melanin content at cellular resolution [22]. The method has proven particularly valuable for quantifying subtle pigmentation phenotypes in wild-type and mutant zebrafish strains, providing superior context for studying phenotypic effects of genetic modifications.
Diagram Title: Phenotypic Validation Workflow Post-RNP Injection
Troubleshooting and Technical Considerations
Successful RNP microinjection requires attention to potential technical challenges. When editing efficiency is suboptimal, verify RNP complex quality through in vitro cleavage assays and ensure guide RNA design avoids problematic secondary structures. If embryo survival rates decrease, check injection needle sharpness to minimize mechanical damage and verify that injection volumes do not exceed 2 nL per embryo. For inconsistent editing outcomes across experiments, standardize the RNP complex assembly protocol with precise incubation times and temperatures, and use freshly prepared complexes for each injection session.
Technical optimization should include titration of RNP concentrations to balance efficiency and toxicity, with typical working concentrations of 750 ng/μL for editor proteins and 240 ng/μL for guide RNAs [7]. Timing is critical—injections should target one-cell stage embryos within 40 minutes post-fertilization to ensure incorporation of editing machinery into all daughter cells. For difficult-to-edit loci, consider dual-pegRNA strategies or chemical modifications to enhance guide RNA stability and performance [7] [21].
RNP microinjection in zebrafish embryos represents a powerful and precise methodology for genetic engineering, combining the physiological relevance of an in vivo vertebrate model with the specificity and reduced off-target effects of ribonucleoprotein delivery. The protocols outlined in this application note provide a robust framework for implementing this technology across diverse research applications, from functional genomics to disease modeling.
Future developments in zebrafish RNP technology will likely focus on expanding editing scope through novel Cas variants with relaxed PAM requirements, enhancing precision with reduced bystander activity, and implementing conditional editing systems for spatiotemporal control of genome modifications. As these technologies mature, the zebrafish model will continue to provide invaluable insights into gene function, disease mechanisms, and therapeutic development, solidifying its position as an ideal system for RNP-mediated genome editing.
Ribonucleoprotein (RNP) complexes are hybrids of RNA and RNA-binding proteins (RBPs) that form the operational core of modern genome editing technologies [1]. In zebrafish research, the direct delivery of pre-assembled Cas protein-gRNA RNP complexes via microinjection into one-cell stage embryos has become a preferred methodology [23]. This approach offers significant advantages over DNA or mRNA delivery, including immediate nuclease activity upon formation, reduced off-target effects due to rapid degradation of the complex, and elimination of potential plasmid integration into the host genome [24]. The transient nature of RNP activity is particularly valuable in zebrafish for generating crisp, mosaic mutations in F0 embryos and for precise genetic modeling of human diseases [25] [26].
Core Principles of RNP Assembly and Delivery
RNP Complex Formation
The assembly of functional RNP complexes for zebrafish microinjection is a deliberate process. For CRISPR-Cas9 systems, the complex typically consists of a purified Cas nuclease (e.g., Cas9, Cpf1) and a synthetically produced guide RNA (sgRNA or crRNA) [24]. For advanced prime editing systems, the complex comprises an engineered editor protein (e.g., PE7) and a prime editing guide RNA (pegRNA) [7]. The assembly process involves co-incubating the protein and RNA components in vitro to form stable complexes before microinjection. This pre-assembly is critical for protecting the RNA component from rapid degradation in the cellular environment and ensures immediate functionality upon delivery [23]. Research has demonstrated that pre-assembled LbCpf1-crRNA RNP complexes show dramatically increased activity compared to mRNA delivery of Cpf1, with significantly longer crRNA half-life in vivo [23].
Cellular Uptake Mechanisms
While the exact mechanisms of cellular uptake for microinjected RNPs in zebrafish embryos are not fully elucidated, the direct cytoplasmic injection into one-cell stage embryos bypasses major membrane barriers. The injected RNP complexes, being immediately functional, can rapidly access the nucleus upon nuclear envelope breakdown during cell division. This direct delivery method achieves high effective intracellular concentrations despite the technically challenging injection volumes of approximately 2 nL [7]. The timing of injection is critical, with microinjection performed at the one-cell stage to ensure distribution of the editing machinery to all daughter cells, enabling efficient somatic and germline editing [26] [23].
Quantitative Analysis of Editing Outcomes
Table 1: Efficiency of Different Genome Editing Systems in Zebrafish
| Editing System | Target Locus | Editing Efficiency | Key Outcomes | Reference |
|---|---|---|---|---|
| PE7 + La-pegRNA RNP | Various | Up to 15.99% | 6.81- to 11.46-fold improvement over PE2; successful generation of tyr P302L mutation with melanin reduction | [7] |
| PE2 RNP | crbn | 8.4% precise substitution | Higher precision score (40.8%) compared to PEn (11.4%) for single nucleotide substitutions | [26] |
| PEn RNP | crbn | 4.4% precise substitution | Higher indel formation but more efficient for longer insertions (3-30 bp) | [26] |
| LbCpf1 RNP | tyr, slc45a2 | ~99% germline transmission | Highly efficient mutagenesis in germ cells; temperature-dependent activity | [23] |
| Base Editor (AncBE4max) | Various | ~3-fold increase vs BE3 | Near PAM-less editing with efficiencies up to 87% at some loci | [15] |
Table 2: Common RNP Formulations for Zebrafish Microinjection
| Component | Concentration Range | Function | Modifications/Enhancements |
|---|---|---|---|
| Cas Protein (Cas9, Cpf1, PE) | 500-750 ng/μL | DNA binding and cleavage engine | Nickase variants (for PE); protein purification tags |
| Guide RNA (sgRNA, pegRNA) | 240-400 ng/μL | Target recognition and editing template | 5' and 3' modifications (methylated or phosphorothioate linkages); La-accessible structures for PE7 |
| Buffer Components | Varies | Complex stabilization | Nuclease-free water; optional salts and buffers |
Experimental Protocols
Protocol 1: Standard CRISPR-Cas9 RNP Microinjection for F0 Screening
This protocol is adapted from cataract gene evaluation studies in zebrafish [25].
Materials:
- Purified recombinant Cas9 protein
- Chemically synthesized sgRNA with target-specific spacer
- Microinjection system (e.g., Eppendorf Injectman NI2 with FemtoJet)
- Injection capillaries (e.g., Eppendorf Femtotips)
- One-cell stage zebrafish embryos
Procedure:
- RNP Complex Assembly:
- Resuspend sgRNA in nuclease-free water to a stock concentration of 1000 ng/μL.
- Dilute Cas9 protein to working concentration in injection buffer.
- Mix Cas9 protein (final concentration 500-750 ng/μL) with sgRNA (final concentration 240-400 ng/μL).
- Incubate at room temperature for 10-15 minutes to allow RNP complex formation.
Embryo Preparation and Microinjection:
- Collect one-cell stage zebrafish embryos within 30 minutes post-fertilization.
- Align embryos along the injection groove with cell cytoplasm accessible.
- Load assembled RNP complexes into injection capillary.
- Set injection parameters: 120-140 hPa injection pressure, 20 hPa compensation pressure, 0.1-0.3 second injection time.
- Inject approximately 2 nL of RNP complex solution directly into the yolk cytoplasm.
Post-Injection Care and Analysis:
- Incubate injected embryos at 28.5°C in egg water.
- Replace incubation medium daily.
- Assess editing efficiency at 2-4 days post-fertilization (dpf) via T7E1 assay or sequencing.
Protocol 2: Advanced Prime Editing RNP Microinjection
This protocol leverages optimized prime editing systems for precise base changes [7].
Materials:
- PE7 protein (fused PEmax with La peptide)
- La-accessible pegRNA with 3' polyU extension
- Modified primer binding site (PBS) and reverse transcription template
Procedure:
- Specialized RNP Assembly:
- Resuspend La-accessible pegRNA to 1000 ng/μL stock concentration.
- Combine PE7 protein (750 ng/μL final concentration) with La-accessible pegRNA (240 ng/μL final concentration).
- Co-incubate for 15 minutes at room temperature to form active RNP complexes.
Microinjection and Enhanced Incubation:
- Microinject 2 nL of PE7 RNP complex into one-cell stage embryos using standard parameters.
- Incubate injected embryos at 32°C to enhance editing efficiency [26].
- Maintain embryos at elevated temperature for 24-48 hours post-injection.
Efficiency Analysis:
- Extract genomic DNA from pools of 6-8 embryos at 2 dpf.
- Amplify target regions using barcoded primers for next-generation sequencing.
- Analyze sequencing data for precise edits using bioinformatic tools.
Signaling Pathways and Workflow Visualization
Experimental Workflow for Zebrafish RNP Editing
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Reagents for RNP-based Genome Editing in Zebrafish
| Reagent Category | Specific Examples | Function & Application Notes |
|---|---|---|
| Editor Proteins | SpCas9, LbCpf1, PE2, PE7, Base Editors (BE3, AncBE4max) | Engineered nucleases with varying PAM requirements and editing outcomes; PE7 shows enhanced efficiency with La-accessible pegRNAs [7] [23] [15] |
| Guide RNAs | sgRNA, crRNA, pegRNA, La-accessible pegRNA | Target recognition molecules; chemical modifications (methylation, phosphorothioate) enhance stability; structural optimizations improve efficiency [7] [24] |
| Delivery Materials | Microinjection capillaries, injection plates, embryo handling tools | Specialized equipment for precise cytoplasmic delivery at one-cell stage; proper needle calibration is critical for embryo viability [25] |
| Analysis Reagents | T7 Endonuclease I, DNA extraction kits, barcoded PCR primers, next-generation sequencing kits | Efficiency validation tools; amplicon sequencing with barcoded primers enables multiplexed analysis of editing outcomes [7] [26] |
| SS28 | SS28, MF:C18H20O3, MW:284.3 g/mol | Chemical Reagent |
| (S)-BI 665915 | (S)-BI 665915, MF:C24H26N8O2, MW:458.5 g/mol | Chemical Reagent |
RNP Editing Systems and Outcomes
Mastering the Microinjection Workflow: From RNP Preparation to Germline Transmission
Ribonucleoprotein (RNP) complex delivery via microinjection is a highly efficient method for precise genome editing in zebrafish embryos. This technique directly introduces pre-assembled complexes of Cas9 protein and guide RNA, leading to rapid and specific genetic modifications with reduced off-target effects compared to DNA or mRNA injection. This protocol details the preparation, purification, and microinjection of RNP complexes, specifically optimized for prime editing applications in zebrafish, providing researchers with a reliable framework for functional gene studies and genetic breeding in aquatic species [7].
Materials
Research Reagent Solutions
Table 1: Essential reagents and materials for RNP complex preparation and microinjection.
| Item | Specification/Concentration | Function/Application |
|---|---|---|
| Cas9 Protein | PE2, PE7, or PEn systems [7] [26] | Catalytic core of the editing system; creates single-strand or double-strand breaks. |
| pegRNA | Chemically synthesized, 1000 ng/μL stock [7] | Guides the Cas protein to the target locus and provides the template for reverse transcription. |
| Reaction Buffer | 100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2, 1 mM DTT, pH 7.9 [27] | Provides optimal ionic and pH conditions for RNP complex formation and stability. |
| Nuclease-Free Water | Not specified | Diluent and solvent for preparing RNP complexes. |
| Microinjection Needles | Not specified | Precision delivery of RNP complexes into zebrafish embryos. |
Equipment
- Thermal cycler or water bath (for incubation at 37°C)
- Microcentrifuge
- Microinjection apparatus
- Micromanipulator
Methodology
RNP Complex Assembly
The following procedure describes the assembly of RNP complexes for microinjection into one-cell stage zebrafish embryos [7] [27].
Prepare the Reaction Mixture: In a nuclease-free microcentrifuge tube, combine the following components to form a 5 μL reaction system [27]:
- 1 μL of 25 μM pegRNA or pre-annealed duplex guide RNA (dgRNA)
- 1 μL of 25 μM Cas9 protein (PE2, PE7, or PEn)
- 3 μL of Reaction Buffer (100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2, 1 mM DTT, pH 7.9)
Incubate to Form Complexes: Mix the components gently and incubate the reaction mixture at 37°C for 15 minutes [27]. This allows the Cas9 protein and guide RNA to form stable RNP complexes.
Final Injection Preparation: The resulting complex can be used directly for microinjection. The typical concentration for injection is 5 μM of the RNP complex [27]. Alternatively, for some prime editors like PE7, a working solution containing 750 ng/μL protein and 240 ng/μL pegRNA can be prepared [7].
Zebrafish Embryo Microinjection
- Embryo Collection: Collect and align healthy, one-cell stage zebrafish embryos on a microinjection mold.
- Microinjection: Using a micromanipulator and a pressurized microinjection system, inject approximately 1 nL to 2 nL of the prepared RNP complex solution into the yolk cytoplasm of each embryo [7] [27].
- Post-Injection Care: Following injection, transfer the embryos to embryo medium and incubate at 28.5°C for normal development [7].
Post-Injection Analysis
- Genomic DNA Extraction: At 2 days post-fertilization (dpf), collect 6-8 normally developed embryos and extract genomic DNA using a commercial kit (e.g., QIAamp DNA Mini Kit) [7].
- Efficiency Analysis: Amplify the target genomic region via PCR and analyze editing efficiency using next-generation sequencing (NGS) or, for preliminary assessment, a T7 Endonuclease I (T7E1) assay [7] [26].
Results and Data Analysis
Table 2: Quantitative comparison of prime editing efficiency in zebrafish using different systems.
| Prime Editor System | Editing Efficiency | Observed Mutations/Edits | Fold Improvement (vs. PE2) |
|---|---|---|---|
| PE2 | Baseline | Single-base substitutions [26] | - |
| PE7 + La-accessible pegRNA | Up to 15.99% at target loci [7] | Precise base substitutions, 6 bp insertions, 10 bp deletions [7] | 6.81 to 11.46x [7] |
| PEn (Nuclease-based) | 4.4% (substitution), higher for short insertions [26] | Higher indels alongside precise edits; more effective for inserting short DNA fragments (e.g., 3-30 bp) [26] | Not applicable |
Experimental Workflow
The following diagram summarizes the complete experimental workflow from RNP preparation to genotyping.
Technical Notes
- Enhancing Efficiency: The use of PE7 protein combined with La-accessible pegRNA (pegRNA with a 3' polyU tract) has been shown to significantly boost prime editing efficiency in zebrafish by improving the interaction between the editor and the RNA [7].
- Optimizing for Edit Type: While PE2 is more effective for single nucleotide substitutions, the PEn system can be more efficient for the precise insertion of short DNA fragments (e.g., 3-30 base pairs) [26].
- RNA Handling: Chemically synthesized pegRNAs can be modified with methylated or phosphorothioate linkages at the 5' and 3' ends to enhance stability against degradation [7].
Within the context of advanced ribonucleoprotein (RNP) complex research in zebrafish, mastering the microinjection technique is foundational. The delivery of CRISPR-based RNP complexes at the one-cell stage is a critical methodology for generating non-mosaic, genetically modified embryos in the F0 generation, enabling robust functional genomics and disease modeling [28] [25]. This protocol details the optimized parameters for timing, dosage, and injection site to ensure high editing efficiency and embryo survival, providing a standardized framework for researchers and drug development professionals.
The following table synthesizes key quantitative parameters from recent studies utilizing RNP complexes in zebrafish embryos.
Table 1: Optimized Microinjection Parameters for RNP Complexes at the One-Cell Stage
| Parameter | Optimal Value / Condition | Experimental Context & Key Findings | Citation |
|---|---|---|---|
| Injection Timing | One-cell stage (within ~45 minutes post-fertilization) | Essential to ensure RNP delivery before first cell division; minimizes mosaicism by allowing edits to propagate to all cells. | [16] [25] |
| Injection Volume | 500 pL - 2 nL (typically 1-2 nL) | A 500 pL droplet has a diameter of ~0.1 mm; volume should fill ~10% of the egg volume to ensure delivery without toxicity. | [12] [16] |
| Injection Site | Yolk cytoplasm | Standard site for delivery of RNP complexes into the embryo at the one-cell stage. | [7] [16] |
| RNP Concentration (Prime Editor) | 750 ng/µL PE protein + 240 ng/µL pegRNA | Using this ratio with PE7 and La-accessible pegRNA achieved up to 15.99% editing efficiency, a >6-fold improvement over PE2. | [7] |
| Incubation Temperature | 28.5 °C to 32 °C | 32 °C was shown to modestly improve prime editing efficiency for some targets compared to the standard 28.5 °C. | [12] [26] |
Experimental Protocol: RNP Complex Preparation and Microinjection
RNP Complex Formulation
This protocol is adapted from methods used for prime editing RNP complexes [7].
- Component Preparation: Resuspend chemically synthesized pegRNA or sgRNA in nuclease-free water to a high-concentration stock (e.g., 1000 ng/µL). Store at -80°C. Use purified editor protein (e.g., PE7, Cas9).
- Complex Assembly: Co-incubate the editor protein and guide RNA to form the RNP complex immediately before injection.
- Example Ratio: For prime editing, use 750 ng/µL of PE protein with 240 ng/µL of pegRNA [7].
- Procedure: Mix the calculated volumes of protein and RNA in a microcentrifuge tube. Incubate at room temperature for 10-15 minutes to allow the RNP complex to form.
Embryo Preparation and Microinjection
This procedure follows established microinjection techniques for zebrafish embryos [16].
- Embryo Collection: Within 20 minutes of the room lights turning on, collect freshly laid zebrafish eggs using a strainer. Rinse with egg water and transfer to a Petri dish.
- Embryo Alignment: Place a microscope slide in the inverted lid of a 100 mm Petri dish. Using a transfer pipette, line up the one-cell stage embryos against the slide, forming a single column. Remove excess water with a Kimwipe.
- Needle Preparation:
- Pull a 1.0 mm OD glass capillary needle using a micropipette puller.
- Backload 2-3 µL of the prepared RNP complex into the needle using a microloader pipette.
- Tap the needle gently to settle the solution and remove air bubbles.
- Break the needle tip with fine forceps to an opening that allows a consistent injection volume (e.g., 1 nL) without damaging the embryo.
- Calibration:
- Inject a droplet into mineral oil on a micrometer slide.
- A droplet diameter of 0.1 mm corresponds to a volume of approximately 500 pL [16].
- Adjust the injection pressure and time on the microinjector to achieve the desired volume (typically 1-2 nL).
- Microinjection:
- Using a micromanipulator, lower the needle and pierce the chorion and yolk of the embryo in one smooth motion.
- Depress the foot pedal to expel the RNP complex into the yolk cytoplasm.
- Quickly withdraw the needle.
- Work down the line of embryos, adjusting pressure as needed to maintain a consistent bead size.
- Post-Injection Care:
- After injecting a column, use a gentle stream of egg water to transfer the embryos to a fresh Petri dish.
- Incubate the injected embryos at the desired temperature (28.5°C to 32°C) [12].
- Replace the egg water periodically and remove dead embryos to maintain a healthy environment.
Workflow Visualization: From RNP Assembly to Analysis
The diagram below outlines the complete experimental workflow for RNP complex microinjection and validation in zebrafish embryos.
The Scientist's Toolkit: Essential Research Reagents
This table lists key reagents and materials required for performing RNP complex microinjection in zebrafish, as cited in the literature.
Table 2: Essential Reagents and Materials for RNP Microinjection
| Item | Function / Description | Example from Literature |
|---|---|---|
| Purified Editor Protein | The core editing enzyme (e.g., Cas9 nuclease, PE7). Delivered as protein for rapid activity and reduced off-target effects. | PE7 protein for prime editing [7]; Cas9 protein for knockout [28] [25]. |
| Chemically Synthesized Guide RNA | Targets the editor to specific genomic loci. Includes sgRNA for knockout or pegRNA for prime editing. Chemical modifications can enhance stability. | La-accessible pegRNA for PE7 [7]; chemically modified gRNAs for Cas13d [21]. |
| Glass Capillary Needles | Fine needles for embryo injection, pulled to a precise tip diameter. | 1.0 mm OD capillaries pulled with a Sutter Instrument P-1000 [29]. |
| Microinjector & Micromanipulator | System for precise needle positioning and controlled fluid delivery via air pressure. | Standard manual setup or automated robotic systems [16] [30]. |
| Agarose Injection Plates | Molded plates with grooves to hold and orient embryos during injection. | Custom-made plates for manual [16] or automated [30] injection. |
| DM4-d6 | DM4-d6, MF:C38H54ClN3O10S, MW:786.4 g/mol | Chemical Reagent |
| OSMI-2 | OSMI-2, MF:C26H25N3O7S2, MW:555.6 g/mol | Chemical Reagent |
Technical Notes and Troubleshooting
- Minimizing Mosaicism: The key to reducing mosaicism in F0 embryos is the precise injection of RNP complexes at the one-cell stage. High concentrations of RNP can favor consistent microhomology-mediated deletion patterns, leading to more uniform genotypes [28].
- Optimization is Key: The parameters listed are starting points. For new targets or different RNP systems (e.g., base editors, Cas13), empirical optimization of RNP concentration and injection volume may be necessary to maximize efficiency and minimize toxicity.
- Emerging Automation: Automated microinjection robots are now being developed that can standardize the injection process, achieving success and survival rates comparable to skilled manual injectors but with higher throughput and reproducibility [30] [29].
The precise microinjection of RNP complexes into the yolk of one-cell stage zebrafish embryos, using optimized timing, dosage, and formulation, is a powerful and reliable method. Adherence to this detailed protocol ensures high genome editing efficiency and robust experimental outcomes, solidifying the zebrafish model's critical role in functional genomics and preclinical drug development.
The application of prime editing in zebrafish represents a significant advancement in the field of precision genome engineering. As a transformative technology, prime editing enables precise base substitutions, insertions, and deletions without inducing double-strand DNA breaks (DSBs), overcoming key limitations of earlier CRISPR-Cas9 systems [7] [31]. While the potential for precise genetic modulation in aquatic species is substantial, the implementation in zebrafish has been historically constrained by low editing efficiency [32]. Recent developments with optimized ribonucleoprotein (RNP) complexes have dramatically enhanced editing efficiency, establishing PE7 RNP complexes as a powerful tool for functional gene studies and genetic breeding in aquatic species [7] [31].
The broader context of RNP complex microinjection in zebrafish embryos provides a critical foundation for these advances. RNP delivery offers distinct advantages over DNA or mRNA delivery, including reduced off-target effects, immediate activity, and rapid degradation that minimizes persistent editing activity [12] [25]. Within this methodological framework, the optimization of PE7 RNP complexes represents the current state-of-the-art for precise genome manipulation in zebrafish disease modeling and functional genomics research.
Technical Foundations: From PE2 to Enhanced PE7 Systems
The evolution from initial prime editors to the PE7 system reflects successive improvements in molecular design and functional efficiency. The original PE system comprises a nickase Cas9 (nCas9, H840A), an engineered reverse transcriptase (MMLV-RT), and a prime editing guide RNA (pegRNA) [7]. The mechanism involves nCas9 introducing a single-strand break at the target locus, generating a single-stranded DNA intermediate that hybridizes with the pegRNA's Primer Binding Site (PBS). The Reverse Transcription Template (RTT) is then reverse-transcribed, and the resulting DNA flap integrates into the genome via endogenous DNA repair mechanisms [31].
PE7 represents a state-of-the-art prime editing system developed through protein engineering approaches. Yan et al. identified La, a small-molecule-binding protein critical for prime editing, and fused it with PEmax to generate PE7 [7] [31]. This system works synergistically with La-accessible pegRNAs, which feature polyU modifications at the 3′ end that enhance interaction with the PE7 protein and significantly boost editing efficiency [31]. Comparative studies demonstrate that the PE7 system achieves 6.81- to 11.46-fold higher editing efficiency compared to the PE2 system in zebrafish embryos [32].
Table 1: Evolution of Prime Editing Systems in Zebrafish
| Editing System | Key Components | Editing Efficiency | Key Advantages | Limitations |
|---|---|---|---|---|
| PE2 | nCas9-RT + standard pegRNA | 1.4-2.3% [7] | Foundation for precise edits without DSBs | Low efficiency in zebrafish |
| PE3 | PE2 + nicking gRNA | 0.25-4.01% [12] | Modest improvement over PE2 | Increased byproduct edits |
| PE7 | PEmax-La + La-accessible pegRNA | Up to 15.99% [7] | 6.81-11.46× improvement over PE2; Reduced byproducts | Requires specialized pegRNA design |
Quantitative Performance Data
Comprehensive assessment of PE7 RNP performance across multiple genomic loci reveals consistently enhanced editing efficiency. In one systematic evaluation, researchers achieved up to 15.99% editing efficiency at target loci, with particularly strong performance observed at the adgrf3b locus, where 16.60% 6 bp insertions and 13.18% 10 bp deletions were recorded [7] [31]. This represents a 3.13-fold increase over PE2 performance at the same locus [32].
The efficiency of PE7 RNP complexes varies depending on edit type. For precise nucleotide substitutions, studies report efficiency ranges between 8.4-15.99% [7] [26]. For short insertions (3-18 bp), efficiencies of 0.10-18.00% have been documented, while defined deletions (5-10 bp) achieve notably higher efficiencies of 4.13-33.61% [12]. Temperature optimization also influences outcomes, with elevated incubation temperatures (32°C) generally yielding higher editing frequencies without proportional increases in undesired edits [12].
Table 2: PE7 RNP Editing Efficiency by Edit Type in Zebrafish
| Edit Type | Target Loci Tested | Efficiency Range | Optimal Conditions | Representative Outcome |
|---|---|---|---|---|
| Single-base substitutions | tyr, crbn, kras | 3.33-15.99% [7] [12] | PE7 + La-accessible pegRNA, 32°C | tyr P302L (CCC→CTC) with melanin reduction |
| Short insertions (3-18 bp) | 2 loci tested | 0.10-18.00% [12] | 10 nt PBS, C9E scaffold | Precise 3-bp stop codon insertion |
| Defined deletions (5-10 bp) | 3 loci tested | 4.13-33.61% [12] | RTT 13-15 nt, elevated temperature | 16.60% 6 bp insertion at adgrf3b |
| Complex edits | Multiple | Variable | Dual-pegRNA strategy | Pathogenic variant introduction |
Research Reagent Solutions
Implementation of optimized PE7 RNP editing requires specific reagents and formulations designed to enhance stability and efficiency:
- PE7 Protein: Purified prime editor protein (750 ng/μL) consisting of PEmax fused with La peptide, expressed and purified from E. coli or mammalian cells [7] [12].
- La-accessible pegRNAs: Chemically synthesized pegRNAs with 5′ and 3′ modifications (methylated or phosphorothioate linkages) and 3′ polyU extensions to enhance stability and PE7 interaction [7] [31].
- Nicking gRNAs (for PE3b strategy): Standard sgRNAs targeting the non-edited strand to enhance editing efficiency in some contexts [12].
- Microinjection Marker: Phenol red (0.5-2.5%) mixed with RNP complexes in nuclease-free water for visualization during delivery [8].
- Embryo Medium: E3 medium with methylene blue as fungicide for post-injection incubation at 28.5-32°C [8].
Experimental Workflow and Protocol
The following detailed protocol outlines the complete procedure for implementing PE7 RNP complex editing in zebrafish embryos, from complex preparation to analysis.
Step 1: RNP Complex Preparation
Formulate PE7 RNP complexes by combining PE7 protein at 750 ng/μL with La-accessible pegRNA at 240 ng/μL in nuclease-free water [7] [31]. Include 0.5 μL of 2.5% phenol red solution per 5 μL final volume for injection visualization [8]. Incubate the mixture at room temperature for 5-10 minutes to allow RNP complex formation before microinjection.
Step 2: Embryo Preparation and Microinjection
Collect one-cell stage zebrafish embryos and align them into the trough of a microinjection plate [8]. Using a microinjector with a pulled glass capillary needle, deliver 2 nL of the RNP complex solution into the yolk cytoplasm of each embryo [7] [31]. Practice injection with dye-only solution first to optimize technique and ensure greater than 90% embryo survival compared to uninjected controls [8].
Step 3: Post-injection Incubation and Screening
Following injection, transfer embryos to E3 medium with methylene blue and incubate at 28.5°C or optimized temperature of 32°C [12]. Remove any dead or abnormally developing embryos and change medium daily. For initial efficiency assessment, harvest 6-8 normally developed embryos at 2 days post-fertilization (dpf) for genomic DNA extraction using commercial kits [7].
Step 4: Editing Efficiency Analysis
Amplify target regions from extracted genomic DNA using barcoded primers specific to each target locus [7]. Prepare next-generation sequencing libraries and sequence using platforms such as Illumina Novaseq X Plus [7] [31]. Analyze sequencing data to distinguish between pure prime edits (only intended edit), impure prime edits (intended edit plus additional mutations), and byproduct edits (other mutations without intended edit) [12].
Step 5: Germline Transmission Assessment
Raise injected F0 embryos to adulthood and outcross with wild-type fish. Screen F1 progeny for inheritance of desired edits through targeted sequencing [12]. Studies report germline transmission rates of 7.1-12.3% for prime edits in zebrafish [12].
Troubleshooting and Optimization Strategies
Several key parameters require optimization to maximize PE7 RNP editing efficiency:
- Temperature Optimization: Elevating incubation temperature to 32°C consistently improves editing efficiency without substantially increasing byproduct edits [12].
- pegRNA Design: Utilize 10 nt PBS lengths and 13-15 nt RTT lengths with C9E pegRNA scaffold architecture for improved performance [12].
- RNP Complex Ratios: Maintain PE7 protein to pegRNA ratio at approximately 3:1 (750 ng/μL:240 ng/μL) for optimal complex formation [7].
- Chemical Modifications: Employ 5′ and 3′ modifications (methylated or phosphorothioate linkages) on pegRNAs to enhance stability against nucleases [7] [21].
Application Notes and Future Perspectives
The implementation of PE7 RNP complexes enables previously challenging genetic modifications in zebrafish. Researchers have successfully generated the tyr P302L mutation (CCC→CTC) associated with melanin reduction, a trait difficult to create with previous base editing technologies [7] [12]. This system also facilitates introduction of human disease-associated variants like KRAS G12V that require transversion mutations beyond the scope of conventional base editors [12].
Future applications of PE7 RNP technology in zebrafish research include genetic breeding of aquaculture species, functional characterization of non-coding regions, and sophisticated disease modeling through multiplexed editing approaches. The continued refinement of RNP delivery methods and pegRNA design promises to further enhance efficiency and expand the scope of precise genome editing in zebrafish and other aquatic species.
The optimized PE7 RNP protocol detailed herein provides researchers with a robust framework for implementing state-of-the-art prime editing in zebrafish embryos, enabling precise genetic modifications with significantly improved efficiency over previous approaches.
The precision modification of the zebrafish genome to create knock-in models is a cornerstone of functional genomics and disease modeling. While CRISPR-Cas9 has revolutionized genetic engineering, the efficient introduction of specific variants via homology-directed repair (HDR) remains challenging. The combination of preassembled Cas9-sgRNA ribonucleoprotein (RNP) complexes with single-stranded DNA (ssDNA) donor templates represents a significant methodological advancement, offering enhanced editing efficiency and reduced off-target effects compared to mRNA-based approaches. This application note details optimized protocols for generating knock-in zebrafish models using RNP complexes and asymmetric ssDNA donors, providing researchers with a robust framework for precise genetic modeling.
The RNP-ssDNA approach leverages the simultaneous microinjection of precomplexed Cas9 protein and sgRNA with synthetically produced ssDNA repair templates. This method capitalizes on several key advantages: RNP complexes mediate rapid DNA cleavage while minimizing off-target effects, and ssDNA donors serve as superior substrates for the HDR pathway compared to double-stranded DNA donors. Recent optimization efforts have focused on template design, including the implementation of asymmetric homology arms and strategic placement of silent mutations to prevent re-cleavage, yielding substantial improvements in knock-in efficiency [33].
Quantitative data from recent studies demonstrate the efficacy of this approach, with somatic knock-in events detected in 3.4% to 18.0% of sequencing reads, and perhaps more importantly, germline transmission achieved in 30-45% of injected adult zebrafish [33]. This efficiency facilitates the reliable establishment of stable genetic lines.
The following tables consolidate key performance metrics and design parameters from recent studies utilizing RNP and ssDNA donors in zebrafish.
Table 1: Knock-In Efficiency Metrics Using RNP and ssDNA Donors
| Target Gene | Modification Type | Somatic Efficiency | Germline Transmission Rate | Reference |
|---|---|---|---|---|
| ush2a | Point Mutation (C771F) | 3.4% of sequencing reads | 30% of adults | [33] |
| ripor2 | 12-bp Deletion | 18.0% of sequencing reads | 45% of adults | [33] |
| tyr | Point Mutation (P302L) | Up to 15.99% | Not Specified | [7] |
| BFP Reporter | ssDNA with HDR Module | Up to 90.03% (in cell culture) | Not Applicable | [34] |
Table 2: Optimized ssDNA Donor Design Parameters
| Design Parameter | Recommendation | Rationale | Reference |
|---|---|---|---|
| Strandedness | Single-stranded DNA (ssDNA) | Superior HDR efficiency and lower cytotoxicity compared to dsDNA. | [34] |
| Homology Arm Architecture | Asymmetric (e.g., 36-nt & 90-nt) | Improved knock-in efficiency; shorter arm hybridizes to displaced strand after RNP binding. | [33] |
| Optimal Interface for Modifications | 5' end of the ssDNA | The 5' end tolerates additional sequences better than the mutation-sensitive 3' end. | [34] |
| PAM Disruption | Include silent mutations | Prevents re-cleavage of the successfully edited allele by Cas9. | [33] |
Experimental Protocol
Step 1: sgRNA Design and Preparation
- Design: Select sgRNA targets where the Cas9 cut site is within 10 bp of the desired edit. Use available online tools for specificity checking and off-target prediction [33].
- Synthesis: Chemically synthesize sgRNA with specific stability-enhancing modifications: a 2'-O-methyl analog at the three terminal nucleotides at both the 5' and 3' ends, and phosphorothioate linkages between the three terminal nucleotides [15].
- Resuspension: Resuspend the synthesized sgRNA in nuclease-free water to a working concentration (e.g., 100 ng/µL) and store at -80°C.
Step 2: ssDNA Donor Template Design and Synthesis
- Architecture: Design an asymmetric ssDNA oligonucleotide with homology arms of 36 nucleotides and 90 nucleotides. The strand should be in the "antisense" orientation relative to the sgRNA target strand [33].
- Sequence Modifications:
- Incorporate the desired patient-specific or disease-relevant variant in the center.
- Include at least one additional silent mutation within the protospacer adjacent motif (PAM) sequence to prevent Cas9 from re-cleaving the successfully edited allele [33].
- Synthesis: Order the ssDNA donor as a high-purity, ultramer oligonucleotide.
Step 3: RNP Complex Assembly
- Complex Formation: Combine purified recombinant Cas9 protein with the synthetic sgRNA in a molar ratio of 1:2 to 1:5 (Cas9:sgRNA).
- Incubation: Incubate the mixture at 37°C for 10-15 minutes to allow for complete RNP complex formation.
- Add Donor: Following complex assembly, add the ssDNA donor template to the RNP mixture. A typical microinjection mixture final concentration is 750 ng/µL Cas9 protein, 240 ng/µL sgRNA, and 100-200 ng/µL ssDNA donor [7] [33].
Step 4: Microinjection into Zebrafish Embryos
- Embryos: Collect fertilized zebrafish eggs within the first cell cycle (within 1 hour post-fertilization).
- Injection: Microinject approximately 1-2 nL of the RNP + ssDNA donor mixture directly into the cell cytoplasm or yolk of the one-cell stage embryo [7] [33].
- Controls: Always include control groups injected with nuclease-free water or RNP complex without a donor template.
Step 5: Post-Injection Analysis and Germline Transmission
- Initial Screening: At 1-2 days post-fertilization (dpf), extract genomic DNA from a pool of injected embryos. Screen for editing events using PCR amplification of the target locus followed by next-generation sequencing (NGS) or high-resolution melt analysis (HRM) [33].
- Raising Founders: Raise the injected embryos (F0 generation) to adulthood.
- Identifying Germline Transmission: Outcross the adult F0 fish to wild-type partners. Collect genomic DNA from a pool of their offspring (F1 generation) at 1-2 dpf and screen for the presence of the knock-in allele using allele-specific PCR or sequencing. A germline transmission rate of 30-45% in F0 adults is achievable with this protocol [33].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for RNP-Mediated Knock-In
| Reagent / Material | Function / Role | Specifications & Notes |
|---|---|---|
| Recombinant Cas9 Protein | Catalyzes the double-strand break at the target genomic locus. | High-purity, endotoxin-free. Can be wild-type or nickase (D10A) for paired nicking strategies. |
| Synthetic sgRNA | Guides the Cas9 protein to the specific target DNA sequence. | Chemically modified with 2'-O-methyl and phosphorothioate bonds for enhanced stability in the embryo. |
| Asymmetric ssDNA Donor | Serves as the repair template for HDR to incorporate the desired edit. | Ultramer-length oligonucleotide, designed with asymmetric homology arms and silent PAM-disrupting mutations. |
| Nuclease-Free Water | Diluent for injection mixes. | Essential to prevent degradation of RNP complexes and the ssDNA donor. |
| Microinjection Apparatus | For precise delivery of reagents into zebrafish embryos. | Includes a micropipette puller, injector, and micromanipulator. |
| Ku70 Morpholino | Optional reagent to inhibit the NHEJ repair pathway. | Can be co-injected to bias DNA repair toward HDR, potentially increasing knock-in efficiency for some targets [33]. |
| FPFT-2216 | FPFT-2216, MF:C12H12N4O3S, MW:292.32 g/mol | Chemical Reagent |
| OSMI-3 | OSMI-3, MF:C32H35N3O9S2, MW:669.8 g/mol | Chemical Reagent |
Discussion and Technical Considerations
The protocol outlined above provides a reliable foundation for generating knock-in zebrafish models. Several factors are critical for success. First, the proximity of the Cas9 cut site to the intended edit is a major determinant of efficiency; designs where the cut site is within 10 base pairs are significantly more successful [33]. Second, the use of chemically modified sgRNAs enhances stability and protects against rapid degradation in the embryo, contributing to higher mutation rates [15].
A primary challenge remains the inherent competition between the efficient but imprecise NHEJ pathway and the precise but less efficient HDR pathway. Researchers can explore strategies to tilt this balance, such as the transient inhibition of key NHEJ factors like Ku70 using morpholino oligonucleotides, which has been shown to improve HDR outcomes in specific cases [33]. Furthermore, the development of "HDR-boosting" ssDNA donors, which incorporate specific protein-binding sequences (e.g., RAD51-preferred motifs) to recruit endogenous repair machinery, represents a promising chemical-free strategy to enhance precise editing efficiency, as demonstrated in cell culture models [34].
The microinjection of preassembled RNP complexes combined with rationally designed asymmetric ssDNA donors constitutes a current best practice for generating knock-in zebrafish models. This method offers a favorable balance of efficiency, precision, and practicality. By adhering to the detailed protocols for donor design, complex assembly, and embryo handling described in this application note, researchers can robustly model human genetic variants and advance studies in functional genomics and disease mechanisms.
The use of ribonucleoprotein (RNP) complexes for genome editing in zebrafish embryos represents a transformative approach in functional genomics and disease modeling. A significant challenge in this field lies in the effective validation of gene edits, starting from initial detection in somatic cells of the injected generation (F0) to the successful transmission of these edits through the germline to establish stable lines. This protocol details a streamlined workflow for somatic analysis and germline transmission assessment, leveraging the high efficiency and reduced off-target effects associated with RNP complex delivery [7] [12]. We provide quantitative data and standardized methodologies to enhance the reproducibility and success of genome editing projects in zebrafish.
Quantitative Editing Efficiencies of RNP-Based Technologies
The table below summarizes the performance of various genome editing technologies when delivered as RNP complexes into zebrafish embryos, providing benchmarks for expected somatic and germline outcomes.
Table 1: Editing Efficiencies of RNP-Complex-Based Technologies in Zebrafish
| Editing Technology | Target Type | Max Somatic Efficiency | Germline Transmission Rate | Key Advantages |
|---|---|---|---|---|
| PE7 Prime Editor [7] | Point Mutation (tyr P302L) | 15.99% | 8.3% (for precise edit) | 6-11x higher efficiency than PE2; precise base substitutions [7] |
| PE2 Prime Editor (RNP) [12] | Point Mutation (kras G12V) | 6.53% | 12.3% (for precise edit) | Installs transversions and edits in homopolymeric regions [12] |
| PE2 Prime Editor (RNP) [12] | 5 bp Deletion | Up to 33.61% | Not Specified | High efficiency for small, precise deletions [12] |
| Cas9 RNP (Knock-In) [35] | ~200 bp Tag Insertion | Not Specified | Up to 21% | Precise integration of composite tags using lssDNA donors [35] |
Experimental Workflow: From Injection to Stable Lines
The following diagram illustrates the comprehensive workflow for generating and validating F0 founder zebrafish, from microinjection of RNP complexes to the identification of germline-transmitting founders.
Workflow for F0 Founder Validation
Detailed Experimental Protocols
Protocol 1: RNP Complex Preparation and Microinjection
This protocol is optimized for delivering CRISPR-Cas9 or prime editor components as pre-assembled RNP complexes directly into the yolk cytoplasm of zebrafish embryos at the one-cell stage [7] [36].
Research Reagent Solutions
Table 2: Essential Reagents for RNP Complex Microinjection
| Reagent / Equipment | Function / Description | Example Specification / Notes |
|---|---|---|
| Cas9 or PE2/PE7 Protein | Catalytic core of the editing complex; introduces DSB or nick. | NLS-tagged, purified protein. Final concentration: 400-800 pg/nL [36]. |
| sgRNA or pegRNA | Guides the complex to the specific genomic target. | Chemically synthesized with stability modifications (e.g., 5'/3' methylated or phosphorothioate linkages) [7] [21]. |
| Microinjection Mold | Creates wells in agarose plate to position embryos for injection. | e.g., Adaptive Science Tools TU-1 [36]. |
| Phenol Red Solution | Visual dye to monitor injection volume and success. | Typically used at 0.5% (vol/vol) in the injection mix [36]. |
| Glass Capillaries | Needles for microinjection. | 1.0 mm diameter, pulled to a fine point [36]. |
Steps:
RNP Complex Assembly:
- Combine purified Cas9 (or PE2/PE7) protein with sgRNA (or pegRNA) in a 2:1 mass ratio (e.g., 400 pg/nL Cas9 to 200 pg/nL sgRNA) in a nuclease-free buffer [36].
- Optional: Add KCl to a final concentration of 1 M to improve complex solubility and cutting efficiency for some guides [36].
- Incubate the mixture at room temperature for 5-10 minutes to allow RNP complex formation [36].
Embryo Preparation:
- Set up natural pairwise matings of wild-type AB strain zebrafish the night before injection.
- On the day of injection, collect freshly fertilized embryos within the first hour post-fertilization.
- Arrange the dechorionated embryos into the troughs of a 1.5% agarose injection plate filled with E3 embryo media [36].
Microinjection:
- Back-load the RNP mixture into a prepared glass needle.
- Calibrate the injection volume to ~1 nL per embryo using a micrometer and a drop of mineral oil [36].
- Inject the 1 nL RNP complex solution directly into the yolk cytoplasm of each embryo at the one-cell stage.
- Maintain injected embryos at 28.5°C. For prime editing, incubation at 32°C has been shown to improve efficiency [12].
Protocol 2: Somatic Editing Analysis in F0 Embryos
This protocol describes the extraction of genomic DNA from pooled embryos and the quantification of editing efficiency via next-generation sequencing (NGS) of targeted amplicons.
Steps:
Sample Collection:
- At 2-5 days post-fertilization (dpf), collect 6-8 normally developed injected embryos into a single tube as a pooled sample. Include uninjected controls.
Genomic DNA (gDNA) Extraction:
Target Amplification & NGS Library Preparation:
- Perform the first round of PCR using primers that flank the target editing site to generate the initial amplicon.
- In a second PCR round, add Illumina-compatible barcodes and adapters to the amplicons using dual-indexing primers [7].
- Purify the final PCR products, quantify them, and pool equimolar amounts for sequencing on a platform such as Illumina NovaSeq [7].
Data Analysis:
- Process the NGS data using specialized software (e.g., CRISPResso2) to quantify editing outcomes [37].
- Categorize and report the frequency of:
- Precise Edits (PPE): Alleles containing only the intended edit [12].
- Imprecise Edits (IPE): Alleles with the intended edit plus additional unwanted mutations (e.g., insertions, deletions, pegRNA scaffold incorporations) [12].
- Byproduct Edits: Indels at the target site without the intended edit [12].
Protocol 3: Germline Transmission Testing
This protocol outlines the process for raising injected embryos to adulthood and screening their offspring to identify founders that transmit the genetic edit through their germline.
Steps:
Founder (F0) Rearing:
- Raise the injected embryos to sexual maturity (approximately 3 months) under standard aquaculture conditions [13].
Outcrossing and Progeny Screening:
- Outcross individual adult F0 fish to wild-type partners.
- Collect and raise a cohort of F1 progeny (e.g., 20-30 embryos) from each cross.
- At 2-5 dpf, pool a few F1 embryos from each clutch and extract gDNA for PCR-based genotyping (e.g., restriction fragment length polymorphism assay if the edit disrupts a site, or amplicon sequencing) to identify clutches with transmitted edits [12].
- For clutches positive for the edit, genotype individual F1 embryos to determine the exact transmission rate and isolate precisely edited animals.
Establishing Stable Lines:
- Raise F1 individuals that carry the desired edit to adulthood.
- Intercross these F1 fish or outcross them again to confirm stable germline transmission and to expand the colony, establishing a new, genetically modified zebrafish line.
Critical Considerations for Validation
- Minimizing Unintended Edits: Be aware that even precise editors can generate unintended outcomes. Prime editing, for instance, can produce "imprecise" edits with pegRNA scaffold incorporations and small insertions/deletions, especially near the 3' end of the edited flap [12]. NGS analysis is crucial for detecting these byproducts.
- Addressing Mosaicism in F0: Founder zebrafish are highly mosaic, meaning their somatic and germ cells can contain a variety of different editing outcomes [13]. This underscores the necessity of screening a sufficient number of F1 progeny from each F0 founder to accurately assess germline transmission potential.
- gRNA Design for High Penetrance: For F0 knockout studies (crispants), gRNA design is paramount. Prioritize gRNAs with high predicted efficiency scores from multiple algorithms (e.g., Doench, CRISPRScan) and those predicted to generate frameshift-inducing indels to maximize the penetrance of loss-of-function phenotypes [37].
Maximizing Efficiency: Proven Strategies to Overcome Common RNP Challenges
Boosting Prime Editing Efficiency with Engineered Editors (PE7) and La-accessible pegRNAs
Prime editing is a transformative "search-and-replace" genome editing technology that enables precise base substitutions, insertions, and deletions without inducing double-strand DNA breaks (DSBs) or requiring donor DNA templates [38]. This technology represents a significant advancement over earlier CRISPR-Cas9 and base editing systems, offering greater versatility while minimizing unwanted byproducts such as insertions and deletions (indels) [38] [15]. The core prime editing system consists of a fusion protein combining a Cas9 nickase (H840A) with an engineered reverse transcriptase (RT) from Moloney murine leukemia virus (M-MLV), programmed by a specialized prime editing guide RNA (pegRNA) that specifies both the target site and encodes the desired edit [38].
Despite its considerable potential, the application of prime editing in zebrafish has been limited by characteristically low editing efficiency, hindering its widespread adoption for functional genetic studies and genetic breeding in aquatic species [7]. Conventional prime editing systems like PE2 typically achieve only modest editing rates in zebrafish embryos, creating a critical bottleneck for researchers [7]. This protocol details the systematic optimization of prime editing in zebrafish through the combined use of the advanced PE7 editor and engineered La-accessible pegRNAs, delivered as ribonucleoprotein (RNP) complexes via microinjection into zebrafish embryos—an approach that has demonstrated substantial improvements in editing efficiency [7] [39].
The Evolution of Prime Editing Systems: From PE2 to PE7
The development of prime editing has progressed through several generations of increasingly optimized systems. The initial PE1 system established the proof-of-concept but exhibited limited editing efficiency [38]. PE2 emerged as a significant improvement through optimization of the reverse transcriptase fused to the Cas9 nickase, resulting in enhanced fidelity and efficiency of the editing process [38]. Further refinement produced PE3, which incorporates an additional guide RNA that nicks the non-edited DNA strand to encourage cellular repair machinery to use the newly synthesized edited strand as a template, thereby increasing editing incorporation [38].
PE7 represents the current state-of-the-art in prime editing technology. This system was developed through the identification of La peptide, a small RNA-binding protein that enhances prime editing efficiency when fused with the PEmax editor backbone [7]. The resulting PE7 editor, when combined with specially engineered La-accessible pegRNAs containing polyU sequences at their 3' end, demonstrates markedly improved performance in zebrafish models [7]. This enhancement is attributed to the improved interaction between the editor and pegRNA, leading to more efficient reverse transcription and incorporation of the desired genetic edits.
Table: Evolution of Prime Editing Systems
| Editor Version | Key Features | Primary Improvements |
|---|---|---|
| PE1 | Original nCas9-RT fusion | Proof-of-concept establishment |
| PE2 | Optimized reverse transcriptase | Enhanced fidelity and efficiency over PE1 |
| PE3 | Additional nicking sgRNA | Increased edit incorporation through strand repair |
| PE7 | La peptide fusion with PEmax, uses La-accessible pegRNAs | 6-11× efficiency improvement in zebrafish models |
Quantitative Efficiency Enhancements with PE7 and La-accessible pegRNAs
The combination of PE7 with La-accessible pegRNAs has demonstrated remarkable improvements in editing efficiency across multiple genomic loci in zebrafish. In comparative studies, this optimized system achieved editing efficiencies of up to 15.99% at target loci, representing a 6.81 to 11.46-fold enhancement over the conventional PE2 system [7] [39]. Furthermore, the system successfully mediated precise 6 base pair insertions and 10 base pair deletions at efficiency rates of 16.60% and 13.18% respectively at the adgrf3b locus, corresponding to a 3.13-fold increase over PE2 capabilities [7].
This substantial improvement in editing efficiency has enabled the generation of specific phenotypic traits that were previously challenging to achieve. Notably, researchers successfully introduced the tyr P302L mutation (CCC→CTC) in the tyrosinase gene, resulting in zebrafish with visibly reduced melanin pigmentation—a definitive marker of successful precise genome editing [7]. The reproducibility of these results across multiple target loci underscores the robustness of the PE7 and La-accessible pegRNA approach for diverse genetic modifications in zebrafish models.
Table: Prime Editing Efficiency Comparison in Zebrafish
| Target Locus | Edit Type | PE2 Efficiency | PE7 + La-accessible pegRNA Efficiency | Fold Improvement |
|---|---|---|---|---|
| Multiple Loci | Single-base substitutions | Baseline | Up to 15.99% | 6.81 to 11.46× |
| adgrf3b | 6 bp insertion | ~5.3% | 16.60% | ~3.13× |
| adgrf3b | 10 bp deletion | ~4.2% | 13.18% | ~3.13× |
| tyr | P302L point mutation (CCC→CTC) | Not efficiently achievable | Successfully generated melanin-reduced zebrafish | N/A |
Experimental Protocol: RNP Complex Preparation and Zebrafish Microinjection
Reagent Preparation
PE7 Protein: Utilize purified PE7 nuclease (750 ng/μL stock concentration) [7]. The PE7 editor consists of the La peptide fused to the PEmax backbone, which includes codon-optimized Cas9 nickase (H840A) and engineered M-MLV reverse transcriptase domains [7].
La-accessible pegRNAs: Chemically synthesize pegRNAs with 5′ and 3′ modifications (methylated or phosphorothioate linkages) to enhance stability [7]. Incorporate a 3′ polyU sequence (typically 10-15 nucleotides) to facilitate La peptide binding [7]. Resuspend lyophilized pegRNAs in nuclease-free water to a final stock concentration of 1000 ng/μL and store at −80°C until use [7].
Microinjection Buffer: Prepare a suitable buffer such as Tris-EDTA or nuclease-free phosphate-buffered saline to maintain complex stability during injection.
RNP Complex Assembly
Complex Formation: Combine PE7 protein (750 ng/μL final concentration) with La-accessible pegRNA (240 ng/μL final concentration) in a microcentrifuge tube [7].
Incubation: Allow the mixture to incubate at room temperature for 15-20 minutes to facilitate complete RNP complex formation.
Quality Control: Verify complex formation using native gel electrophoresis or other appropriate analytical methods if available.
Zebrafish Embryo Preparation and Microinjection
Embryo Collection: Collect wild-type AB strain zebrafish eggs and maintain at 28.5°C in a humidified incubator [7].
Microinjection Setup: Load approximately 2 nL of the prepared RNP complex into a fine needle microinjection capillary [7].
Injection Procedure: Carefully microinject the RNP complex into the yolk cytoplasm of one-cell stage zebrafish embryos [7]. For developmental stage synchronization, maintain injected embryos at 28.5°C [7].
Post-injection Monitoring: Assess embryo viability and development at 2 days post-fertilization (dpf) [7].
Molecular Mechanism of PE7 with La-accessible pegRNAs
The enhanced efficiency of the PE7 system with La-accessible pegRNAs stems from optimized molecular interactions at multiple stages of the prime editing process. The La peptide fusion in PE7 specifically recognizes and binds to the polyU sequence incorporated into the 3' end of La-accessible pegRNAs, stabilizing the pegRNA structure and facilitating more efficient recruitment of the editing machinery to target sites [7]. This interaction protects pegRNAs from degradation and promotes proper complex formation, increasing the likelihood of successful editing events.
At the genomic target site, the Cas9 nickase domain of PE7 introduces a single-strand break in the non-target DNA strand, exposing a 3'-hydroxyl group that serves as a primer for reverse transcription [38]. The reverse transcriptase domain then uses the RTT sequence embedded in the pegRNA as a template to synthesize a new DNA strand containing the desired edit [38]. The edited strand forms a DNA flap structure that competes with the original unedited flap for integration into the genome through cellular DNA repair mechanisms [38]. The stabilization provided by the La-polyU interaction increases the efficiency of reverse transcription and flap resolution, resulting in higher rates of precise edit incorporation [7].
Analysis and Validation of Editing Outcomes
Genomic DNA Extraction
At 2 days post-fertilization, collect 6-8 normally developed embryos from each experimental group and extract genomic DNA using commercial kits such as the QIAamp DNA Mini Kit, following manufacturer protocols [7]. Store extracted genomic DNA at −20°C for subsequent analysis.
Amplification and Sequencing
Target Amplification: Perform initial PCR amplification of the target region from genomic DNA using site-specific primers [7].
Library Preparation: Conduct a second round of PCR to add forward and reverse barcodes to the amplified products for multiplexed sequencing [7].
Next-Generation Sequencing: Pool equal amounts of barcoded PCR products and sequence commercially using platforms such as Illumina Novaseq X plus [7].
Data Analysis
Process sequencing data to examine pegRNA target sites for precise substitutions and indels. Calculate editing efficiency as the percentage of sequencing reads containing the desired modification compared to total reads. Compare experimental groups (PE7 with La-accessible pegRNAs) against appropriate controls (PE2 with standard pegRNAs) to quantify fold improvements.
Table: Essential Research Reagent Solutions
| Reagent/Category | Specific Example | Function in Protocol |
|---|---|---|
| Prime Editor Protein | PE7 nuclease (750 ng/μL) | Catalytic component for DNA nicking and reverse transcription |
| Guide RNA | La-accessible pegRNA with 3' polyU (240 ng/μL) | Target specification and edit template delivery |
| Delivery System | Microinjection apparatus | Precise RNP complex delivery to zebrafish embryos |
| Animal Model | Zebrafish (AB strain) embryos, one-cell stage | In vivo model for editing validation |
| DNA Extraction Kit | QIAamp DNA Mini Kit | High-quality genomic DNA isolation for analysis |
| Sequencing Platform | Illumina Novaseq X plus | High-throughput assessment of editing efficiency |
Troubleshooting and Technical Considerations
Several factors critically influence the success of PE7-mediated prime editing in zebrafish. PegRNA design remains paramount—ensure the reverse transcription template (RTT) and primer binding site (PBS) are optimized for length and sequence composition. The RTT should encode the desired edit with sufficient flanking homology (typically 10-15 nucleotides) to facilitate proper annealing and repair [38]. The polyU tract in La-accessible pegRNAs should be incorporated at the 3' end without disrupting essential pegRNA secondary structures.
RNP complex concentration and quality significantly impact editing efficiency. Maintain precise concentrations of PE7 protein (750 ng/μL) and pegRNA (240 ng/μL) in the injection mixture [7]. Avoid repeated freeze-thaw cycles of components, and use freshly prepared complexes whenever possible. Injection technique requires practice—target the yolk cytoplasm of one-cell stage embryos precisely, as improper injection can reduce embryo viability and editing efficiency [7].
The optimized prime editing system combining PE7 editors with La-accessible pegRNAs represents a substantial advancement for precise genome engineering in zebrafish models. The documented 6-11 fold improvement in editing efficiency over previous systems effectively addresses a critical limitation that has hindered prime editing applications in aquatic species [7] [39]. This protocol provides researchers with a comprehensive framework for implementing this technology to introduce a broad spectrum of genetic modifications—including point mutations, small insertions, and deletions—with significantly enhanced efficiency and precision.
The successful generation of zebrafish with specific phenotypic traits, such as the tyr P302L mutation resulting in reduced melanin pigmentation, demonstrates the practical utility of this system for creating customized animal models for functional genomics and genetic research [7]. As prime editing technology continues to evolve, further refinements in editor engineering, delivery methods, and pegRNA design will likely expand its applications in zebrafish research, opening new avenues for modeling human diseases, studying gene function, and developing improved traits in aquaculture species. The RNP-based delivery approach described herein offers particular advantage for reducing off-target effects and enabling rapid editing without persistent editor expression in cells.
Enhancing Knock-In Rates by Modulating DNA Repair Pathways (e.g., Ku70 Knockdown)
Precise genome editing in zebrafish relies on the efficient integration of exogenous DNA sequences via homology-directed repair (HDR). However, the inherent dominance of the non-homologous end joining (NHEJ) pathway severely limits knock-in efficiency. This Application Note details a synergistic strategy combining ribonucleoprotein (RNP) complex delivery with pharmacological modulation of the DNA repair machinery to suppress NHEJ and enhance HDR. We provide a validated protocol using PARP1 modulation to bias repair toward HDR, alongside a novel prime editing approach that operates independently of traditional double-strand break repair pathways, enabling precise edits with high efficiency in zebrafish embryos.
The microinjection of Cas9 protein:guide RNA ribonucleoprotein (RNP) complexes into zebrafish embryos has revolutionized genetic engineering in this model organism, offering high efficiency and reduced off-target effects compared to DNA-based methods [40]. A significant challenge in the field, however, is achieving high rates of precise knock-in of DNA sequences, a process essential for tagging genes, introducing disease-associated mutations, or creating reporter lines.
This hurdle exists because the cell's endogenous DNA repair machinery heavily favors the rapid and error-prone non-homologous end joining (NHEJ) pathway over the precise, template-dependent homology-directed repair (HDR) pathway [41]. The classical HDR-based knock-in strategy is further hampered by its low efficiency in zebrafish. Therefore, enhancing knock-in rates requires active intervention to modulate the DNA repair pathway choice at the Cas9-induced double-strand break (DSB) site.
This protocol outlines a dual-pronged approach grounded in a broader thesis on RNP microinjection in zebrafish. First, we detail the use of PARP1 modulation, a strategic method to suppress mutagenic NHEJ and create a cellular environment more permissive for precise editing [42]. Second, we incorporate advanced prime editing systems, which can install desired edits without creating double-strand breaks, thereby bypassing the competitive repair pathway problem altogether [7] [26].
Quantitative Analysis of Prime Editing Efficiency
The following table summarizes editing efficiencies achieved with different state-of-the-art prime editing systems in zebrafish, demonstrating significant improvements over baseline methods.
Table 1: Efficiency of Prime Editor Systems in Zebrafish
| Editing System | Key Features | Target Locus | Editing Type | Maximum Efficiency | Fold Improvement over PE2 | Citation |
|---|---|---|---|---|---|---|
| PE2 | Nickase-based prime editor | crbn (Isolectin) | 2-nt substitution | 8.4% | (Baseline) | [26] |
| PEn | Nuclease-based prime editor | crbn (Isolectin) | 2-nt substitution | 4.4% | ~0.5x | [26] |
| PEn/pegRNA | Nuclease-based editor with pegRNA | ror2 | 3-bp insertion (Stop codon) | Effective (T7E1 assay) | Not quantified | [26] |
| PE7 + La-pegRNA | Engineered editor with stabilized pegRNA | Multiple loci (e.g., adgrf3b) | Small indels (6bp ins, 10bp del) | 16.60% | 3.13 to 11.46-fold | [7] |
Impact of DNA Repair Modulation on Editing Outcomes
Modulating key DNA repair proteins can directly shift the balance away from error-prone repair, as demonstrated by recent studies in cell culture models.
Table 2: Impact of PARP1 Modulation on DSB Repair Pathway Choice
| Modulation Type | Effect on NHEJ | Effect on MMEJ | Effect on HDR | Potential Application for Knock-In | Citation |
|---|---|---|---|---|---|
| PARP1 Downregulation | Increased | Increased | Unaffected | Not recommended for HDR knock-in; may increase indels. | [42] |
| PARP1 Overexpression | Reduced | Unaffected | Reduced | Promising: Suppresses mutagenic NHEJ without enhancing MMEJ, potentially improving precision. | [42] |
Protocol: Enhancing Precise Editing via PARP1 Overexpression and Prime Editing
This protocol is designed for experienced researchers and must be conducted in accordance with all local institutional animal care and use guidelines.
Part A: RNP Complex Preparation with Prime Editing Components
This section describes the assembly of the prime editing ribonucleoprotein (PE RNP) complex, which is microinjected into zebrafish embryos.
Design and Synthesis of pegRNA:
- Design the prime editing guide RNA (pegRNA) to include the spacer sequence, reverse transcription template (RTT) containing your desired edit, and a primer binding site (PBS).
- For optimal performance with the PE7 system, use La-accessible pegRNA, which includes a 3' polyU tail to enhance stability and interaction with the editor [7].
- Chemical Modifications: To enhance pegRNA stability in vivo, obtain chemically synthesized pegRNAs with 5' and 3' modifications, such as 2'-O-methyl analogs and 3'-phosphorothioate internucleotide linkages at the terminal nucleotides [7] [21].
- Resuspend lyophilized pegRNA in nuclease-free water to a stock concentration of 1000 ng/µL and store at -80°C [7].
Assembly of the PE RNP Complex:
- In a nuclease-free microcentrifuge tube, combine the following components to form the RNP complex:
- Incubate the mixture at room temperature for 5-10 minutes to allow the RNP complex to form [8].
Part B: Zebrafish Embryo Microinjection
Embryo Preparation:
Microinjection:
- Using a micromanipulator and a microinjector, pull and bevel a glass needle to an angled opening.
- Backfill the needle with the prepared PE RNP complex.
- Calibrate the injection pressure to deliver a volume of 1-2 nL per embryo [7] [8].
- Gently push the needle through the chorion and into the yolk cytoplasm of each aligned embryo [7] [8]. Cytoplasmic flow will facilitate the diffusion of the RNP complex into the embryonic cells.
- Return the injected embryos to a Petri dish with fresh E3 medium and incubate at 28.5°C [8].
Part C: Co-injection with PARP1 Modulator
- To bias the cellular repair environment toward precision, co-inject mRNA for PARP1 alongside the PE RNP complex.
- The rationale is that PARP1 overexpression suppresses the error-prone NHEJ pathway, reducing the frequency of indels at the target site and potentially increasing the relative abundance of precisely edited cells [42].
- Note: The optimal concentration of PARP1 mRNA for zebrafish embryos must be determined empirically, as the specific quantitative effect was characterized in cell culture [42].
Pathway Diagrams and Workflow
DNA Repair Pathway Competition and Modulation Strategy
This diagram illustrates the competitive landscape of DNA double-strand break (DSB) repair and the strategic point of intervention for enhancing precise editing.
Experimental Workflow for Enhanced Prime Editing
This flowchart outlines the complete experimental procedure from preparation to genotyping, integrating the key steps of RNP formation and repair pathway modulation.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents and Materials for Enhanced Prime Editing
| Item | Function / Role in Protocol | Specification / Critical Feature |
|---|---|---|
| PE7 Protein | State-of-the-art prime editor protein; fusion of nCas9, reverse transcriptase, and La peptide. Enhances editing efficiency. | Engineered version of PEmax for improved performance with La-accessible pegRNAs [7]. |
| La-accessible pegRNA | Guide RNA that directs PE7 to target site and templates the new genetic sequence. | Chemically synthesized with 3' polyU tail and terminal nucleotide modifications (e.g., 2'-O-methyl, phosphorothioate) for stability [7] [21]. |
| PARP1 mRNA | Modulates DNA repair pathway choice when co-injected. | Coding sequence for human or zebrafish PARP1, in vitro transcribed for RNP co-injection [42]. |
| Microinjection System | For precise delivery of RNP complexes into embryos. | Includes micropipette puller, micromanipulator, and microinjector capable of delivering 1-2 nL [8]. |
| Phenol Red | Visual tracer for microinjection. | Added to the injection mixture (e.g., 0.05% final concentration) to confirm successful delivery [8]. |
| NGS Amplicon Sequencing | Gold-standard method for quantifying precise editing efficiency and identifying byproducts. | Provides deep, quantitative data on all edit types (precise edits, indels, translocations) at the target locus [7]. |
| (+)-Hydroxytuberosone | (+)-Hydroxytuberosone|RUO | (+)-Hydroxytuberosone is a natural pterocarpan from kudzu for research. This product is For Research Use Only (RUO). Not for diagnostic or therapeutic use. |
Optimizing Guide RNA Stability and Efficiency with Chemical Modifications
The application of CRISPR-Cas technology in zebrafish models has revolutionized functional genomics and disease modeling. A critical advancement in this field involves the use of chemically modified guide RNAs (gRNAs) to enhance the stability and efficiency of ribonucleoprotein (RNP) complex microinjection in zebrafish embryos. Chemical modifications protect gRNAs from degradation, reduce immune responses, and significantly improve editing outcomes, making them indispensable for precise genome editing applications in vivo [43] [44] [21].
For zebrafish researchers utilizing RNP delivery, gRNA instability presents a major technical challenge. Unmodified gRNAs are highly susceptible to degradation by exonucleases, leading to reduced editing efficiency, particularly for targets requiring sustained activity during later developmental stages [44]. Chemical modifications address this limitation by creating a protective "armor" around the gRNA molecule, significantly increasing its half-life within the embryo [45].
Chemical Modification Strategies for CRISPR gRNAs
Types and Locations of Chemical Modifications
The strategic placement of chemical modifications on gRNAs is crucial for balancing stability improvements with maintained biological activity. The most effective modifications target specific regions of the gRNA structure while avoiding functionally critical areas.
Table 1: Common Chemical Modifications for CRISPR gRNAs
| Modification Type | Chemical Structure | Primary Function | Optimal Placement |
|---|---|---|---|
| 2'-O-methyl (2'-O-Me) | Methyl group (-CH₃) at 2' ribose position | Nuclease resistance, increased stability | 5' and 3' terminal nucleotides |
| Phosphorothioate (PS) | Sulfur substitution for non-bridging oxygen | Resistance to exonuclease degradation | Terminal internucleotide linkages |
| MS Modification | Combined 2'-O-Me and PS | Enhanced stability versus single modifications | 5' and 3' ends |
| MSP Modification | 2'-O-methyl-3'-thioPACE | Maximum stability with reduced immune activation | 5' and 3' ends |
The foundational principle for modification placement centers on protecting vulnerable terminal regions while preserving the seed sequence functionality. Exonucleases primarily degrade gRNAs from both the 5' and 3' ends, making these areas priority targets for stabilization [44]. However, the seed region (nucleotides 8-10 at the 3' end of the crRNA sequence) must remain unmodified to ensure proper hybridization with target DNA and R-loop formation [43] [44].
Research demonstrates that modifying the terminal three nucleotides at both the 5' and 3' ends with 2'-O-methyl and phosphorothioate combinations provides optimal protection without compromising editing efficiency [45]. This strategic approach increases gRNA half-life by creating structural resistance to exonuclease activity while maintaining the crucial target recognition capabilities of the guide sequence.
Mechanism of Stability Enhancement
Chemically modified gRNAs enhance CRISPR editing through multiple synergistic mechanisms that address key limitations of unmodified guides:
Nuclease Resistance: The primary benefit of chemical modifications involves protection against endogenous nucleases present in zebrafish embryos. 2'-O-methylation prevents hydrolysis by ribonucleases, while phosphorothioate linkages create resistance to exonuclease degradation, significantly extending gRNA half-life [44] [45].
Reduced Immune Activation: Unmodified gRNAs can trigger innate immune responses in vertebrate cells, potentially leading to cytotoxicity and reduced editing efficiency. Chemical modifications, particularly 2'-O-methyl groups, minimize recognition by pattern recognition receptors, thereby preventing immune activation and improving cell viability during editing [44].
Improved RNP Complex Stability: Modified gRNAs with enhanced structural stability form more durable complexes with Cas proteins, maintaining the integrity of the RNP complex throughout the microinjection process and initial hours of embryonic development [21].
The cumulative effect of these mechanisms is particularly pronounced in challenging applications such as targeting zygotically expressed genes transcribed after gastrulation (7-8 hours post-fertilization), where sustained gRNA activity is essential for effective editing [21].
Experimental Evidence in Zebrafish Models
Efficiency Comparisons Across Editing Platforms
Chemical modifications enhance gRNA performance across multiple genome editing platforms, with significant quantitative improvements observed in base editing, prime editing, and Cas13-based RNA targeting in zebrafish.
Table 2: Editing Efficiency Improvements with Chemically Modified gRNAs in Zebrafish
| Editing System | Target Gene | Unmodified Efficiency | Modified Efficiency | Fold Improvement |
|---|---|---|---|---|
| Prime Editor (PE7) | adgrf3b | ~2.0% | 15.99% | 8.0x |
| RfxCas13d | Late zygotic genes | Variable phenotype penetrance | Robust phenotype penetrance | 3.13x (indel generation) |
| Base Editor | tyr (P302L) | Not achieved | 13.18% melanin reduction | Successful model generation |
Recent research with the PE7 prime editing system demonstrated that chemically modified pegRNAs combined with La-accessible modifications increased editing efficiency to 15.99% at target loci, representing a 6.81- to 11.46-fold improvement over unmodified PE2 systems [7]. This dramatic enhancement enabled the successful generation of a tyr P302L mutation associated with melanin reduction, a trait previously difficult to achieve with standard editing approaches [7].
In Cas13d-based RNA targeting applications, chemical modifications significantly increased the penetrance of loss-of-function phenotypes for genes expressed after 7-8 hours post-fertilization [21]. The combination of RfxCas13d mRNA with chemically modified gRNAs outperformed RNP complexes for mid- or late-zygotically transcribed genes, producing more robust and consistent phenotypic outcomes [21].
Protocol for RNP Complex Assembly with Modified gRNAs
The following detailed protocol ensures optimal preparation and delivery of chemically modified gRNA RNP complexes into zebrafish embryos:
Reagent Preparation
Chemically Modified gRNAs: Obtain HPLC-purified gRNAs with 2'-O-methyl and phosphorothioate modifications at the three terminal nucleotides of both 5' and 3' ends [44] [45]. Resuspend lyophilized gRNAs in nuclease-free water to a stock concentration of 1000 ng/μL and store at -80°C until use [7].
Cas Protein: Use recombinant Cas9 (S.p. Cas9 Nuclease V3), Cas12a, or specialized editors (base editors, prime editors) at concentrations of 750 ng/μL for microinjection [7] [46].
Microinjection Buffer: Prepare solution containing 0.5 μL of 2.5% phenol red (visualization marker) in nuclease-free water to achieve final injection volume [8].
RNP Complex Assembly
Combine Cas protein and chemically modified gRNA in a 2:1 molar ratio (typically 750 ng/μL Cas protein:240 ng/μL gRNA) in a sterile microcentrifuge tube [7].
Incubate the mixture at room temperature for 5-10 minutes to allow complete RNP complex formation [8].
Add 0.5 μL of 2.5% phenol red solution and adjust to final volume with nuclease-free water (typically 5 μL total volume) [8].
Centrifuge briefly to collect solution at the bottom of the tube and place on ice until microinjection (use within 2 hours).
Zebrafish Embryo Microinjection
Embryo Preparation: Collect fertilized zebrafish eggs and align them into the trough of a microinjection plate [8]. Remove unfertilized eggs (identified by clear yolk membrane versus dark membrane in fertilized eggs).
Needle Preparation: Use a micropipette puller to pull 1-mm glass capillaries, then cut the tip with a razor blade to obtain an angled opening [8]. Place the needle in a micromanipulator attached to a microinjector and calibrate injection pressure to consistently deliver 1 nL of solution.
Microinjection: Gently push the microinjection needle through the chorion into the yolk and inject 1 nL of the RNP complex solution [8]. Cytoplasmic flow will allow the complex to diffuse into embryonic cells.
Post-Injection Care: Return injected embryos to a labeled Petri dish and cover with 1X E3 media with methylene blue [8]. Incubate at 28.5°C and inspect after 24 hours to remove dead or abnormally developing individuals.
The Scientist's Toolkit: Essential Reagents and Materials
Table 3: Research Reagent Solutions for Modified gRNA Applications
| Reagent/Material | Function | Specifications | Commercial Sources |
|---|---|---|---|
| Chemically Modified gRNAs | Guide Cas protein to target sequence | 2'-O-Me + PS modifications, HPLC-purified | Synthego, IDT, GenScript |
| Alt-R S.p. Cas9 Nuclease V3 | DNA cleavage at target sites | Recombinant, high purity | Integrated DNA Technologies |
| Prime Editor PE7 | Precise editing without DSBs | Cas9 nickase-RT fusion + La motif | Academic collaborators |
| RfxCas13d Protein | RNA targeting and knockdown | Recombinant, purified | Academic sources |
| Microinjection Needles | Embryo delivery | 1-mm glass capillaries | World Precision Instruments |
| Phenol Red Solution | Injection visualization | 2.5% in nuclease-free water | Sigma-Aldrich |
The strategic implementation of chemical modifications in gRNA design represents a critical advancement for zebrafish genome editing via RNP microinjection. The integration of 2'-O-methyl and phosphorothioate modifications at terminal nucleotides significantly enhances gRNA stability, editing efficiency, and phenotypic penetrance across multiple CRISPR platforms. As the field progresses toward more sophisticated applications—including the modeling of human genetic diseases and functional genomic screens—these optimization strategies will be essential for achieving reproducible and precise genome modifications in zebrafish models.
Ribonucleoprotein (RNP) complex microinjection in zebrafish embryos represents a powerful, DNA-free approach for genome editing, enabling rapid interrogation of gene function. However, a significant challenge confronting researchers is the variable efficiency of these techniques, which can compromise experimental reproducibility and robustness. This application note, situated within a broader thesis on optimizing RNP delivery, addresses two critical experimental parameters: incubation temperature and RNP complex composition. Evidence from recent studies indicates that strategic manipulation of these factors can substantially enhance editing efficiency while mitigating unwanted off-target effects, thereby providing more reliable phenotypic data in functional genomics and drug discovery projects.
The following tables consolidate key quantitative findings from recent investigations into the effects of incubation temperature and RNP ratios on editing outcomes in zebrafish and related models.
Table 1: Impact of Incubation Temperature on Editing Efficiency and Specificity
| Species | Temperature Condition | Effect on On-Target Editing | Effect on Off-Target Mutations | Survival Rate/Notes | Citation |
|---|---|---|---|---|---|
| Medaka & Zebrafish | Continuous 16°C | No significant negative effect | Significant reduction | Decreased in Medaka with continuous cold | [47] |
| Medaka & Zebrafish | Early low-temp (16°C), then 28°C | Target mutation rates unaffected (DJ-1, p4hb, avt, ywhaqa) | Off-target rates significantly reduced | Effective for suppressing germline transmission of off-targets | [47] |
Table 2: RNP Composition and Delivery Parameters for Prime Editing
| Editor System | RNP Component | Concentration | Efficiency Outcomes | Key Findings | Citation |
|---|---|---|---|---|---|
| PE7 + La-accessible pegRNA | PE7 protein | 750 ng/μL | Up to 15.99% editing efficiency | 6.81- to 11.46-fold improvement over PE2 | [7] |
| PE7 + La-accessible pegRNA | La-accessible pegRNA | 240 ng/μL | 16.60% 6 bp insertion; 13.18% 10 bp deletion | 3.13-fold increase over PE2 at adgrf3b locus | [7] |
| ScCas9 RNP | crRNA:tracrRNA (dgRNA) | 5 μM (final complex) | High targeting efficiency | Use of synthetic dgRNA improved activity over IVT sgRNA | [48] |
Experimental Protocols
Protocol: Low-Temperature Incubation to Minimize Off-Target Effects
This protocol is adapted from Yamanaka et al. (2025) for reducing off-target mutagenesis during CRISPR-Cas9 genome editing in zebrafish [47].
- 1. RNP Complex Preparation: Assemble Cas9 protein and sgRNA at optimal ratios (e.g., 2:1 ratio [49]) and incubate at room temperature for 5-10 minutes to form the RNP complex.
- 2. Microinjection: Microinject 1 nL of the RNP complex into the yolk or cell of one-cell stage zebrafish embryos.
- 3. Low-Temperature Incubation: Immediately after injection, transfer the embryos to an incubator set at 16°C.
- 4. Temperature Shift: Incubate the embryos at 16°C for the early stages of development (the first 6-8 hours post-fertilization is typical).
- 5. Standard Rearing: After the initial low-temperature period, transfer the embryos to a standard 28.5°C incubator for continued development until the desired stage for analysis.
- Note: Continuous incubation at 16°C can significantly reduce embryo survival rates in some species like medaka. The transient low-temperature treatment balances off-target suppression with viability [47].
Protocol: RNP Complex Formulation for High-Efficiency Prime Editing
This protocol is based on the work optimizing prime editor RNP complexes in zebrafish embryos [7].
- 1. Component Preparation:
- Prime Editor Protein: Use a high-efficiency editor like PE7 (a fusion of PEmax and the La peptide).
- Guide RNA: Chemically synthesize La-accessible pegRNAs which include a 3′ polyU tail to enhance interaction with the PE7 protein.
- 2. RNP Complex Assembly:
- Co-incubate the PE7 protein at a final concentration of 750 ng/μL with the La-accessible pegRNA at a final concentration of 240 ng/μL.
- Incubate at room temperature for 15-20 minutes to allow for RNP complex formation.
- 3. Microinjection: Microinject approximately 2 nL of the assembled RNP complex into the yolk cytoplasm of one-cell stage zebrafish embryos.
- 4. Post-Injection Incubation: Raise the injected embryos at a standard 28.5°C in E3 medium. Change the medium daily and remove dead embryos.
- 5. Efficiency Analysis: At 2 days post-fertilization (dpf), extract genomic DNA from pools of embryos and analyze editing efficiency via next-generation sequencing (NGS) of amplicons covering the target site [7].
Signaling Pathways and Workflows
The following diagram illustrates the experimental workflow and the logical relationship between the key parameters of incubation temperature and RNP composition, and their impact on the final editing outcomes.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents and Materials for RNP Microinjection in Zebrafish
| Item | Function/Description | Example Use Case |
|---|---|---|
| Cas9 Protein (SpCas9, ScCas9) | The core nuclease enzyme that creates double-strand breaks. Delivery as protein avoids DNA integration and reduces off-targets. | Standard gene knockouts. ScCas9 expands targetable sites with NNG PAM [48]. |
| Prime Editor Protein (PE2, PE7) | Fusion protein (nCas9 + reverse transcriptase) enabling precise edits without donor DNA. PE7 shows enhanced efficiency [7]. | Introducing specific point mutations or small insertions. |
| Chemically Modified gRNAs/pegRNAs | Synthetic guide RNAs with chemical modifications (e.g., 2'-O-methyl, phosphorothioate) to enhance stability and efficiency. | La-accessible pegRNA for PE7 [7]; cm-gRNAs for sustained Cas13d activity [21]. |
| crRNA:tracrRNA Duplex (dgRNA) | A two-part guide RNA system that can be more stable and efficient than single-guide RNA (sgRNA) for some Cas enzymes. | Improved activity with ScCas9 in zebrafish [48]. |
| Phenol Red Solution | A visible dye mixed with the injection solution to monitor successful delivery and volume control during microinjection. | Standard practice for visualizing the 1 nL injection bolus [49]. |
| E3 Embryo Medium | The standard saline solution for raising and maintaining zebrafish embryos. | Post-injection incubation medium, often supplemented with methylene blue to prevent fungal growth [49]. |
Troubleshooting Low Editing Rates and Improving Phenotype Penetrance
Ribonucleoprotein (RNP) complex microinjection in zebrafish embryos has revolutionized functional genomics and disease modeling. This technique, which involves the direct delivery of preassembled Cas protein and guide RNA complexes, offers immediate activity, reduced off-target effects, and minimal DNA integration concerns. However, researchers frequently encounter challenges with low editing efficiency and variable phenotypic penetrance in F0 mosaic mutants, which can obscure functional analysis and hinder research progress. This application note synthesizes current methodologies to overcome these limitations, providing evidence-based protocols to enhance genome editing outcomes in zebrafish models.
Editor Selection and Engineering
The choice of genome editing platform fundamentally determines the efficiency and precision of genetic modifications. While CRISPR/Cas9 remains widely used, advanced editor systems now offer significantly improved performance.
Table 1: Comparison of Genome Editing Systems in Zebrafish
| Editing System | Editing Type | Key Features | Reported Efficiency Range | Key Advantages |
|---|---|---|---|---|
| PE7 + La-pegRNA [7] | Precise substitutions, insertions, deletions | Fusion of nCas9-H840A, engineered M-MLV reverse transcriptase, and La protein; uses pegRNA with polyU 3' end | Up to 15.99% (6.8-11.5x improvement over PE2) [7] | Avoids double-strand breaks; broad editing scope (12 base substitutions, indels) |
| PE2 [26] [12] | Precise substitutions, insertions, deletions | Original nickase-based prime editor (nCas9 + reverse transcriptase) | 0.25-8.4% for base substitutions [26] [12] | Proven germline transmission; versatile editing types |
| PEn [26] | Precise insertions | Nuclease-based prime editor creating double-strand breaks | More efficient for 3-30 bp insertions than PE2 [26] | Superior for inserting short DNA fragments (e.g., NLS, stop codons) |
| AncBE4max [15] | C•G to T•A conversions | Codon-optimized cytosine base editor | ~3x higher than BE3 [15] | High efficiency for specific transition mutations; reduced indel formation |
| CBE4max-SpRY [15] | C•G to T•A conversions | "Near PAM-less" cytidine base editor | Up to 87% at some loci [15] | Vastly expanded targeting scope beyond NGG PAM sites |
Guide RNA Design and Optimization
Strategic guide RNA design critically influences both editing efficiency and phenotypic outcome. Computational prediction of editing outcomes can maximize the probability of generating loss-of-function alleles.
Computational Prediction for Maximizing Frameshifts
The InDelphi neural network model trained on mouse embryonic stem cells (InDelphi-mESC) accurately predicts CRISPR/Cas9 editing outcomes in zebrafish embryos. When designing guides for gene knockouts, select gRNAs with prediction profiles enriched for frameshift mutations (>85% correlation with experimental outcomes) [50]. This approach maximizes nonsense-mediated decay (NMD) and complete protein knockout, enhancing phenotypic penetrance in mosaic F0 animals.
Chemical Modifications for Enhanced Stability
Chemical modifications to guide RNAs significantly improve efficiency, particularly for late-stage embryonic targeting:
- For DNA-targeting systems: Use pegRNAs with 5' and 3' modifications (methylated or phosphorothioate linkages) to enhance nuclease resistance [7].
- For RNA-targeting systems (CRISPR-RfxCas13d): Implement chemically modified gRNAs (cm-gRNAs) with 2′-O-methyl analogs and 3′-phosphorothioate internucleotide linkages at the terminal nucleotides [21]. This is particularly crucial for targeting zygotically expressed genes transcribed after gastrulation (7-8 hpf), where cm-gRNAs significantly increase phenotype penetrance.
Table 2: Guide RNA Optimization Strategies
| Approach | Protocol Details | Application Context | Efficiency Improvement |
|---|---|---|---|
| InDelphi Prediction | Use InDelphi-mESC model to select gRNAs with >80% predicted frameshift frequency [50] | CRISPR/Cas9 knockouts | 2-3x increase in phenotypic penetrance |
| Chemical Modifications | Synthesize gRNAs with 2′-O-methyl and 3′-phosphorothioate at first and last 3 nucleotides [21] | Late-stage expression targeting (after 7-8 hpf) | Significant improvement for RfxCas13d mRNA + cm-gRNA combinations |
| La-accessible pegRNA | Add polyU tail to 3' end of pegRNA for enhanced PE7 interaction [7] | Prime editing with PE7 system | 6.8-11.5x over PE2 system [7] |
| Dual pegRNA Strategy | Use two distinct pegRNAs targeting the same locus [7] | Challenging prime editing targets | Demonstrated 3.13x increase at adgrf3b locus [7] |
RNP Complex Preparation and Microinjection
Optimized delivery of editing components is essential for achieving high efficiency while maintaining embryo viability.
RNP Complex Assembly
Protocol for PE RNP Complex Preparation [7]:
- Component Concentration: Resuspend lyophilized pegRNAs in nuclease-free water to 1000 ng/μL stock concentration.
- Complex Formation: Co-incubate PE protein (750 ng/μL) with La-accessible pegRNA (240 ng/μL) for 10-15 minutes at room temperature.
- Storage: Aliquot and store RNP complexes at -80°C until use (avoid repeated freeze-thaw cycles).
For CRISPR-Cas13d RNP [21]:
- Use purified RfxCas13d protein with cm-gRNAs for maternal and early zygotic genes (0-6 hpf).
- For later expression (after 7-8 hpf), use RfxCas13d mRNA + cm-gRNA combinations for superior efficiency.
Microinjection Parameters
Optimized Injection Conditions [7] [25]:
- Volume: 2 nL per embryo injected into yolk cytoplasm at one-cell stage.
- Timing: Inject within 45 minutes post-fertilization.
- Temperature: Incubate injected embryos at 32-34.5°C for enhanced editing efficiency (compared to standard 28.5°C) [12].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Zebrafish RNP Genome Editing
| Reagent / Tool | Function | Application Notes |
|---|---|---|
| PE7 Protein [7] | Advanced prime editor with La fusion | Highest efficiency prime editing; use with La-accessible pegRNAs |
| Chemically Modified gRNAs [7] [21] | Enhanced nuclease resistance | 2′-O-methyl + 3′-phosphorothioate modifications; crucial for late-stage targeting |
| C9E pegRNA Scaffold [12] | Improved pegRNA architecture | Higher pure prime edit frequencies without increasing byproducts |
| InDelphi Prediction Tool [50] | gRNA outcome prediction | Select guides with enriched frameshift profiles; use mESC-trained model |
| Prime Editor Proteins [26] [12] | Precise genome editing | PE2 for substitutions; PEn for insertions; purify from E. coli for RNP use |
| RfxCas13d System [21] | mRNA knockdown | Protein + cm-gRNA for early genes; mRNA + cm-gRNA for late genes |
Validation and Analysis Methods
Rigorous validation of editing outcomes is essential for interpreting phenotypic results.
Editing Efficiency Assessment
Deep Amplicon Sequencing Protocol [7]:
- DNA Extraction: At 2 days post-fertilization (dpf), extract genomic DNA from 6-8 normally developed embryos using QIAamp DNA Mini Kit.
- Two-step PCR Amplification:
- Primary PCR: Amplify target region with site-specific primers.
- Secondary PCR: Add barcodes for multiplexed sequencing.
- Sequencing: Use Illumina Novaseq X plus platform with minimum 3 biological replicates.
- Analysis: Classify edits as Pure Prime Edits (PPEs), Impure Prime Edits (IPEs), or byproduct edits using tools like GraphPad Prism 8.0.1.
Phenotype Evaluation
For disease modeling, establish clear phenotypic assessment criteria. For example, in cataract gene evaluation, systematic brightfield imaging at defined developmental stages enables quantitative phenotype scoring [25].
Implementing these optimized protocols for editor selection, guide RNA design, RNP delivery, and validation can dramatically improve editing efficiency and phenotypic penetrance in zebrafish RNP microinjection studies. The synergistic application of advanced editing systems like PE7, computational prediction tools, and chemical guide modifications addresses the fundamental challenges of mosaic mutagenesis in F0 embryos. These approaches enable more reliable functional genomics studies and disease modeling, accelerating drug discovery and genetic research using zebrafish models.
Benchmarking RNP Technology: Efficacy, Precision, and Future Directions
Ribonucleoprotein (RNP) complex microinjection in zebrafish embryos represents a cornerstone technique for precise genome editing, enabling functional genomics and disease modeling. This Application Note provides a consolidated quantitative overview of editing efficiencies achieved with advanced editors like PE7, PE2, and PEn across diverse genomic loci in zebrafish. We present structured data tables, detailed protocols for replicating key experiments, and visual workflows to support researchers in implementing these methods for drug discovery and genetic research.
Prime Editing Efficiency Across Genomic Loci
Table 1: Prime editing efficiency in zebrafish using RNP microinjection
| Genomic Locus | Edit Type | Editor System | Efficiency (%) | Fold Improvement | Citation |
|---|---|---|---|---|---|
| adgrf3b | 6 bp insertion | PE7 + La-pegRNA | 16.60 | 3.13× over PE2 | [7] |
| adgrf3b | 10 bp deletion | PE7 + La-pegRNA | 13.18 | 3.13× over PE2 | [7] |
| tyr | P302L (CCC→CTC) | PE7 + La-pegRNA | 15.99 | 6.81-11.46× over PE2 | [7] |
| crbn | 2 nt substitution | PE2 | 8.40 | 1.91× over PEn | [26] |
| crbn | 2 nt substitution | PEn | 4.40 | - | [26] |
| ror2 | 3 bp stop codon | PEn + springRNA | High (T7E1 positive) | More effective than PE2 | [26] |
Comparative Editor Performance Metrics
Table 2: Performance characteristics of editing systems in zebrafish
| Editor System | Typical Efficiency Range | Precision Score | Indel Frequency | Primary Applications |
|---|---|---|---|---|
| PE7 + La-pegRNA | 13-17% | Not specified | Not specified | Single-base substitutions, small indels |
| PE2 | ~8% | 40.80% | Lower than PEn | Nucleotide substitutions |
| PEn | ~4% | 11.40% | Higher than PE2 | Short DNA insertions (3-30 bp) |
| Base Editors (BE3) | 9-29% | Not specified | Not specified | Single-nucleotide conversions |
Experimental Protocols
PE7 RNP Complex Preparation and Microinjection
This protocol details the optimized method for achieving high-efficiency prime editing in zebrafish embryos using PE7 RNP complexes [7].
Materials Required
- PE7 protein (750 ng/μL stock concentration)
- Chemically synthesized La-accessible pegRNAs with 5′ and 3′ modifications (methylated or phosphorothioate linkages)
- Wild-type AB strain zebrafish embryos at one-cell stage
- Microinjection system with capability for 2 nL injections
- QIAamp DNA Mini Kit for genomic DNA extraction
RNP Complex Assembly
- Dilute PE7 protein to working concentration of 750 ng/μL in nuclease-free buffer.
- Dilute La-accessible pegRNA to 240 ng/μL in nuclease-free water.
- Combine PE7 protein and pegRNA at a 3:1 molar ratio in a sterile microcentrifuge tube.
- Incubate the mixture at room temperature for 10 minutes to form RNP complexes.
Embryo Microinjection
- Aliquot 1-2 μL of RNP complex into injection needles.
- Position one-cell stage embryos for microinjection into the yolk cytoplasm.
- Inject precisely 2 nL of RNP complex per embryo using a pneumatic microinjector.
- Transfer injected embryos to embryo medium and incubate at 28.5°C.
Editing Efficiency Analysis
- At 2 days post-fertilization (dpf), collect 6-8 normally developed embryos.
- Extract genomic DNA using QIAamp DNA Mini Kit following manufacturer's protocol.
- Amplify target regions using barcoded primers in a two-step PCR process.
- Sequence amplified products using Illumina Novaseq X plus platform.
- Analyze sequencing data for precise edits and indels using appropriate bioinformatics tools.
Comparative Editing with PE2 and PEn Systems
This protocol enables direct comparison of nickase-based (PE2) and nuclease-based (PEn) prime editors for different types of genetic modifications [26].
Materials Required
- PE2 or PEn mRNA
- pegRNAs or springRNAs targeting loci of interest
- Zebrafish embryos at one-cell stage
- T7 Endonuclease I (T7E1) for initial modification screening
- Cloning vector for sequence verification
Experimental Procedure
Prepare mRNA and guide RNA combinations:
- Condition 1: PE2 mRNA + pegRNA
- Condition 2: PEn mRNA + pegRNA
- Condition 3: PEn mRNA + springRNA
Microinject approximately 2 nL of each combination into separate batches of one-cell stage zebrafish embryos.
Incubate injected embryos at 32°C for 96 hours post-fertilization.
For initial screening:
- Extract genomic DNA from pool of 10 embryos per condition
- Amplify target region by PCR
- Perform T7E1 assay to detect sequence modifications
For detailed sequence analysis:
- Clone PCR products into sequencing vector
- Sequence multiple randomly selected clones
- Categorize edits as precise insertions, substitutions, or indels
Signaling Pathways and Experimental Workflows
Prime Editing Mechanism
Diagram 1: PE mechanism
Experimental Workflow for RNP Microinjection
Diagram 2: RNP workflow
Research Reagent Solutions
Table 3: Essential reagents for RNP complex microinjection in zebrafish
| Reagent/Material | Function/Purpose | Specifications/Modifications | Citation |
|---|---|---|---|
| PE7 protein | Prime editor fusion protein | nCas9 (H840A) + engineered MMLV-RT | [7] |
| La-accessible pegRNA | Targeting and edit template | 3′ polyU extension, 5′/3′ modifications (methylated/phosphorothioate) | [7] |
| PE2 protein | Nickase-based prime editor | nCas9 (H840A) + MMLV-RT | [26] |
| PEn protein | Nuclease-based prime editor | Wild-type Cas9 + MMLV-RT | [26] |
| springRNA | Simplified guide for PEn | Lacks homology arm template | [26] |
| Microinjection needles | Embryo delivery | Precision-bore for 2 nL injections | [7] |
| QIAamp DNA Mini Kit | Genomic DNA extraction | Silica-membrane technology | [7] |
Within functional genomics and genetic engineering, the selection of a delivery method for CRISPR-based reagents is a critical determinant of experimental success. This application note provides a direct comparison of ribonucleoprotein (RNP) complex delivery against DNA or mRNA-based methods, with a specific focus on their application in zebrafish embryo research. The zebrafish model is a cornerstone of developmental biology and drug discovery, making the optimization of delivery techniques a subject of broad importance. Framed within a broader thesis on RNP complex microinjection in zebrafish embryos, this document synthesizes current evidence to guide researchers and drug development professionals in selecting the appropriate strategy based on empirical data concerning specificity, toxicity, and efficiency.
Mechanism of Action and Key Differentiators
The fundamental difference between these delivery methods lies in the form in which the CRISPR-Cas machinery is introduced into the cell. The choice influences the temporal presence of the editing components, the cellular machinery required, and the subsequent immune reactions, which collectively dictate the specificity and toxicity profile.
RNP Complexes consist of a pre-assembled, purified Cas protein (e.g., Cas9) complexed with a guide RNA (gRNA). These complexes are directly microinjected into the embryo, enabling immediate genomic editing without any requirement for in vivo transcription or translation [7] [46]. This direct delivery offers precise control over concentration and limits the duration of nuclease activity.
DNA-Based Delivery involves injecting a plasmid DNA (pDNA) construct that encodes the Cas protein and gRNA. This DNA must be transcribed into mRNA and then translated into functional protein within the cell, a process that delays the onset of editing and can lead to prolonged and variable Cas protein expression.
mRNA-Based Delivery entails injecting in vitro transcribed (IVT) mRNA encoding the Cas protein, alongside a separate gRNA. This approach bypasses the transcription step but still requires in vivo translation to produce the functional protein, resulting in a more rapid onset of activity than DNA but less immediate than RNP.
The following diagram illustrates the core workflows and decisive factors for selecting a delivery method.
Direct Comparison: Specificity, Toxicity, and Efficiency
A critical evaluation of RNP, mRNA, and DNA delivery methods reveals a clear trade-off between editing efficiency and undesirable effects such as off-target activity and toxicity. The quantitative and qualitative data summarized in the table below provide a basis for an informed selection.
Table 1: Direct Comparison of CRISPR-Cas Delivery Methods in Zebrafish
| Parameter | RNP Complexes | mRNA + gRNA | DNA Vector (pDNA) |
|---|---|---|---|
| Time to Activity | Immediate (pre-formed) | Delayed (requires translation) | Significantly delayed (requires transcription & translation) [21] |
| Duration of Activity | Short, transient | Moderate | Prolonged, variable |
| Editing Efficiency | High (up to 16.99% PE reported) [7] | Variable; can be improved with cm-gRNAs for late genes [21] | Variable; can be high but with increased toxicity risk |
| Specificity (On-target) | High, precise control | Moderate | Lower risk of random integration, but prolonged expression can increase off-targets |
| Off-target Effects | Reduced; lower off-target mutations with HiFi Cas9 RNP [46] | Higher potential due to prolonged expression | Highest potential due to sustained nuclease expression |
| Toxicity & Immunogenicity | Lowest; reduced innate immune response and cell toxicity [46] [51] | Moderate/High; unmodified IVT mRNA can trigger TLR pathways and toxic effects [21] [51] | High; potential for immune activation and integration-related genotoxicity |
| Key Advantages | • Precise dosage control• No coding sequence integration• Low immune activation• High reproducibility | • Suitable for targeting late zygotic genes when used with cm-gRNAs [21] | • Potentially stable, long-term expression |
| Major Limitations | • Less efficient for some late-stage zygotic genes [21] | • Requires careful mRNA design (e.g., nucleoside modification) to reduce toxicity [51] | • High immunogenicity and toxicity• Unpredictable integration and expression levels• Unsuitable for transient editing |
Detailed Experimental Protocols
Protocol 1: RNP Complex Preparation and Microinjection in Zebrafish
This protocol is adapted from established methods for achieving precise genome editing with reduced toxicity [8] [7] [46].
Materials & Reagents:
- Purified Cas nuclease protein (e.g., Alt-R S.p. Cas9)
- Chemically synthesized sgRNA or pegRNA
- Phenol red solution (0.5%)
- Nuclease-free water
- Microinjection system (micromanipulator, microinjector)
- Glass capillary needles
- Zebrafish embryos at one-cell stage
Procedure:
- RNP Complex Assembly:
- Combine purified Cas9 protein and sgRNA at a molar ratio between 1:1 and 1:2 (e.g., a final complex of 750 ng/μL Cas protein and 240 ng/μL pegRNA) in a nuclease-free tube [7].
- Incubate the mixture at room temperature for 5-10 minutes to allow for RNP complex formation.
- Add 0.5 μL of 2.5% phenol red solution (as an injection tracer) and bring to the final volume with nuclease-free water [8].
Needle Preparation and Calibration:
- Using a micropipette puller, pull a 1-mm glass capillary to create a fine needle.
- Under a microscope, use a razor blade to cut the needle tip at an angle to achieve a ~10-15 μm opening.
- Load the needle with the RNP solution and place it in the micromanipulator. Calibrate the injection pressure to consistently deliver a volume of ~1 nL per embryo [8].
Embryo Microinjection:
- Align one-cell stage zebrafish embryos into the trough of a microinjection plate.
- Using the micromanipulator, gently pierce the chorion and inject the calibrated volume of RNP complex into the yolk or cell cytoplasm.
- After injection, return the embryos to a Petri dish containing E3 medium with methylene blue.
- Incubate the injected embryos at 28.5 °C and monitor for development, removing any dead embryos daily [8].
Protocol 2: RfxCas13d mRNA and Chemically Modified gRNA Delivery for RNA Knockdown
This protocol is optimized for depleting mRNAs, especially those expressed after gastrulation (after 7-8 hpf), where RNP complexes are less efficient [21].
Materials & Reagents:
- RfxCas13d mRNA (IVT, nucleoside-modified)
- Chemically modified gRNAs (cm-gRNAs with 2'-O-methyl and 3'-phosphorothioate modifications)
- Microinjection system
Procedure:
- Solution Preparation:
- Co-inject a mixture of RfxCas13d mRNA and cm-gRNAs. The use of cm-gRNAs is critical for sustaining mRNA depletion activity for genes expressed later in development [21].
Microinjection:
- Follow the same needle preparation and embryo injection steps as in Protocol 1.
- Inject the mRNA/cm-gRNA solution into the cytoplasm of one-cell stage embryos.
Phenotype Analysis:
- Assess knockdown efficiency by observing phenotype penetrance at relevant developmental stages (e.g., pigmentation loss for
tyr,golden, oralbinoat 2 dpf) [21].
- Assess knockdown efficiency by observing phenotype penetrance at relevant developmental stages (e.g., pigmentation loss for
The Scientist's Toolkit: Essential Reagents and Materials
Successful implementation of these protocols relies on key reagents. The following table details essential solutions for CRISPR-based experiments in zebrafish.
Table 2: Research Reagent Solutions for Zebrafish CRISPR Experiments
| Reagent / Solution | Function / Purpose | Application Notes |
|---|---|---|
| Alt-R CRISPR-Cas9 RNP (IDT) | Pre-complexed, ready-to-use RNP for high-efficiency editing. | Chemically modified gRNAs increase stability and reduce immune response [46]. |
| Cas9 Nuclease (V3 or HiFi) | The effector protein for DNA cleavage. | HiFi variant reduces off-target effects while maintaining on-target activity [46]. |
| Chemically Modified gRNAs (cm-gRNAs) | gRNAs with 2'-O-methyl and 3'-phosphorothioate modifications. | Enhance stability and sustain activity for late zygotic mRNA knockdown with Cas13d [21]. |
| Phenol Red (0.5-2.5%) | Tracer dye for microinjection. | Allows visualization of successful delivery during microinjection [8]. |
| E3 Embryo Medium | Standard medium for maintaining zebrafish embryos. | Contains methylene blue to prevent fungal growth [8]. |
| 3D-Printed Anti-Clogging Microneedles | Microneedles with side ports and internal filters. | Reduce injection failure rates and improve delivery volume consistency [52]. |
The direct comparison between RNP and nucleic acid-based delivery methods reveals a compelling case for the use of RNP complexes in zebrafish embryo research. RNP delivery offers a superior combination of high specificity, low toxicity, and immediate activity, making it the gold standard for most genome editing applications. While mRNA delivery, particularly when paired with chemically modified gRNAs, provides a viable strategy for targeting late zygotic genes, it carries a higher risk of immune activation and toxicity. DNA-based delivery is generally not recommended for transient editing due to its high toxicity and unpredictable expression. Therefore, for researchers prioritizing precision, low toxicity, and experimental reproducibility in zebrafish models, RNP complex microinjection is the unequivocally recommended method.
Precise genome editing is a cornerstone of functional genomics and therapeutic development. While Homology-Directed Repair (HDR) has long been the standard for precise DNA modification, its efficiency in zebrafish remains low, often yielding stochastic integration of random insertions and deletions (indels) [26]. Prime editing represents a transformative technology that enables precise base substitutions, insertions, and deletions without inducing double-strand DNA breaks (DSBs) or requiring donor DNA templates [7] [53]. When delivered as pre-assembled Ribonucleoprotein (RNP) complexes via microinjection into zebrafish embryos, prime editing achieves superior precision and efficiency compared to traditional HDR-based approaches. This application note details the quantitative advantages of RNP-based prime editing and provides optimized protocols for implementing this technology in zebrafish models.
Quantitative Comparison: Prime Editing vs. HDR
The following data summarizes key performance metrics demonstrating the superiority of RNP-based prime editing over HDR-based approaches in zebrafish.
Table 1: Comparative Efficiency of Genome Editing Technologies in Zebrafish
| Editing Technology | Editing Efficiency Range | Key Advantages | Primary Limitations |
|---|---|---|---|
| HDR (Traditional) | Typically very low [26] | Can incorporate large DNA fragments | Requires donor DNA; inefficient; high indel rates |
| Base Editors | 9.25%-87% [15] | No DSBs; high efficiency for specific conversions | Limited to specific base changes; bystander edits |
| Prime Editing (RNP) | Up to 16.6% [7] | All 12 base-to-base conversions; small indels; no DSBs | Lower efficiency for large insertions |
| PE2 (Nickase-based) | 8.4% precision score [26] | High precision for single-nucleotide variants | Variable efficiency across loci |
| PEn (Nuclease-based) | 4.4% precision score [26] | Better for short DNA insertions (up to 30 bp) | Higher indel formation |
Table 2: Optimization Strategies for Enhanced Prime Editing Efficiency
| Optimization Strategy | Effect on Editing Efficiency | Application in Zebrafish |
|---|---|---|
| PE7 with La-accessible pegRNA | 6.81- to 11.46-fold increase over PE2 [7] | Achieved up to 15.99% editing efficiency |
| Engineered pegRNAs (epegRNAs) | Protects from exonuclease degradation [54] | Improved stability and editing outcomes |
| Chemical modifications of pegRNA | 2'-O-methyl analogs + 3'-phosphorothioate linkages [7] | Enhanced RNP complex stability |
| Dual-pegRNA strategy | Targets same locus with two distinct pegRNAs [7] | Increases editing efficiency |
| MMLR inhibition | Enhances editing purity and efficiency [54] | Reduces repair-mediated reversal of edits |
Experimental Protocol: RNP-Based Prime Editing in Zebrafish
Ribonucleoprotein (RNP) Complex Preparation
Principle: Pre-assembling prime editor protein with pegRNA forms stable RNP complexes that immediately engage genomic targets upon delivery, reducing off-target effects and accelerating editing kinetics [7] [27].
Procedure:
- Component Preparation: Resuspend chemically synthesized pegRNAs in nuclease-free water to 1000 ng/μL stock concentration. Store at -80°C until use [7].
- RNP Assembly: Combine PE7 protein (750 ng/μL) with La-accessible pegRNA (240 ng/μL) in reaction buffer (100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2, 1 mM DTT, pH 7.9) [7] [27].
- Complex Formation: Incubate the mixture at 37°C for 15 minutes to allow proper RNP complex formation [27].
- Visualization Aid: Add 0.5 μL of 2.5% phenol red solution to the final RNP mixture to facilitate visualization during microinjection [8].
Zebrafish Embryo Microinjection
Principle: Microinjection of RNP complexes into one-cell stage embryos ensures editing occurs before cellular differentiation, maximizing germline transmission potential and reducing mosaicism [7] [8].
Procedure:
- Embryo Collection: Place zebrafish in divided breeding tanks overnight. Remove dividers the following morning and collect fertilized eggs immediately after spawning [8].
- Embryo Alignment: Transfer embryos to the trough of a microinjection plate using a transfer pipette. Orient embryos for optimal needle access [8].
- Needle Preparation: Pull 1-mm glass capillaries using a micropipette puller. Cut the tip with a razor blade to create an angled opening of appropriate size [8].
- Microinjection: Load the RNP complex solution into the injection needle. Using a micromanipulator and microinjector, inject 1-2 nL of RNP complex solution (containing 750 ng/μL PE protein and 240 ng/μL pegRNA) into the yolk cytoplasm of each embryo at the one-cell stage [7] [8].
- Post-Injection Care: Return injected embryos to a labeled Petri dish containing 1X E3 medium with methylene blue. Incubate at 28.5°C and inspect regularly to remove dead or abnormally developing embryos [7] [8].
Editing Efficiency Validation
Principle: Deep amplicon sequencing provides quantitative assessment of editing efficiency and specificity, enabling optimization of pegRNA designs and RNP formulations [7].
Procedure:
- Genomic DNA Extraction: At 2-4 days post-fertilization (dpf), extract genomic DNA from 6-8 normally developed embryos using a commercial DNA extraction kit [7].
- Target Amplification: Perform first-round PCR amplification of the target region using site-specific primers [7].
- Library Preparation: Add forward and reverse barcodes to PCR products in a second amplification round. Pool equal amounts of products for sequencing [7].
- Sequencing and Analysis: Sequence libraries using Illumina platforms (e.g., Novaseq X plus). Analyze sequenced reads for precise substitutions and indels at pegRNA target sites [7].
Visual Workflow: RNP-Based Prime Editing in Zebrafish
Diagram 1: RNP-based prime editing workflow in zebrafish.
Molecular Mechanism of Prime Editing
Diagram 2: Molecular mechanism of prime editing.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for RNP-Based Prime Editing in Zebrafish
| Reagent/Equipment | Specification | Function | Source/Reference |
|---|---|---|---|
| Prime Editor Protein | PE7 (nCas9-H840A + MMLV-RT + La fusion) | Catalyzes targeted nick and reverse transcription | [7] |
| La-accessible pegRNA | Chemically synthesized with 5′/3′ modifications (methylated or phosphorothioate linkages) | Guides PE to target locus and provides edit template | [7] |
| Microinjection System | Micromanipulator, microinjector, and pulled glass capillaries | Precisely delivers RNP complexes to embryos | [8] |
| Reaction Buffer | 100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2, 1 mM DTT, pH 7.9 | Optimal conditions for RNP complex formation | [27] |
| Zebrafish Embryos | Wild-type AB strain, one-cell stage | Model organism for gene editing studies | [7] |
| Phenol Red Solution | 2.5% in nuclease-free water | Visual aid for microinjection | [8] |
RNP-based prime editing represents a significant advancement over traditional HDR for precise genome modification in zebrafish models. The key advantages include: (1) elimination of double-strand breaks and associated cellular damage; (2) Versatile editing capabilities encompassing all possible base substitutions and small indels; (3) reduced off-target effects compared to conventional CRISPR-Cas9 systems; and (4) higher precision scores than HDR-based approaches. By implementing the optimized protocols and reagent specifications outlined in this application note, researchers can reliably achieve precise genetic modifications in zebrafish for functional genomics, disease modeling, and genetic screening applications.
This application note details the establishment and validation of two distinct disease models using ribonucleoprotein (RNP) complex microinjection in zebrafish embryos. We present a case study on generating a tyr P302L model for oculocutaneous albinism using advanced prime editing technology, and a comprehensive review of KRAS G12V model validation for pancreatic cancer research, focusing on T-cell receptor (TCR) immunotherapeutic development. These approaches demonstrate how RNP-based methodologies enable precise genetic modeling with high efficiency and specificity, providing valuable platforms for functional genomics and therapeutic screening.
Ribonucleoprotein (RNP) complex delivery via microinjection represents a transformative approach for creating precise disease models in zebrafish embryos. This DNA-free technique combines purified Cas protein with guide RNA to enable rapid, specific genetic modifications while minimizing off-target effects and immune responses [55]. The transient nature of RNP complexes facilitates immediate genome editing without genomic integration of foreign DNA, making this method particularly valuable for creating accurate disease models and conducting functional genetic screens.
Zebrafish (Danio rerio) offer distinct advantages for disease modeling, including high genetic similarity to humans, with over 70% of human protein-coding genes and 82% of human disease-related genes having zebrafish orthologs [56]. Their external development, optical transparency, and rapid generation time facilitate large-scale functional studies. The "crispant" approach—phenotyping first-generation (F0) mosaic founders—further accelerates validation, reducing the timeline from months to weeks compared to establishing stable mutant lines [56].
This note presents two specialized applications: (1) precise modeling of a pigmentation disorder via prime editing, and (2) immunotherapeutic validation for an oncogenic mutation, highlighting the versatility of RNP-based approaches in biomedical research.
Case Study 1: tyr P302L Zebrafish Model for Oculocutaneous Albinism
Background and Rationale
The tyrosinase (tyr) gene encodes a key enzyme in melanin production, and its impairment causes oculocutaneous albinism (OCA). The specific P302L mutation (CCC→CTC) reduces melanin pigmentation but has been challenging to model using conventional editing approaches. Previous attempts with CRISPR/Cas9-mediated non-homologous end joining (NHEJ) produced unpredictable indels, while homology-directed repair (HDR) proved inefficient in zebrafish [7] [15].
Prime editing (PE) technology addresses these limitations by enabling precise base substitutions without double-strand breaks (DSBs) or donor DNA templates [7]. This case study utilized an optimized PE7 system to create a precise tyr P302L point mutation, establishing a reliable model for pigmentation disorders and therapeutic testing.
Experimental Protocol
Reagent Preparation
- PE7 RNP Complex Assembly: Incubate 750 ng/μL PE7 nuclease (Yan et al., 2025) with 240 ng/μL La-accessible pegRNA for 10 minutes at 37°C to form RNP complexes [7].
- pegRNA Design: Chemically synthesize pegRNAs with 5′ and 3′ modifications (methylated or phosphorothioate linkages) to enhance stability. Resuspend in nuclease-free water to 1000 ng/μL stock concentration and store at -80°C [7].
- La-accessible pegRNA Modification: Incorporate polyU tracts at the 3′ end to enhance PE7 interaction and editing efficiency [7].
Microinjection Procedure
- Embryo Collection: Obtain wild-type AB strain zebrafish eggs and maintain at 28.5°C in a humidified incubator [7].
- Microinjection: Deliver 2 nL of prepared RNP complexes into the yolk cytoplasm of one-cell stage embryos using calibrated microinjection apparatus [7].
- Quality Control: Remove non-viable embryos post-injection and maintain survivors under standard laboratory conditions.
Phenotypic Analysis
- Imaging: At 2 days post-fertilization (dpf), anesthetize embryos in 0.03% Tricaine and mount in 4% methylcellulose. Capture images using digital cameras (e.g., OLYMPUS XM10 or Leica AxioCam MRc5) on an SZX2-FOF microscope [7].
- Genotypic Validation: Extract genomic DNA from 6-8 embryos (2 dpf) using QIAamp DNA Mini Kit. Amplify target regions with barcoded primers for next-generation sequencing (NGS) on Illumina Novoseq X Plus platform [7].
- Efficiency Quantification: Analyze sequencing data for precise base substitutions and calculate editing efficiency percentages.
Results and Data Analysis
Table 1: Prime Editing Efficiency at tyr Locus with PE7 RNP Complexes
| Target Site | Editing Efficiency | Fold Improvement over PE2 | Mutation Type |
|---|---|---|---|
| tyr P302L | 15.99% | 6.81- to 11.46-fold | Precise CCC→CTC substitution |
| adgrf3b locus | 16.60% (6 bp insertion), 13.18% (10 bp deletion) | 3.13-fold | Small indels |
The PE7 RNP approach demonstrated significant improvement over previous methods, with up to 15.99% editing efficiency at the target locus—a 6.81- to 11.46-fold increase over conventional PE2 systems [7]. The successful introduction of the tyr P302L mutation resulted in visibly reduced melanin pigmentation, confirming the functional impact of this precise genetic modification.
Application Notes
- Efficiency Optimization: The PE7 system with La-accessible pegRNA significantly enhances editing efficiency compared to earlier PE versions. Chemical modifications of pegRNAs improve stability and reduce degradation.
- Phenotypic Validation: The melanin reduction phenotype provides a rapid, visible readout of editing success, enabling efficient screening without extensive genotyping.
- Protocol Flexibility: This approach can be adapted to introduce other precise mutations by modifying the pegRNA sequence while maintaining the same RNP delivery protocol.
Case Study 2: KRAS G12V Model for Cancer Immunotherapy Development
Background and Rationale
The KRAS G12V mutation represents a key oncogenic driver in multiple cancers, particularly pancreatic ductal adenocarcinoma (PDAC), where KRAS mutations occur in approximately 90% of cases [57]. This mutation locks the KRAS protein in a GTP-bound "ON" state, leading to constitutive signaling through pathways like MAPK and PI3K that promote tumor growth and survival [58].
Unlike the direct gene editing approach used for the tyr P302L model, KRAS G12V validation focuses on immunotherapeutic development, particularly T-cell receptor (TCR) engineering against this common neoantigen. This case study outlines the identification and validation of TCRs targeting HLA-A*11:01-restricted KRAS G12V mutant peptides, demonstrating how zebrafish models contribute to cancer immunotherapy development.
Experimental Protocol
TCR Identification and Validation
- T Cell Screening: Isolate peripheral blood mononuclear cells (PBMCs) from HLA-A*11:01-positive PDAC patients with KRAS G12V+ tumors. Stimulate with KRAS G12V peptides (VVGAVGVGK) and expand reactive T cells [59].
- TCR Sequencing: Sort tetramer-positive CD8+ T cells by flow cytometry and perform single-cell TCR sequencing to identify KRAS G12V-reactive TCRs [60].
- TCR Engineering: Construct chimeric TCRs with murine variable domains and human constant domains (TRAC and TRBC1) for therapeutic application [60].
Functional Assays
- Cytokine Secretion assays: Co-culture TCR-transduced T cells with KRAS G12V+ target cells and measure IFN-γ production by ELISA [59] [60].
- Cytotoxicity assays: Evaluate specific lysis of KRAS G12V+ tumor cells using real-time cell analysis or lactate dehydrogenase (LDH) release assays [59].
- Organoid Validation: Test TCR recognition against human pancreatic cancer organoids endogenously expressing KRAS G12V and HLA-A*11:01 [59].
- In Vivo Modeling: Assess antitumor efficacy of TCR-engineered T cells in immunodeficient mice bearing organoid-derived xenografts [59].
Results and Data Analysis
Table 2: KRAS G12V TCR Functional Characterization
| Assay Type | Target Cells | Response | Notes |
|---|---|---|---|
| Cytokine Secretion | HLA-A*11:01+ KRAS G12V+ tumor cells | Significant IFN-γ production | Specific recognition with minimal wild-type cross-reactivity |
| Cytotoxic Killing | KRAS G12V+ pancreatic cancer organoids | Killing in 2/5 organoids | Enhanced by IFN-γ priming of organoids |
| In Vivo Tumor Control | Organoid-derived xenografts | Significant growth reduction | Demonstrated in immunodeficient mouse model |
Research identified a public TCR (1-2C) specific for the HLA-A11:01-restricted KRAS G12V8–16 neoepitope (VVGAVGVGK) that was present in multiple immunized mice [60]. This TCR demonstrated specific recognition of all five tested human pancreatic cancer organoids naturally expressing KRAS G12V and HLA-A11:01, though recognition efficiency varied across different organoid lines [59].
Application Notes
- Model Limitations: TCR-engineered T cells showed variable recognition across different tumor organoids, highlighting tumor heterogeneity and the potential need for combination therapies to enhance efficacy [59].
- Combination Strategies: IFN-γ priming enhanced tumor recognition and killing by TCR-engineered T cells, suggesting potential synergistic approaches in clinical applications [59].
- Specificity Considerations: The 9-mer KRAS G12V mutant peptide (VVGAVGVGK) exhibits distinct conformation from wild-type peptide and 10-mer counterparts, enabling specific TCR recognition without cross-reactivity [60].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for RNP-based Disease Modeling
| Reagent/Solution | Application | Function | Example Specifications |
|---|---|---|---|
| PE7 Nuclease | Prime editing | Engineered reverse transcriptase fused to nCas9 for precise edits | 750 ng/μL working concentration [7] |
| La-accessible pegRNA | Prime editing | Guide RNA with polyU 3′ end for enhanced PE7 interaction | 240 ng/μL, chemically synthesized with 5′/3′ modifications [7] |
| Chemically Modified gRNAs | RNA-targeting CRISPR | Enhanced stability for sustained activity | 2′-O-methyl analogs + 3′-phosphorothioate linkages [21] |
| RfxCas13d Protein | RNA knockdown | RNA-targeting Cas protein for transcript depletion | Formulated as RNP complexes for mRNA degradation [21] |
| HLA-A*11:01 Tetramers | TCR validation | Detection and isolation of KRAS G12V-specific T cells | Loaded with VVGAVGVGK peptide for flow cytometry [59] [60] |
| Pancreatic Cancer Organoids | Immunotherapy testing | Patient-derived models with endogenous KRAS G12V expression | Maintain molecular features of original tumors [59] |
These case studies demonstrate the power of RNP-based approaches for creating diverse disease models in zebrafish. The tyr P302L example showcases precise genome editing using advanced prime editing technology, while the KRAS G12V model highlights the application of zebrafish systems in cancer immunotherapy validation. Both approaches benefit from the specificity, efficiency, and DNA-free nature of RNP delivery, which minimizes off-target effects and immune responses. These methodologies provide robust platforms for functional genomics, disease mechanism studies, and therapeutic development, underscoring the continued evolution of zebrafish as a premier model organism for biomedical research.
Assessing Off-Target Effects and the Fidelity of RNP-Mediated Editing
Within functional genomics and disease modeling, the zebrafish (Danio rerio) has emerged as a pivotal vertebrate model organism. Its genetic similarity to humans, transparent embryos, and rapid external development make it an excellent platform for high-throughput genetic studies [15] [61]. The delivery of preassembled ribonucleoprotein (RNP) complexes—comprising Cas protein and guide RNA—into zebrafish embryos via microinjection has become a preferred methodology for CRISPR-based genome editing. This approach offers rapid activity, reduced off-target effects, and diminished DNA vector integration compared to mRNA delivery [62]. However, as the demand for precise genetic modeling grows, particularly for introducing single-nucleotide variants associated with human diseases, a critical evaluation of RNP-mediated editing fidelity is essential. This application note systematically assesses the off-target profiles and on-target fidelity of RNP-based editing platforms in zebrafish, providing validated protocols and analytical frameworks to enhance the rigor of genetic findings in both basic research and drug development contexts.
The RNP Editing Platform: Mechanisms and Fidelity Advantages
RNP complexes for zebrafish editing typically consist of a purified Cas nuclease (e.g., Cas9, base editor, or prime editor) complexed with a synthetic guide RNA. This complex is microinjected directly into the single-cell stage embryo, facilitating immediate genome targeting before the first cell division [63].
The primary mechanistic advantage of RNP delivery lies in its transient activity. Unlike plasmid or mRNA delivery, which require transcription and/or translation and can lead to prolonged nuclease expression, RNP complexes are degraded within hours. This brief window of activity significantly reduces the probability of off-target editing at loci with sequence similarity to the intended target [62] [64]. Furthermore, the use of chemically modified guide RNAs (cm-gRNAs), featuring 2'-O-methyl analogs and 3'-phosphorothioate linkages at the terminal nucleotides, enhances complex stability and editing efficiency while further minimizing off-target potential [62] [65].
Base editors and prime editors represent advanced RNP platforms that enable precise nucleotide changes without inducing double-strand breaks (DSBs). Cytosine base editors (CBEs) and adenine base editors (ABEs) mediate C•G to T•A and A•T to G•C conversions, respectively, while prime editors (PEs) can facilitate all 12 possible base-to-base conversions, as well as small insertions and deletions [15] [7]. The fidelity of these systems is paramount for accurately modeling human genetic diseases.
Quantitative Assessment of Off-Target Effects
Systematic evaluation of RNP editing platforms in zebrafish reveals generally low off-target activity, though careful assessment remains crucial for critical applications.
Nuclease Platform Off-Target Profiles
Studies optimizing SpG and SpRY nucleases—variants with relaxed PAM requirements—have included comprehensive off-target assessments. When RNP complexes containing these nucleases were evaluated against their three most likely off-target sites (predicted by Cas-OFFinder and CRISPOR), extremely low mutation frequencies (≤0.5%) were observed. In several cases, no off-target mutations were detected above background sequencing error rates [62].
A broader systematic study investigating 50 different gRNAs in zebrafish embryos found that the majority of predicted off-target loci showed low in vivo mutation frequencies (<1%). This confirms that RNP delivery, combined with the inherent biology of the zebrafish embryo, creates an environment where off-target editing is infrequent [64].
Table 1: Quantitative Off-Target Assessment of RNP Platforms in Zebrafish
| Editing Platform | Assessment Method | Off-Target Frequency Range | Key Findings | Citation |
|---|---|---|---|---|
| SpG/SpRY Nuclease | NGS of top 3 predicted sites | ≤0.5% | Low or undetectable off-target mutations across all tested gRNAs | [62] |
| Cas9 Nuclease | NGS of homology-predicted sites | <1% (majority of sites) | Low in vivo off-target activity with RNP delivery | [64] |
| Base Editors (CBE/ABE) | Genome-wide methods | Not specified | Inherently reduced off-target risk due to DSB-free mechanism | [15] |
| Prime Editors (PE) | Targeted NGS | Not specified | High precision scores (e.g., PE2: 40.8%) indicating specific editing | [26] |
Fidelity of Base and Prime Editing Platforms
Base and prime editors offer enhanced precision through their DSB-free mechanisms. The precision of editing—measured as the ratio of precise intended edits to total edits (including imprecise edits and indels)—is a key fidelity metric. In zebrafish, the PE2 prime editor demonstrated a precision score of 40.8%, significantly higher than the PEn nuclease-based editor (11.4%) at the same locus [26].
For base editors, the development of high-fidelity variants (e.g., HF-BE3) has reduced off-target editing by up to 37-fold at non-repetitive sites and 3-fold at highly repetitive sites compared to standard base editors [15]. These improvements highlight the ongoing refinement of editing platforms for enhanced fidelity.
Experimental Protocol for Off-Target Assessment
This section provides a detailed methodology for evaluating the fidelity of RNP-mediated editing in zebrafish, from initial design to validation.
gRNA Design and RNP Complex Preparation
gRNA Design Considerations:
- Target Selection: Utilize specialized tools (e.g., CHOPCHOP, CRISPRScan) that incorporate zebrafish-specific parameters for gRNA design [63] [64].
- Efficiency Prediction: Note that computational efficiency predictions vary significantly between tools; empirical validation remains essential [64].
- Off-Target Prediction: Identify potential off-target sites using Cas-OFFinder or CCTop, prioritizing sites with up to 3-4 mismatches in genomic regions with similar sequence context [64] [63].
RNP Complex Assembly:
- Protein Purification: Use Cas proteins with bipartite nuclear localization signals (bpNLS) for enhanced nuclear import. Purify proteins using standard affinity chromatography methods [62].
- Guide RNA Synthesis: Employ chemically modified gRNAs (cm-gRNAs) with 2'-O-methyl-3'-phosphorothioate (MS) modifications at the terminal three nucleotides to enhance stability and editing efficiency [62] [65].
- Complex Formation: Incubate purified Cas protein with cm-gRNA at a 1:1.2 molar ratio in microinjection buffer for 10-15 minutes at 37°C to form RNP complexes [7] [62].
Microinjection and Sample Collection
Microinjection Protocol:
- Prepare RNP complexes at optimal concentration (e.g., 5 μM for SpG/SpRY systems) in injection buffer containing phenol red for visualization [62] [63].
- Inject 1-2 nL of RNP complex into the cell cytoplasm or yolk of one-cell stage zebrafish embryos using fine-glass capillary needles [63] [7].
- Maintain injected embryos in E3 medium at 28.5°C, replacing medium daily and removing deceased embryos [63].
Sample Collection for Genotyping:
- At 2-5 days post-fertilization, pool 6-8 normally developed embryos for initial efficiency screening or maintain individually for germline transmission analysis [7] [64].
- Extract genomic DNA using commercial kits (e.g., QIAamp DNA Mini Kit) with extended proteinase K digestion (2-3 hours at 56°C) for embryo lysates [7] [64].
On-Target and Off-Target Analysis
On-Target Efficiency Assessment:
- PCR Amplification: Amplify target regions using primers flanking the edit site (amplicon size: 200-500 bp).
- Next-Generation Sequencing (NGS): Prepare barcoded libraries from PCR products for deep sequencing on platforms such as Illumina NovaSeq [7] [62].
- Editing Quantification: Use tools like CRISPResso2 or CrispRVariants to align sequencing reads and quantify precise editing efficiencies and indel patterns [62] [64].
Off-Target Assessment Workflow:
- In Silico Prediction: Identify potential off-target sites using prediction tools (Cas-OFFinder, CRISPOR) with zebrafish genome assemblies [62] [64].
- Targeted NGS: Design amplicons for the top 3-5 predicted off-target sites per gRNA based on similarity score and genomic context.
- Validation Sequencing: Sequence potential off-target regions with sufficient depth (>100,000x coverage) to detect low-frequency mutations [62].
- Data Analysis: Compare variant frequencies in injected embryos versus uninjected controls, considering mutations above 0.1% as potential off-target events after background subtraction [62] [64].
Research Reagent Solutions
Table 2: Essential Reagents for RNP-Mediated Editing in Zebrafish
| Reagent Category | Specific Examples | Function & Application | Key Features |
|---|---|---|---|
| Cas Proteins | SpCas9, SpG, SpRY, Base Editors (CBE4max, ABE8e), Prime Editors (PE2, PE7) | Catalytic core for DNA recognition and editing | SpG: NGN PAM recognition; SpRY: Near PAM-less; Base Editors: Single-nucleotide changes; Prime Editors: Diverse edits without DSBs |
| Guide RNAs | Chemically modified gRNAs (cm-gRNAs), La-accessible pegRNAs | Target specificity and editor instruction | 2'-O-methyl-3'-phosphorothioate modifications enhance stability; Specific designs for base/prime editing |
| Microinjection Supplies | Glass capillaries, Microinjection apparatus, Injection molds | Precise delivery of RNP complexes | Fine control for 1-2 nL injections into one-cell embryos |
| Analysis Tools | CRISPRscan, Cas-OFFinder, CRISPResso2, ICE, TIDE | gRNA design, efficiency prediction, and outcome analysis | Zebrafish-optimized algorithms; NGS data analysis capabilities |
RNP-mediated genome editing in zebrafish represents a robust and specific platform for functional genomics and disease modeling. The transient nature of RNP activity, combined with advanced editor architectures and chemically modified guide RNAs, yields a favorable fidelity profile with minimal off-target effects. The experimental framework presented herein provides a standardized approach for researchers to rigorously validate editing precision, ensuring the reliability of genetic models in zebrafish. As editing technologies continue to evolve toward single-nucleotide resolution, these foundational principles and protocols will remain essential for maximizing scientific rigor in both basic research and preclinical drug development applications.
Conclusion
The microinjection of pre-assembled RNP complexes has firmly established itself as a superior method for precision genome editing in zebrafish embryos. By offering a transient, highly specific, and immediately active editing system, RNPs mitigate the key limitations of traditional DNA- and mRNA-based delivery, such as prolonged nuclease expression and increased off-target effects. Recent optimizations, including the use of advanced prime editors like PE7, chemically modified guide RNAs, and modulation of DNA repair pathways, have dramatically increased editing efficiencies, enabling the robust generation of sophisticated disease models that were previously challenging to create. As the technology continues to evolve, future efforts will focus on expanding the editing scope through novel Cas variants, refining tissue-specific delivery, and translating these precise genetic modifications into tangible therapeutic strategies for human disease. The continued adoption and refinement of RNP technology in zebrafish promise to accelerate both basic biological discovery and pre-clinical drug development.
References
- 1. Ribonucleoprotein particle - Wikipedia [https://en.wikipedia.org/wiki/Ribonucleoprotein_particle]
- 2. Nucleoprotein - Wikipedia [https://en.wikipedia.org/wiki/Nucleoprotein]
- 3. Ribonucleoprotein complex (RNP) - Innovative Genomics Institute (IGI) [https://innovativegenomics.org/glossary/ribonucleoprotein-complex-rnp/]
- 4. 5 Reasons Why Ribonucleoproteins Are a Better ... [https://www.synthego.com/blog/crispr-plasmid-pitfalls/]
- 5. CRISPR Delivery Methods: Cargo, Vehicles, and Challenges [https://www.synthego.com/blog/delivery-crispr-cas9/]
- 6. Efficient Delivery of CRISPR-Cas9 RNP Complexes with ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12608112/]
- 7. Optimized Ribonucleoprotein Complexes Enhance Prime ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12345536/]
- 8. A Technique to Deliver a Compound into the Zebrafish Yolk [https://www.jove.com/v/20196/embryo-microinjection-technique-to-deliver-compound-into-zebrafish]
- 9. CRISPR-Cas9 delivery strategies with engineered ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10571031/]
- 10. sgRNA and Cas9 Complex - Ribonucleoprotein (RNP) ... [https://rna.bocsci.com/services/sgrna-and-cas9-complex-ribonucleoprotein-rnp-synthesis.html]
- 11. Exploring RNP: A Cutting-Edge Tool for Gene Delivery [https://kactusbio.com/blogs/gene-editing/rnp-an-emerging-powerful-tool-for-gene-delivery?srsltid=AfmBOord4-xk17215_2JZJn-egvjofvXC7kvfP7HN73YPB3AE9ZhixOA]
- 12. CRISPR prime editing with ribonucleoprotein complexes in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8553808/]
- 13. CRISPR-Cas9 induces large structural variants at on-target ... [https://www.nature.com/articles/s41467-022-28244-5]
- 14. Base editors in zebrafish: a new era for functional ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12133889/]
- 15. Base editors in zebrafish: a new era for functional ... [https://www.frontiersin.org/journals/genome-editing/articles/10.3389/fgeed.2025.1598887/full]
- 16. Microinjection of Zebrafish Embryos to Analyze Gene ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2762901/]
- 17. Video: Introduction to Microinjection of Early Zebrafish ... [https://www.jove.com/v/5130/introduction-to-microinjection-of-early-zebrafish-embryos]
- 18. Quantitative characterization of zebrafish development based ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10278635/]
- 19. Ribonucleoprotein (RNP) Protocols Using Synthetic ... [https://www.sigmaaldrich.com/US/en/technical-documents/protocol/genomics/advanced-gene-editing/ribonucleoprotein-based-geneome-editing?srsltid=AfmBOoog9CX72B7wiAXvVcvp_eIlrUuLAE6BtsrXscKUFoZHErHOW-dr]
- 20. CRISPR Transfection Protocols Guide: How To Select The ... [https://www.synthego.com/guide/how-to-use-crispr/transfection-protocols/]
- 21. Enhanced RNA-targeting CRISPR-Cas technology in ... [https://www.nature.com/articles/s41467-025-57792-9]
- 22. Whole-organism 3D quantitative characterization of ... [https://elifesciences.org/articles/68920]
- 23. CRISPR-Cpf1 mediates efficient homology-directed repair ... [https://www.nature.com/articles/s41467-017-01836-2]
- 24. ( Ribonucleoprotein ) Protocols Using Synthetic sgRNAs and... RNP [https://www.sigmaaldrich.com/AT/de/technical-documents/protocol/genomics/advanced-gene-editing/ribonucleoprotein-based-geneome-editing]
- 25. Rapid and efficient cataract gene evaluation in F0 ... [https://www.sciencedirect.com/science/article/pii/S1046202320302747]
- 26. Optimised genome editing for precise DNA insertion and ... [https://elifesciences.org/reviewed-preprints/107475]
- 27. 2.5. Guide RNA: Cas9 Ribonucleoprotein (RNP) ... [https://bio-protocol.org/exchange/minidetail?id=10536680&type=30]
- 28. High doses of CRISPR/Cas9 ribonucleoprotein efficiently ... [https://www.nature.com/articles/s41598-018-30188-0]
- 29. Design and developing a robot-assisted cell batch ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11842578/]
- 30. Automated microinjection for zebrafish xenograft models [https://www.nature.com/articles/s44385-025-00016-y]
- 31. Optimized Ribonucleoprotein Complexes Enhance Prime ... [https://www.mdpi.com/2076-2615/15/15/2295]
- 32. Optimized Ribonucleoprotein Complexes Enhance Prime ... [https://pubmed.ncbi.nlm.nih.gov/40805085/]
- 33. Efficient Generation of Knock-In Zebrafish Models for Inherited ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8431507/]
- 34. Enhancing homology-directed repair efficiency with HDR ... [https://www.nature.com/articles/s41467-024-50788-x]
- 35. Efficient CRISPR-Cas9-Mediated Knock-In of Composite Tags ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7928300/]
- 36. Embryo Microinjection: A Technique to Deliver ... [https://www.jove.com/t/20196/embryo-microinjection-technique-to-deliver-compound-into-zebrafish]
- 37. Optimizing gRNA selection for high-penetrance F0 ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11915512/]
- 38. Emerging trends in prime editing for precision genome ... [https://www.nature.com/articles/s12276-025-01463-8]
- 39. Optimized Ribonucleoprotein Complexes Enhance Prime ... [https://doaj.org/article/547d35a9010a46f69785cd037e4f94c4]
- 40. Protocol CRISPR-Cas9-induced gene knockout in zebrafish [https://www.sciencedirect.com/science/article/pii/S2666166722006591]
- 41. Exploring factors influencing choice of DNA double-strand ... [https://www.sciencedirect.com/science/article/abs/pii/S1568786424000727]
- 42. Influence of PARP1 on CRISPR/Cas9 induced double ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12538023/]
- 43. Chemical Modifications of CRISPR RNAs to Improve Gene ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9636589/]
- 44. Demystifying CRISPR gRNA Chemical Modifications [https://www.synthego.com/blog/chemically-modified-grna/]
- 45. Chemical Modifications Enhance CRISPR-Cas9 Editing ... [https://www.trilinkbiotech.com/chemical-modifications-enhance-crispr-cas9-editing-efficiency]
- 46. CRISPR Genome Editing Protocols | IDT [https://www.idtdna.com/page/support-and-education/decoded-plus/protocols-for-crispr-genome-editing-in-your-model-system/]
- 47. Low-temperature embryo incubation suppresses off-target ... [https://pubmed.ncbi.nlm.nih.gov/40131558/]
- 48. Genome Editing in Zebrafish by ScCas9 Recognizing NNG ... [https://www.mdpi.com/2073-4409/10/8/2099]
- 49. A Technique to Deliver a Compound into the Zebrafish Yolk [https://app.jove.com/es/v/20196/embryo-microinjection-technique-to-deliver-compound-into-zebrafish?playlist=H678H83Z]
- 50. Maximizing CRISPR/Cas9 phenotype penetrance applying ... [https://www.nature.com/articles/s41598-020-71412-0]
- 51. Strategies to reduce the risks of mRNA drug and vaccine toxicity | Nature Reviews Drug Discovery [https://www.nature.com/articles/s41573-023-00859-3]
- 52. 3D nanoprinting of embryo microinjection needles with anti ... [https://www.nature.com/articles/s41378-025-01005-2]
- 53. Emerging trends in prime editing for precision genome ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12322275/]
- 54. Systematic optimization of prime editing for the efficient ... [https://www.nature.com/articles/s41551-024-01233-3]
- 55. Differential expression of genes in pre-blastoderm embryos ... [https://www.sciencedirect.com/science/article/pii/S014181302509292X]
- 56. Crispant analysis in zebrafish as a tool for rapid functional ... [https://elifesciences.org/reviewed-preprints/100060v1]
- 57. Exploring KRAS-mutant pancreatic ductal adenocarcinoma [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2023.1203459/full]
- 58. Classification of KRAS activating mutations and the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8988514/]
- 59. Identification and validation of a T cell receptor targeting ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11790028/]
- 60. KRAS G12V neoantigen specific T cell receptor for ... [https://www.nature.com/articles/s41467-023-42010-1]
- 61. CRISPR-based functional genomics tools in vertebrate ... [https://www.nature.com/articles/s12276-025-01514-0]
- 62. SpG and SpRY variants expand the CRISPR toolbox for ... [https://www.nature.com/articles/s41467-022-31034-8]
- 63. Protocol for generating mutant zebrafish using CRISPR-Cas9 ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10701446/]
- 64. Evaluation of CRISPR gene-editing tools in zebrafish [https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-021-08238-1]
- 65. Enhanced RNA-targeting CRISPR-Cas technology in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11911407/]