Optimizing Tyramide Signal Amplification FISH for Planarian Research: A Comprehensive Guide from Principles to Practice
This article provides a comprehensive resource for researchers applying Tyramide Signal Amplification FISH (TSA-FISH) in the planarian model system.
Optimizing Tyramide Signal Amplification FISH for Planarian Research: A Comprehensive Guide from Principles to Practice
Abstract
This article provides a comprehensive resource for researchers applying Tyramide Signal Amplification FISH (TSA-FISH) in the planarian model system. It covers the foundational principles of TSA-FISH and its critical role in enhancing the detection of low-abundance transcripts in planarian stem cell and regeneration studies. The content details optimized methodological protocols, including specialized steps for mucus removal, pigment bleaching, and autofluorescence quenching unique to planarians. Furthermore, it presents systematic troubleshooting guidance and explores advanced validation techniques and emerging multiplexing technologies. This guide is tailored for scientists and drug development professionals seeking to leverage the high sensitivity of TSA-FISH to elucidate gene expression patterns in regeneration, stem cell biology, and tissue homeostasis.
Understanding TSA-FISH and Its Critical Role in Planarian Biology
Core Principles of Tyramide Signal Amplification
Tyramide Signal Amplification (TSA) is a powerful enzyme-mediated detection method that utilizes the catalytic activity of horseradish peroxidase (HRP) to achieve high-density labeling of target proteins or nucleic acids in situ [1]. This technology, also referred to as Catalyzed Reporter Deposition (CARD), provides a significant increase in detection sensitivity compared to conventional methods [2].
The fundamental TSA reaction involves three key stages. First, a specific probe binds to the target via immunoaffinity for proteins or hybridization for nucleic acids, followed by detection with an HRP-labeled conjugate [1]. Second, in the presence of hydrogen peroxide (Hâ‚‚Oâ‚‚), the HRP enzyme catalyzes the oxidation of tyramide derivatives, converting them into highly reactive, short-lived radicals [3] [2]. Finally, these activated tyramide radicals form covalent bonds primarily with the electron-rich phenol moiety of tyrosine residues on proteins in the immediate vicinity of the HRP reaction site [1]. This deposition results in the situ accumulation of numerous reporter molecules at the target site, enabling detection of low-abundance targets that would otherwise remain undetectable [3].
The signal amplification conferred by the turnover of multiple tyramide substrates per peroxidase label can enhance detection sensitivity up to 100-fold compared to conventional avidin-biotinylated enzyme complex (ABC) procedures [1]. This covalent labeling ensures excellent spatial resolution and high signal intensity with minimal diffusion-related loss of signal localization [1].
TSA Applications in Planarian FISH Research
In planarian research, TSA technology enables highly sensitive detection of low-abundance transcripts crucial for studying stem cell dynamics and regeneration mechanisms. The exceptional sensitivity of TSA is particularly valuable for visualizing spatially restricted gene expression patterns and rare cell populations in this model organism.
Key Advantages for Planarian Research
- Detection of Low-Abundance Transcripts: TSA facilitates the visualization of weakly expressed fate-specifying transcription factors (FSTFs) that direct neoblast differentiation into specific lineages [4] [5].
- Spatial Mapping of Gene Expression: Enables precise localization of mRNA expression patterns within the complex tissue architecture of planarians, such as identifying ectopic pharynx neurons (EPNs) in the brain of
roboA(RNAi)animals [5]. - Multiplexing Capabilities: Sequential TSA labeling with different fluorophores allows simultaneous detection of multiple targets within a single tissue section, enabling comprehensive analysis of intricate biological systems [3] [6].
Experimental Protocols for TSA-Based FISH in Planarian Research
Basic TSA-FISH Protocol for Planarian Tissue Sections
This protocol adapts standard TSA methodology for detecting low-abundance mRNAs in planarian tissues, with specific considerations for planarian biology.
Day 1: Sample Preparation and Hybridization
- Fixation: Fix planarians in 4% formaldehyde in PEM (pH 7.4) for 1-2 hours at room temperature [5].
- Dehydration: Gradually dehydrate samples through an ethanol series (50%, 80%, 100%) and embed in paraffin.
- Sectioning: Cut 5-8 μm thick sections using a microtome and mount on positively charged slides.
- Deparaffinization and Rehydration: Remove paraffin with xylene and rehydrate through a descending ethanol series to PBS.
- Proteinase Digestion: Treat sections with 10 μg/mL proteinase K in PBS for 15 minutes at 37°C to expose target mRNA.
- Pre-hybridization: Pre-hybridize with hybridization buffer for 1 hour at the appropriate hybridization temperature.
- Hybridization: Apply digoxigenin-labeled riboprobes specific to target mRNA (diluted 1:100-1:500 in hybridization buffer) and incubate overnight at 55-60°C.
Day 2: Washes and Signal Detection
- Stringency Washes: Perform serial washes with SSC solutions of decreasing concentration (2× SSC to 0.2× SSC) at hybridization temperature.
- Blocking: Incubate sections with TSA blocking reagent for 1 hour to minimize non-specific binding [1].
- Antibody Incubation: Apply anti-digoxigenin-HRP Fab fragments (1:100-1:500) for 1 hour at room temperature.
- TSA Reaction: Prepare fluorochromized tyramide working solution (1:50-1:500 in amplification diluent) and apply to sections for 2-10 minutes [7] [1].
- Reaction Termination: Stop the reaction by washing with PBS or PBS-Tween.
- Counterstaining and Mounting: Counterstain nuclei with DAPI and mount with anti-fade mounting medium.
Advanced Multiplex TSA-FISH Protocol
For simultaneous detection of multiple mRNA targets, the basic protocol can be extended with sequential labeling rounds:
- Complete the basic TSA-FISH protocol for the first target.
- HRP Inactivation: Incubate sections with 2% sodium azide (NaN₃) in PBS for 30 minutes to completely quench HRP activity from the first round [7].
- Antibody Elution: Perform heat-induced epitope retrieval (HIER) using citrate buffer (pH 6.0) at 95-100°C for 20 minutes to remove antibodies while leaving deposited tyramide intact [6].
- Repeat the hybridization and TSA detection steps for the second target using a different fluorochromized tyramide.
- Repeat steps 2-4 for additional targets as needed.
Table 1: TSA Reagent Dilution and Incubation Parameters
| Reagent | Recommended Dilution | Incubation Time | Temperature |
|---|---|---|---|
| Primary Riboprobe | 1:100 - 1:500 | Overnight | 55-60°C |
| Anti-Digoxigenin-HRP | 1:100 - 1:500 | 1 hour | Room Temperature |
| Fluorochromized Tyramide | 1:50 - 1:500 | 2-10 minutes | Room Temperature |
| HRP Quencher (NaN₃) | 2% in PBS | 30 minutes | Room Temperature |
Research Reagent Solutions for Planarian TSA-FISH
Table 2: Essential Reagents for TSA-Based FISH in Planarian Research
| Reagent Category | Specific Examples | Function in TSA-FISH |
|---|---|---|
| Tyramide Reagents | Alexa Fluor Tyramides [1], Cyanine 3 Tyramide [8], iFluor Styramides [3] | Enzyme-activated substrates that deposit fluorescent markers near target sites |
| Enzyme Conjugates | HRP-conjugated anti-digoxigenin [1], HRP-streptavidin | Catalyze tyramide activation and deposition |
| Probe Labeling Systems | Digoxigenin-labeled riboprobes, Biotinylated probes | Provide target-specific recognition with haptens for detection |
| Signal Amplification Kits | TSA Vivid Fluorophore Kits [8], FT-GO System [7] | Optimized reagent systems for enhanced sensitivity and multiplexing |
| HRP Quenching Reagents | Sodium azide, Hydrogen peroxide | Inactivate peroxidase between sequential labeling rounds |
| Blocking Agents | TSA blocking reagent [1], casein [9] | Reduce non-specific background signal |
Workflow Visualization and Signaling Pathways
TSA-FISH Experimental Workflow
TSA-FISH Experimental Workflow
Molecular Mechanism of Tyramide Signal Amplification
Molecular Mechanism of TSA
Technical Considerations and Optimization
Critical Parameters for Successful TSA-FISH
Tyramide Concentration and Incubation Time: Optimal results require careful titration of tyramide concentration (typically 1:50-1:500 dilution) and reaction time (2-10 minutes) to maximize signal while minimizing background [7] [1]. Excessive tyramide or prolonged incubation can increase non-specific staining.
HRP Quenching Efficiency: Complete inactivation of HRP between sequential labeling rounds is essential for successful multiplexing. Sodium azide treatment (2% for 30 minutes) effectively quenches residual peroxidase activity without damaging tissue antigens or previously deposited tyramides [7].
Probe Design and Specificity: For planarian research, riboprobes should target unique gene regions with minimal homology to other transcripts. Specificity can be verified using RNAi-treated controls and BLAST analysis against the planarian genome database.
Troubleshooting Common Issues
High Background Signal: Reduce tyramide concentration, shorten development time, or increase blocking agent concentration. Ensure thorough washing between steps.
Weak or No Signal: Check HRP enzyme activity, increase probe concentration, or extend tyramide incubation time. Verify target expression in positive control samples.
Incomplete Multiplexing: Ensure complete HRP quenching between rounds by verifying sodium azide freshness and concentration. Consider incorporating heat-induced antibody elution for more complete removal of HRP conjugates.
The implementation of TSA technology in planarian FISH research provides unprecedented sensitivity for detecting low-abundance transcripts, enabling detailed analysis of gene expression patterns in stem cell biology and regeneration studies. The protocols and reagents outlined here offer a foundation for adapting this powerful signal amplification method to specific research applications in planarian and other model systems.
Why Planarians? Unique Challenges in Flatworm Gene Expression Analysis
Planarians, particularly Schmidtea mediterranea, have emerged as a powerful model system for studying regeneration, stem cell biology, and evolutionary developmental processes. Their remarkable capacity for whole-body regeneration [10] [11], driven by adult pluripotent stem cells called neoblasts [11], provides unparalleled opportunities for investigating fundamental biological questions. However, this same regenerative capability presents unique and significant challenges for gene expression analysis that complicate molecular studies. The planarian field lacks the comprehensive antibody libraries available for traditional model organisms, making RNA in situ hybridization (ISH) not merely an alternative but the principal method for visualizing gene expression patterns in these animals [12].
The technical obstacles are substantial. Planarians possess complex, thick, and highly autofluorescent tissues that impede clear signal detection [12]. Furthermore, their rapidly regenerating tissues require specialized fixation and permeabilization approaches to preserve morphology while allowing probe penetration [13]. Perhaps most challenging is the planarian genome itself, which exhibits a strong AT bias (>70% A/T), abundant repetitive elements including giant >30 kb Burro retroelements, and significant heterozygosity that complicates assembly and probe design [14]. These genomic peculiarities directly impact gene expression studies by creating potential for non-specific probe binding and complicating the identification of unique target sequences. Additionally, researchers must account for the dynamic nature of gene expression during regeneration and the continuous tissue turnover occurring even during homeostasis, requiring methods with sufficient sensitivity to detect subtle spatial and temporal expression patterns.
Fundamental Challenges in Planarian Gene Expression Studies
Genomic and Structural Barriers
The very genetic architecture of planarians presents foundational challenges for gene expression analysis. Recent comparative genomic analyses reveal that planarian genomes undergo rapid structural evolution, including frequent retrotransposon-associated chromosomal inversions and interchromosomal translocations [14]. Remarkably, planarians and other flatworms have experienced nearly complete loss of ancestral metazoan synteny, suggesting their genomes evolve without the structural constraints observed in most other animal groups [14]. This structural plasticity has direct implications for gene regulation studies, as conserved regulatory elements may be located at varying genomic positions even between closely related species.
The planarian transcriptome is equally complex, with improved genome annotations revealing approximately 58,000 gene loci and 21,000 high-confidence genes [14]. This genetic expansion necessitates highly specific probes to distinguish between paralogous genes, while the high AT-content complicates probe design for standard ISH methods. Additionally, the extensive heterozygosity in planarian populations, particularly maintained through a large chromosomal inversion on Chromosome 1 that resists inbreeding [14], means that effective probes must target both haplotypes to avoid false negatives in expression analysis.
Technical Limitations in Detection Methodology
Conventional RNA in situ hybridization approaches face multiple limitations when applied to planarian tissues. The gold standard methods of enzyme-catalyzed reporter deposition, including alkaline phosphatase (AP) colorimetric ISH and horseradish peroxidase (HRP) tyramide signal amplification (TSA) fluorescent ISH (FISH) [12], often prove inadequate for detecting low-abundance transcripts that play crucial roles in regeneration and stem cell maintenance. These technical constraints directly impact the study of critical biological processes in planarians, including the characterization of rare cell populations identified through single-cell RNA sequencing [15].
The autofluorescent properties of planarian tissues present another significant hurdle, particularly for fluorescent detection methods. This natural autofluorescence can mask specific signals, leading to both false negatives and false positives. Traditional probe systems also lack modularity, requiring researchers to design, validate, and optimize separate probes for each detection method they employ. This inflexibility consumes valuable time and resources, particularly when studying dynamic processes like regeneration that may require both colorimetric and fluorescent approaches at different stages of analysis.
Solution: An Integrated Approach with OneSABER and TSA-FISH
The OneSABER Platform: Unified Probe Design
The OneSABER platform represents a significant advancement for planarian gene expression studies, functioning as a modular "one probe fits all" approach that connects commonly used ISH techniques within a unified framework [12]. This system addresses the core challenge of probe compatibility by using a single type of DNA probe that can be adapted to multiple signal detection methods, thereby reducing both development time and resource investment.
At the heart of the OneSABER system is a pool of 15-30 custom short single-stranded DNA oligonucleotides (35-45 nucleotides) complementary to the target RNA [12]. Each probe contains a specific 9-nucleotide 3' initiator sequence that enables in vitro extension through a primer exchange reaction (PER) to generate long concatemerized probes. The length of these concatemers, which directly determines signal amplification strength, can be precisely controlled by adjusting reaction time [12]. These concatemers then serve as universal landing platforms for short secondary oligonucleotide probes modified according to the chosen detection method (colorimetric or fluorescent).
Table: OneSABER Platform Components and Functions
| Component | Description | Function in Assay |
|---|---|---|
| Primary Oligonucleotides | 15-30 short ssDNA (35-45 nt) with initiator sequence | Target-specific binding to RNA of interest |
| Primer Exchange Reaction (PER) | Catalytic DNA hairpin with strand-displacing polymerase | Generates long concatemers for signal amplification |
| Secondary Probes | Short ssDNA (20 nt) with hapten labels | Adapts concatemers to various detection methods |
| Concatemer Landing Pads | Repeated sequences on extended probes | Enables binding of multiple secondary probes |
Enhanced TSA-FISH for Planarians
Tyramide Signal Amplification combined with Fluorescent in situ Hybridization (TSA-FISH) provides the necessary sensitivity for detecting low-abundance transcripts in planarians. The TSA system operates through a horseradish peroxidase (HRP)-catalyzed deposition of fluorescent tyramide derivatives that form covalent bonds with electron-rich amino acids in nearby proteins, resulting in substantial signal amplification at the site of hybridization [12] [13].
Critical modifications to standard TSA-FISH protocols have been developed specifically to address planarian-specific challenges. A short bleaching step in formamide dramatically enhances signal intensity by reducing tissue autofluorescence [13]. Copper sulfate quenching provides further reduction of autofluorescence, while iterative rounds of tyramide signal amplification boost sensitivity for low-expression targets [13]. For regenerating planarians, a heat-induced antigen retrieval step improves the balance between tissue permeabilization and preservation of delicate regenerating structures [13]. Between development rounds in multicolor FISH experiments, azide effectively quenches peroxidase activity to prevent cross-reaction [13].
Research Reagent Solutions for Planarian Studies
Table: Essential Research Reagents for Planarian Gene Expression Analysis
| Reagent/Category | Specific Examples | Function in Planarian Research |
|---|---|---|
| Probe Systems | OneSABER concatemers, SABER-FISH probes | Universal probe platform for multiple detection methods |
| Hapten Labels | Digoxigenin (DIG), Fluorescein (FITC) | Antibody-recognizable labels for signal detection |
| Detection Enzymes | Horseradish peroxidase (HRP), Alkaline phosphatase (AP) | Enzyme conjugates for colorimetric and fluorescent detection |
| Signal Amplification | Tyramide Signal Amplification (TSA) kits | Signal enhancement for low-abundance targets |
| Tissue Processing | Formamide, Proteinase K, Heat-induced antigen retrieval | Tissue permeabilization and autofluorescence reduction |
| Microscopy Mounting | Copper sulfate solution, Azide compounds | Autofluorescence quenching and peroxidase inactivation |
Detailed Experimental Protocol: OneSABER with TSA-FISH for Planarians
Probe Design and Preparation
Target Selection: Identify unique target sequences within your gene of interest using the most recent planarian genome assemblies (S3h1 or S3h2) [14] to avoid cross-reactivity with repetitive elements or paralogous genes.
Oligonucleotide Design: Design 15-30 single-stranded DNA oligonucleotides (35-45 nucleotides each) complementary to the target RNA. Add a specific 9-nucleotide 3' initiator sequence (e.g., from Kishi et al., 2019) to each oligonucleotide.
Primer Exchange Reaction: Perform PER extension using catalytic DNA hairpin and strand-displacing polymerase to generate concatemerized probes:
- Assemble reaction with 1µM of each oligonucleotide, 1µM catalytic hairpin, and 0.5U/µL Bst DNA polymerase
- Incubate at 37°C for 30-120 minutes (longer times yield longer concatemers)
- Heat-inactivate at 80°C for 20 minutes
Secondary Probe Labeling: Design 20-nucleotide secondary probes complementary to the concatemer repeat sequence. Label these probes with haptens (DIG, FITC) appropriate for your detection method.
Sample Preparation and Hybridization
Planarian Fixation: Fix planarians in 4% formaldehyde in 1X PBS for 45 minutes at room temperature. For regenerating specimens, reduce fixation time to 20-30 minutes to preserve tissue integrity [13].
Autofluorescence Reduction:
Heat-Induced Antigen Retrieval: For regenerating tissues or difficult-to-penetrate specimens, perform heat-induced antigen retrieval by incubating in pre-warmed 10mM sodium citrate (pH 6.0) at 85°C for 20 minutes [13].
Hybridization:
- Permeabilize with 1µg/mL Proteinase K in 1X PBS with 0.1% Tween-20 for 10-20 minutes
- Pre-hybridize in hybridization buffer for 2-4 hours at 55-60°C
- Hybridize with OneSABER concatemer probes (10-100nM) in hybridization buffer overnight at 55-60°C
Signal Detection and Amplification
Secondary Probe Hybridization:
- Wash 4×15 minutes in SSCT (0.3-0.5X SSC) at 55-60°C
- Hybridize with hapten-labeled secondary probes (10-50nM) in hybridization buffer for 2-4 hours at room temperature
Antibody Binding:
- Block in 5% heat-inactivated goat serum in 1X PBS with 0.1% Triton X-100 for 2-4 hours
- Incubate with HRP-conjugated anti-hapten antibodies (1:1000-1:2000) overnight at 4°C
Tyramide Signal Amplification:
- Wash 6×20 minutes in 1X PBS with 0.1% Triton X-100
- Develop with appropriate fluorescent tyramide substrate (1:100-1:500) for 10-30 minutes
- For multiplexing, quench peroxidase activity with 0.1% sodium azide in 1X PBS for 30-60 minutes between rounds [13]
Imaging and Analysis:
- Mount in anti-fading mounting medium
- Image using confocal or epifluorescence microscopy with appropriate filter sets
- For multicolor experiments, establish spectral unmixing parameters using single-labeled controls
Applications in Planarian Research
The integrated OneSABER with TSA-FISH approach enables critical advancements across multiple domains of planarian biology. In regeneration studies, this method has revealed species-specific differences in wound site expression of the head determinant notum, explaining why narrow fragments of Girardia sinensis frequently regenerate as double-headed animals while Schmidtea mediterranea maintains regeneration specificity [10]. The technique has proven essential for characterizing the role of the nucleosome remodeling and deacetylase (NuRD) complex in planarian stem cell differentiation, where it drives the transition from progenitor cells to somatic cells in epidermis and intestine [11].
In planarian neuroscience, enhanced FISH has enabled the mapping of mechanosensory neurons marked by the transcription factor pou4-2, which regulates the regeneration of ciliated sensory cells that detect water flow [16]. Single-cell RNA sequencing studies of planarian allometry have relied on FISH for validating cell type proportions that change with body size, particularly revealing dynamic responses in epidermal cells and gut basal cells [15]. The method has also proven valuable for optimizing RNAi protocols, where it can visually confirm knockdown efficiency and determine that a single feeding of double-stranded RNA induces phenotypes lasting up to 11 weeks [17].
These applications demonstrate how overcoming the technical challenges of planarian gene expression analysis provides deeper insights into the remarkable biology of these regenerating organisms, from stem cell regulation to whole-body regeneration and tissue polarity maintenance.
The Problem of Low-Abundance Transcripts in Stem Cell and Regeneration Studies
In the field of stem cell and regeneration research, accurately profiling transcriptional activity is fundamental to understanding mechanisms of self-renewal, differentiation, and tissue repair. A significant technical challenge in these studies is the reliable detection of low-abundance transcripts. These transcripts, which include key fate-specifying transcription factors (FSTFs) and regulatory genes, are often expressed at low levels but play critical roles in determining stem cell fate [4] [18]. In planarian research, which utilizes organisms like Schmidtea mediterranea as powerful models for regeneration, this problem is acute. The complex microenvironment and the vast diversity of specialized cell types generated from neoblasts mean that crucial low-copy-number mRNAs can be masked by more abundant RNAs, such as those for housekeeping genes or structural proteins [19]. This application note, framed within a broader thesis on tyramide signal amplification FISH (TSA-FISH) in planarian research, details the problem of low-abundance transcripts and provides validated protocols to overcome it, enabling deeper insights into stem cell biology.
Technical Challenges and Quantitative Impact
The inability to detect low-abundance transcripts can lead to incomplete or biased cellular taxonomy and a flawed understanding of regulatory networks. In planarians, for instance, neoblasts choose among over 125 fates, a process guided by the expression of specific FSTFs [18]. If these FSTF transcripts go undetected, the mechanisms of fate specification remain opaque.
Recent research quantifies the extent and impact of this issue. One study systematically evaluated the effect of highly abundant transcripts on single-cell RNA sequencing (scRNA-seq) libraries. It found that these abundant RNAs can dominate sequencing reads, effectively obscuring the detection of less prevalent but biologically critical mRNAs [19]. The table below summarizes key quantitative findings on the effects of masking by abundant transcripts and the efficacy of a novel CRISPR-Cas9 cleaning method.
Table 1: Impact and Mitigation of Low-Abundance Transcript Masking
| Parameter | Value / Finding | Experimental Context |
|---|---|---|
| Read Redistribution | ~50% of sequencing reads redistributed to low-abundance transcripts after cleaning [19]. | scRNA-seq libraries following CRISPR-Cas9 removal of abundant RNAs. |
| Transcript Removal | <1% of highly abundant transcripts targeted and removed [19]. | CRISPR-Cas9 targeting of abundant transcripts (e.g., ribosomal, mitochondrial). |
| Detection Enhancement | Enhanced biological signature detection in both heterogeneous and homogeneous cell populations [19]. | Application in immune cells and vascular smooth muscle cells. |
| Sequencing Efficiency | Maintained sequencing quality at half the depth, reducing costs [19]. | Comparison of cleaned vs. standard libraries from human intestinal tissue. |
| Fate Specification Location | Specialized neoblasts are spatially intermingled, not clustered by fate [18]. | MERFISH spatial mapping of 61 FSTFs in planarians. |
| Distance to Target Tissue | Body-wall muscle-specialized neoblasts averaged ~72 microns from target tissue [18]. | Spatial analysis of neoblast proximity to differentiated tissues. |
Spatial context adds another layer of complexity. Studies using multiplexed error-robust FISH (MERFISH) have revealed that neoblasts specifying different fates are highly intermingled throughout the planarian body, with neighbors often committing to divergent lineages [18]. This spatial distribution means that low-resolution or low-sensitivity techniques risk averaging out critical signals or missing them entirely, failing to capture the true complexity of stem cell decision-making in situ.
Methodological Approaches for Enhanced Detection
Several advanced methodologies have been developed to overcome the challenge of low-abundance transcripts. The choice of method depends on whether the goal is to profile all transcripts within a cell population or to visualize the spatial location of specific targets.
- CRISPR-Cas9-mediated Transcript Depletion (scCLEAN): This wet-lab method uses CRISPR-Cas9 to selectively fragment and remove pervasive non-variable RNAs (e.g., ribosomal, mitochondrial) from single-cell RNA-seq libraries before sequencing. This physical removal of abundant species allows the sequencing budget to be reallocated to low-abundance transcripts, significantly enhancing their detection without increasing sequencing depth [19].
- Multiplexed Error-Robust Fluorescence In Situ Hybridization (MERFISH): MERFISH is a single-molecule FISH methodology that enables the simultaneous detection of hundreds to thousands of distinct RNA species within tissue sections with subcellular resolution. Its high multiplexing capability and error-robust encoding scheme make it exceptionally powerful for creating spatial maps of gene expression, including for low-abundance FSTFs, within complex tissues like planarians [18].
- Tyramide Signal Amplification FISH (TSA-FISH): TSA-FISH is a catalytic signal amplification method that greatly enhances the sensitivity of traditional FISH. It is particularly well-suited for detecting low-copy-number mRNAs in whole-mount planarian specimens, making it an accessible and powerful tool for many laboratories [18].
Detailed Protocol: TSA-FISH for Low-Abundance Transcripts in Planarians
This protocol is optimized for the detection of low-abundance transcripts, such as FSTFs, in intact planarians, providing high signal-to-noise ratio for clear visualization.
Materials and Reagents
Table 2: Key Research Reagent Solutions
| Item | Function / Explanation |
|---|---|
| Fixative (4% Paraformaldehyde) | Preserves tissue morphology and immobilizes RNA transcripts in situ. |
| Proteinase K | Permeabilizes the fixed tissue by digesting proteins, allowing probe access. |
| Digoxigenin (DIG)-labeled RNA Probe | A target-specific RNA probe labeled with DIG, which serves as a hapten for antibody binding. |
| Anti-DIG-Horseradish Peroxidase (HRP) | A conjugate antibody that binds to the DIG label on the hybridized probe. |
| Tyramide-Fluorophore Conjugate | The substrate for HRP. Upon reaction, it deposits numerous fluorescent tyramide molecules at the site of probe hybridization, providing massive signal amplification. |
| Blocking Reagent (e.g., from Roche) | Reduces nonspecific binding of antibodies to the tissue, minimizing background noise. |
Step-by-Step Procedure
Sample Preparation and Fixation:
- Anesthetize planarians in ice-cold water.
- Fix specimens in 4% paraformaldehyde in PBS for 2 hours at room temperature or overnight at 4°C with gentle agitation.
- Wash 3 x 10 minutes in PBS.
Permeabilization and Pre-hybridization:
- Treat fixed planarians with Proteinase K (e.g., 10 µg/mL in PBS) for a duration optimized for your sample size (typically 5-15 minutes). Critical: Do not over-digest.
- Post-fix in 4% PFA for 10 minutes to stabilize the tissue after proteinase treatment.
- Wash in PBS.
- Pre-hybridize in a hybridization buffer containing formamide for at least 2 hours at the hybridization temperature (e.g., 56-60°C).
Probe Hybridization:
- Dilute the DIG-labeled riboprobe (1-2 µg/mL) in fresh hybridization buffer.
- Incubate planarians in the probe solution for 16-40 hours (overnight to two nights) at the hybridization temperature in a dark, humidified chamber.
Stringency Washes:
- Remove unbound probe with a series of stringent washes:
- Wash in a solution of 50% formamide, 2x SSC, 0.1% Tween-20 at hybridization temperature.
- Follow with washes in 2x SSC/0.1% Tween-20 and then 0.2x SSC/0.1% Tween-20 at the same temperature.
- Finally, wash in a solution of MAB (Maleic Acid Buffer) or PBS with 0.1% Tween-20.
- Remove unbound probe with a series of stringent washes:
Signal Amplification with Tyramide:
- Block nonspecific sites by incubating in a blocking solution (e.g., 2% Blocking Reagent in MAB or PBS) for 2-4 hours.
- Incubate with anti-DIG-HRP conjugate (typically 1:500-1:2000 dilution in blocking solution) overnight at 4°C.
- Wash extensively (e.g., 6 x 30 minutes) with PBS/0.1% Tween-20 to remove all unbound antibody.
- Immediately before development, prepare the fluorophore-tyramide working solution according to the manufacturer's instructions (e.g., 1:100 dilution in amplification buffer).
- Incubate planarians in the tyramide working solution for 10-30 minutes in the dark. Note: The reaction time must be optimized empirically for each probe to balance signal strength and background.
- Stop the reaction by washing in PBS/0.1% Tween-20.
Imaging and Analysis:
- Counterstain nuclei with DAPI if desired.
- Mount samples for microscopy and image using a fluorescence or confocal microscope.
The following workflow diagram illustrates the key steps and logical progression of the TSA-FISH protocol.
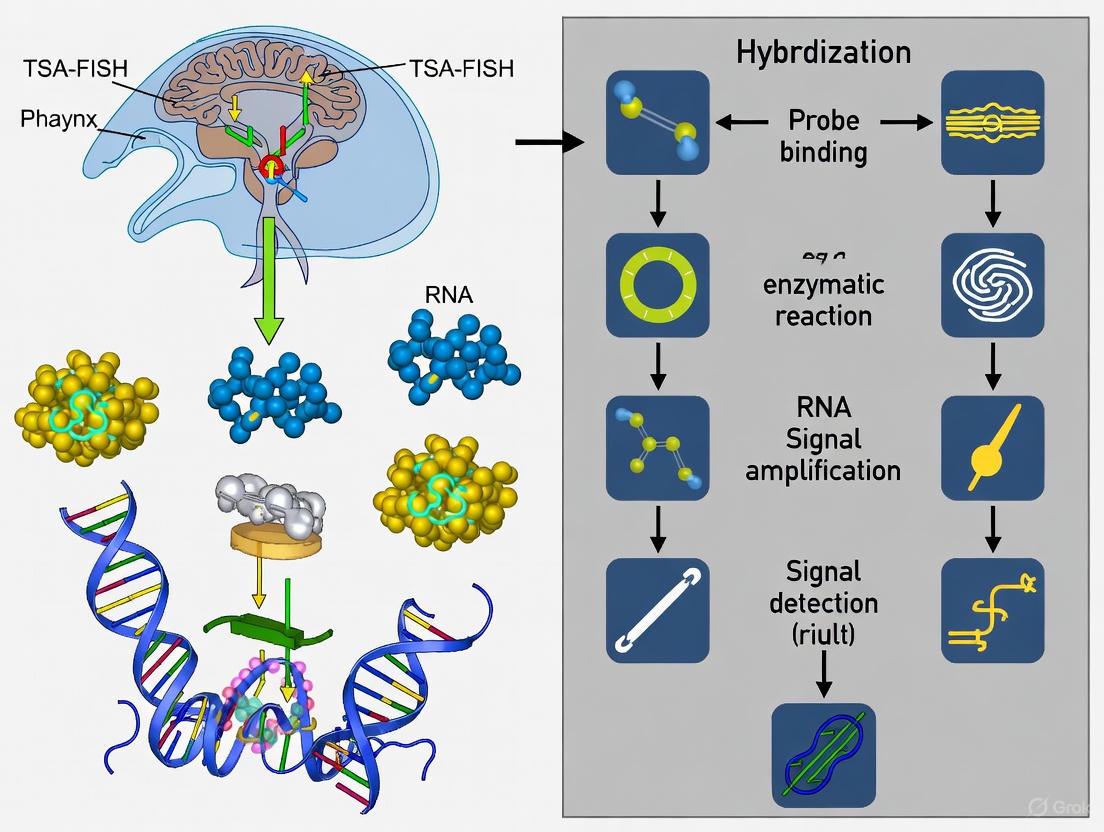
Figure 1: TSA-FISH experimental workflow for detecting low-abundance transcripts.
TSA Amplification Mechanism
The core of TSA-FISH's sensitivity lies in the catalytic reaction driven by Horseradish Peroxidase (HRP). The following diagram details the mechanism of signal amplification at the target transcript site.
Figure 2: TSA signal amplification mechanism for sensitive mRNA detection.
Tyramide Signal Amplification Fluorescent In Situ Hybridization (TSA-FISH) represents a powerful methodological synergy that combines the precise genetic targeting of FISH with the exceptional signal enhancement of TSA. This technique is particularly vital in research organisms like planarians, where it enables the detection of low-abundance mRNA transcripts critical for understanding stem cell biology and regenerative mechanisms [20] [21]. The core principle hinges on utilizing horseradish peroxidase (HRP) to catalyze the deposition of fluorescently-labeled tyramide substrates at the site of probe hybridization, resulting in signal amplification that can reach up to 100-fold compared to conventional FISH methods [22] [23]. This application note details a standardized TSA-FISH workflow, contextualized for planarian research to support drug discovery scientists in studying gene expression dynamics.
The TSA-FISH procedure integrates sequential molecular and enzymatic steps to achieve highly sensitive detection. The workflow begins with sample preparation and probe design, followed by hybridization of hapten-labeled nucleic acid probes to target RNA sequences. Subsequently, an HRP enzyme conjugate binds to the hapten, catalyzing the activation and covalent deposition of tyramide molecules onto nearby tyrosine residues [23] [8]. This deposition creates a dense fluorescent signal localized precisely at the target site.
A key advantage of TSA-FISH is its compatibility with multiplexing. By performing sequential rounds of HRP inactivation and re-probing with different tyramide dyes, researchers can visualize multiple RNA transcripts simultaneously within the same sample [24] [25]. This capability is invaluable for analyzing complex gene regulatory networks in planarian stem cells (neoblasts) and their progeny [21].
Research Reagent Solutions
Selecting appropriate reagents is fundamental to a successful TSA-FISH experiment. The table below outlines essential components and their functions within the workflow.
Table 1: Key Reagents for TSA-FISH
| Reagent Category | Specific Examples | Function & Role in Assay |
|---|---|---|
| Haptens for Probe Labeling | Biotin, Digoxigenin, Fluorescein, DNP [24] [23] | Incorporated into nucleic acid probes for subsequent detection via HRP-conjugates. |
| HRP Conjugates | Streptavidin-HRP, Anti-Digoxigenin-HRP, Anti-Fluorescein-HRP, Anti-Hapten Antibody-HRP [24] [23] | Binds to hapten on hybridized probe and catalyzes tyramide activation. |
| Fluorescent Tyramides | Alexa Fluor tyramides (e.g., 488, 546, 647), Cyanine dye tyramides (e.g., Cy3, Cy5) [22] [8] | HRP substrate; activated form binds covalently to tyrosine residues for signal amplification. |
| Amplification Buffer | Commercially available buffers (e.g., from Biotium, Thermo Fisher) with Hâ‚‚Oâ‚‚ [22] [23] | Provides optimal chemical environment (pH, Hâ‚‚Oâ‚‚ concentration) for the HRP-tyramide reaction. |
Detailed TSA-FISH Protocol
Probe Design and Preparation
Effective probe design is the foundation for specific target detection. For planarian research, this often involves targeting transcripts of interest, such as Smed-pou4-2, a transcription factor critical for mechanosensory neuron regeneration [21].
- Design Parameters: Design RNA or DNA probes complementary to your target mRNA, typically 200-300 bases in length for optimal tissue penetration [24].
- Labeling: Incorporate a hapten (e.g., digoxigenin or biotin) during probe synthesis via in vitro transcription or PCR labeling methods.
- Fragmentation (Optional): For some probes, perform limited alkaline hydrolysis to reduce fragment size and improve signal-to-noise ratio [24].
- Incubate probe in 2X carbonate buffer (pH 10.2) at 42°C for 30-60 minutes.
- Precipitate fragmented probe with ethanol and sodium acetate, then resuspend in hybridization buffer to a final concentration of 5-50 ng/µL. [24]
Sample Preparation and Fixation
Proper tissue preservation is critical for maintaining RNA integrity and morphology. For planarians:
- Fixation: Anesthetize planarians and fix whole animals or tissue fragments in 4% paraformaldehyde (PFA) in PBS for several hours to overnight at 4°C [21].
- Permeabilization: Treat fixed samples with a proteinase K solution (e.g., 10 µg/mL in PBT for 5-7 minutes) to expose target mRNA. This step must be empirically optimized to avoid over-digestion [24].
- Pre-hybridization: Equilibrate samples in hybridization solution for at least 1 hour at the hybridization temperature (e.g., 55°C) [24].
Hybridization and Post-Hybridization Washes
This phase ensures specific binding of the probe to its target sequence.
- Hybridization: Denature the probe at 65°C for 10 minutes, then add to samples in sufficient hybridization buffer. Incubate for 20-24 hours at the optimized hybridization temperature (e.g., 55°C) [24].
- Stringency Washes: Remove non-specifically bound probe through a series of post-hybridization washes:
- Wash 2-3 times with pre-warmed hybridization solution at the hybridization temperature.
- Perform a graded series of washes with a mixture of hybridization solution and PBT.
- Complete with several washes in PBT alone at room temperature [24].
Tyramide Signal Amplification
The amplification step dramatically enhances detection sensitivity.
- Blocking: Incubate samples in a blocking solution (e.g., PBT with 2% normal goat serum and 2% BSA) for 30-60 minutes to minimize non-specific binding [24].
- HRP Conjugate Incubation: Apply the appropriate HRP conjugate (e.g., streptavidin-HRP for biotinylated probes or anti-digoxigenin-HRP) diluted in blocking buffer. Incubate for 2 hours at room temperature, protected from light [24] [23].
- Tyramide Reaction:
- Prepare the tyramide working solution by diluting fluorescent tyramide reagent (1:100 to 1:500) in a dedicated amplification buffer, adding 0.01% Hâ‚‚Oâ‚‚ to initiate the reaction [24] [25].
- Incubate samples in the tyramide solution for 1-2 hours at room temperature, protected from light.
- Stop the reaction by washing several times with PBST to remove unbound tyramide [24].
Signal Detection and Multiplexing
- Imaging: Mount samples in an anti-fading mounting medium and image using a fluorescence microscope equipped with appropriate filter sets. The amplified signal is typically bright and photostable [22].
- Multiplexing: For detecting multiple targets, the HRP activity from the first round can be inactivated by treating with a peroxidase quenching buffer (e.g., 3% Hâ‚‚Oâ‚‚). The entire process, starting with a second primary probe hybridization, can then be repeated using a tyramide conjugated to a different fluorophore [8].
Table 2: Troubleshooting Common TSA-FISH Issues
| Problem | Potential Cause | Solution |
|---|---|---|
| High Background | Incomplete washing, over-amplification, non-specific probe binding. | Optimize wash stringency; titrate down probe concentration and tyramide incubation time; ensure adequate blocking. |
| Weak or No Signal | Low target abundance, inefficient hybridization, inactive HRP. | Use TSA for amplification [22]; check probe quality and hybridization conditions; verify HRP conjugate activity. |
| Poor Tissue Morphology | Over-fixation or excessive proteinase K treatment. | Optimize fixation time; titrate proteinase K concentration and incubation time. |
| Incomplete Permeabilization | Insufficient proteinase K or detergent. | Increase proteinase K concentration or incubation time empirically [24]. |
Quantitative Data and Performance
TSA-FISH provides significant advantages in sensitivity and signal quality over conventional methods, as quantified in recent studies.
Table 3: Performance Metrics of TSA-FISH
| Performance Metric | TSA-FISH Performance | Comparison to Conventional FISH |
|---|---|---|
| Signal Amplification | Up to 100-fold increase [22] [23] | Baseline (1x) |
| Signal-to-Noise Ratio | ~2-fold improvement reported in tissue clearing studies [25] | Standard |
| Signal Stability | Excellent; signals remain stable for months in cleared tissues [25] | Variable; often prone to fading |
| Dynamic Range | ~3x broader for single-EV staining [26] | Limited dynamic range |
| Multiplexing Capacity | High; enabled by sequential staining and HRP inactivation [24] [8] | Limited by antibody host species |
The TSA-FISH workflow provides an exceptionally powerful tool for visualizing gene expression with high sensitivity and spatial resolution, making it indispensable for modern planarian research and drug discovery. The key to success lies in meticulous optimization of each step—from probe design and tissue preparation to the precise control of tyramide amplification. By enabling the detection of low-abundance transcripts and facilitating multiplexed analyses, this protocol empowers researchers to decipher complex genetic programs governing stem cell dynamics and regeneration, thereby accelerating the identification and validation of novel therapeutic targets.
Planarian neoblasts, the adult pluripotent stem cells responsible for the remarkable regenerative capabilities of these organisms, are a central focus of regenerative biology research [27]. The detection of gene expression in these cells, particularly for low-abundance transcripts, presents significant technical challenges that can obscure critical biological insights. Whole-mount in situ hybridization (WISH) and fluorescent in situ hybridization (FISH) are indispensable methods for determining gene expression patterns in planarians, yet established protocols often fail to provide sufficient signal intensity for particularly scarce mRNAs [28] [29]. This limitation becomes especially problematic when studying neoblast subpopulations or the subtle gene expression changes that occur during stem cell differentiation.
The application of Tyramide Signal Amplification (TSA) technology, also known as Catalyzed Reporter Deposition (CARD), has emerged as a powerful solution to these sensitivity limitations [30] [22]. TSA utilizes horseradish peroxidase (HRP) to catalyze the deposition of labeled tyramide, resulting in high-density labeling near the target of interest and providing up to 100-fold higher detection sensitivity compared to conventional methods [22]. This exceptional sensitivity makes TSA-FISH particularly well-suited for studying planarian neoblasts, where detecting weakly expressed genes can reveal critical aspects of stem cell biology, including fate specification, pluripotency maintenance, and differentiation pathways.
Technical Foundations: Principles of Enhanced Detection
Tyramide Signal Amplification Mechanism
Tyramide Signal Amplification operates through a simple yet powerful enzymatic reaction. When horseradish peroxidase reacts with hydrogen peroxide, it activates the phenolic functional group of tyramine, creating a reactive, radical-containing quinone-like structure [30]. This activated tyramine molecule covalently binds to tyrosine residues of nearby proteins, resulting in dense deposition of the label at the site of interest [30] [22]. The amplification occurs because a single HRP enzyme can activate numerous tyramide molecules, leading to significant signal enhancement without increasing non-specific background.
For fluorescence in situ hybridization applications, this process allows for the detection of low-abundance intracellular targets that would otherwise remain undetectable with conventional staining methods [30]. The exceptional sensitivity of TSA has been shown to improve resolution by up to 30-fold, enabling researchers to identify functional heterogeneity within cell populations by detecting slight variations in protein expression [30]. In the context of planarian neoblasts, this sensitivity is crucial for distinguishing subtle differences in gene expression between stem cell subpopulations or during the early stages of differentiation.
Critical Modifications for Planarian Systems
Planarians present several unique challenges for FISH detection, including a protective mucous layer that must be removed prior to fixation, "sticky" tissues that promote non-specific antibody binding, broad tissue autofluorescence leading to poor signal-to-noise ratios, and the fragility of regenerating tissues that necessitates careful balancing of permeabilization conditions [28]. Research by the planarian community has yielded specific modifications that address these challenges while dramatically enhancing signal detection.
A key innovation is the replacement of overnight peroxide bleaching in methanol with a short peroxide bleaching step in formamide, which dramatically enhances signal intensity for both WISH and FISH [28]. This modification not only improves signal strength but also enhances tissue permeability, particularly in densely packed regions like the prepharyngeal area where neoblasts are concentrated. The optimal bleaching time was found to be between 1 to 2 hours in formamide bleaching solution, with longer incubations providing no additional benefit and potentially leading to more diffuse signal [28].
Additional critical improvements include the optimization of blocking conditions using Roche Western Blocking Reagent (RWBR), which dramatically reduces background without compromising signal intensity, and the incorporation of 0.3% Triton X-100 in blocking and wash solutions to further improve signal specificity [28]. For fluorescent applications, a copper sulfate quenching step effectively eliminates planarian autofluorescence, while azide proves most effective for quenching peroxidase activity between rounds of development in multicolor FISH experiments [28].
Enhanced FISH Protocol for Neoblast Detection
Sample Preparation and Fixation
- Mucous Removal: Transfer planarians to a solution of 5% N-acetyl-cysteine in PBS for 5-10 minutes to remove the protective mucous layer [28].
- Fixation: Fix planarians in 4% formaldehyde in PBS for 20-45 minutes at room temperature.
- Bleaching: Incubate fixed specimens in formamide bleaching solution (1.5% Hâ‚‚Oâ‚‚, 0.5% Triton X-100, 5% formamide in PBS) for 1-2 hours at room temperature to enhance tissue permeability and signal intensity [28].
- Dehydration: Gradually dehydrate samples through a methanol series (25%, 50%, 75% in PBSTx) and store in 100% methanol at -20°C.
Hybridization and Signal Amplification
- Rehydration and Permeabilization: Rehydrate samples through a methanol/PBSTx series and incubate in proteinase K (10 μg/mL) for 5-15 minutes.
- Hybridization: Incubate with digoxigenin-, fluorescein-, or dinitrophenol-labeled riboprobes in hybridization buffer at 56°C for 16-40 hours.
- Post-hybridization Washes: Perform stringent washes with 50% formamide/2× SSC and 2× SSC at 56°C, followed by room temperature washes with 75% 2× SSC/25% PBSTx and PBSTx.
- Blocking: Incubate samples in blocking solution (5% Roche Western Blocking Reagent, 0.3% Triton X-100 in PBSTx) for 2-4 hours to minimize non-specific binding [28].
- Antibody Incubation: Incubate with peroxidase-conjugated anti-hapten antibodies (e.g., anti-DIG-POD, anti-FAM-POD) diluted in blocking solution overnight at 4°C.
- Tyramide Signal Amplification: Develop signal using fluorescent tyramide substrates (1:100-1:500 dilution in amplification buffer) for 30-60 minutes [22].
- Peroxidase Quenching: For multicolor FISH, incubate with 0.1% sodium azide in PBSTx for 1-2 hours to quench residual peroxidase activity between rounds of amplification [28].
- Autofluorescence Reduction: Incubate samples in 1-10 mM copper sulfate in 50 mM ammonium acetate buffer (pH 5.0) for 30-60 minutes to reduce planarian autofluorescence [28].
Table 1: Key Reagents for Enhanced Planarian FISH
| Reagent | Function | Optimal Concentration/Type | Citation |
|---|---|---|---|
| Formamide Bleaching Solution | Enhances tissue permeability and signal intensity | 1.5% Hâ‚‚Oâ‚‚, 0.5% Triton X-100, 5% formamide | [28] |
| Blocking Reagent | Reduces non-specific antibody binding | 5% Roche Western Blocking Reagent | [28] |
| Detergent | Improves antibody penetration and reduces background | 0.3% Triton X-100 in blocking and wash buffers | [28] |
| Tyramide Substrates | Signal amplification | TyraMax dyes or CF Dye tyramides | [22] |
| Copper Sulfate | Quenches tissue autofluorescence | 1-10 mM in ammonium acetate buffer, pH 5.0 | [28] |
| Sodium Azide | Quenches peroxidase activity between TSA rounds | 0.1% in PBSTx | [28] |
Workflow Visualization
Diagram 1: Enhanced FISH workflow for planarian neoblast detection, highlighting critical modifications including formamide bleaching, specialized blocking, and TSA amplification with optional multiplexing cycles.
Research Reagent Solutions for Planarian FISH
Table 2: Essential Research Reagents for TSA-FISH in Planarians
| Category | Specific Product/Type | Key Features | Application in Planarian Research |
|---|---|---|---|
| Tyramide Substrates | TyraMax Amplification Dyes [22] | Wide selection (blue to near-IR), photostable, chemically stable in buffer | Superior sensitivity for low-abundance neoblast markers |
| Blocking Reagents | Roche Western Blocking Reagent [28] | Dramatically reduces background without signal loss | Essential for clean detection in "sticky" planarian tissues |
| Detergents | Triton X-100 [28] | Improves antibody penetration and reduces non-specific binding | Used in bleaching, blocking, and wash solutions (0.3%) |
| Peroxidase Substrates | Hydrogen Peroxide [30] | Required for HRP-mediated tyramide activation | Used at low concentrations in TSA reaction |
| Quenching Agents | Sodium Azide [28] | Effectively quenches peroxidase activity | Critical for sequential TSA rounds in multicolor FISH |
| Autofluorescence Reducers | Copper Sulfate [28] | Virtually eliminates planarian autofluorescence | Incubation in CuSOâ‚„/NHâ‚„Ac buffer, pH 5.0 |
| Permeabilization Enhancers | Formamide [28] | Improves tissue permeability when used in bleaching | Key component of enhanced bleaching solution |
Applications in Planarian Neoblast Research
The enhanced FISH protocol with TSA amplification has enabled significant advances in planarian neoblast research. This methodology has proven particularly valuable for studying the expression patterns of low-abundance transcripts that were previously undetectable with conventional methods [28]. The dramatically improved signal-to-noise ratio facilitates precise cellular localization of gene expression, which is essential for understanding neoblast heterogeneity and function.
In evolutionary and developmental studies, these sensitive detection methods have revealed how changes in neoblast behavior contribute to species-specific characteristics. For example, research on the cave planarian Girardia multidiverticulata utilized fluorescence in situ hybridization to demonstrate that the reduced eye size in this species results from a decreased rate of stem cell fate specification to eye progenitors, rather than a complete absence of eye formation [31]. This finding was enabled by the ability to detect and quantify photoreceptor neurons despite their reduced numbers, showcasing the power of sensitive detection methods for uncovering fundamental mechanisms of stem cell biology.
Similarly, comparative regeneration studies across multiple planarian species have relied on robust detection methods to analyze expression patterns of key regulatory genes like notum and components of the Wnt signaling pathway [10] [27] [32]. These investigations have revealed species-specific differences in regeneration robustness and the underlying molecular mechanisms, providing insights into the evolution of regenerative abilities. The ability to detect subtle differences in gene expression patterns between species has been instrumental in understanding how stem cell regulation contributes to regenerative diversity.
Troubleshooting and Optimization Guidelines
Despite the significant improvements offered by the enhanced FISH protocol, researchers may encounter specific challenges during implementation. The following troubleshooting guidelines address common issues:
High Background Staining: Ensure fresh hydrogen peroxide is used in the TSA reaction, as degraded peroxide can increase background. Optimize blocking conditions by testing different concentrations of Roche Western Blocking Reagent (3-5%) and extend blocking time to 4-6 hours if necessary [28].
Weak or Absent Signal: Verify that the formamide bleaching step does not exceed 2 hours, as extended bleaching can diminish signal. Increase tyramide substrate concentration or extension time, but avoid excessive development that can increase background. Check antibody concentrations and consider using tyramide substrates with higher brightness, such as TyraMax 555/565 or TyraMax 647/670 [22].
Tissue Damage or Morphology Loss: Reduce proteinase K concentration or incubation time, particularly for regenerating specimens. For fragile regenerating tissues, employ a heat-induced antigen retrieval step instead of proteinase K treatment to better balance permeabilization with tissue preservation [28].
Incomplete Autofluorescence Quenching: Increase copper sulfate concentration up to 10 mM or extend incubation time to 60 minutes. Ensure the pH of the ammonium acetate buffer is precisely 5.0 for optimal quenching efficiency [28].
Incomplete Peroxidase Quenching in Multiplex FISH: Extend azide incubation time to 2 hours or include multiple azide washes between TSA rounds. Verify that the tyramide reaction has been thoroughly washed out before applying the next primary antibody [28].
The enhanced FISH protocol with TSA amplification represents a significant advancement for studying planarian neoblasts and their progeny. By dramatically improving signal sensitivity while maintaining excellent tissue morphology, this methodology enables researchers to address previously intractable questions in stem cell biology and regeneration.
A Step-by-Step Protocol for Planarian TSA-FISH
Fixation and permeabilization are critical foundational steps for successful tyramide signal amplification fluorescence in situ hybridization (TSA-FISH) in planarian research. The freshwater planarian Schmidtea mediterranea presents unique challenges for these techniques due to its mucous-covered epidermis, highly regenerative tissues, and widespread autofluorescence. The delicate nature of the regeneration blastema—the unpigmented tissue that forms after amputation—is particularly vulnerable to damage from harsh chemical treatments, potentially compromising morphological preservation and experimental outcomes [33] [34].
Recent methodological advances have addressed these challenges through innovative fixation protocols that better preserve tissue integrity while enabling effective probe penetration. This application note details optimized sample preparation methods specifically validated for TSA-FISH in planarian studies, providing researchers with standardized protocols that ensure superior tissue preservation, enhanced signal-to-noise ratio, and increased compatibility with multiplexed detection approaches [33] [34].
Comparative Analysis of Fixation and Permeabilization Methods
Table 1: Quantitative comparison of fixation and permeabilization methods for planarian TSA-FISH
| Method | Tissue Preservation | Signal Intensity | Autofluorescence Reduction | Compatibility with Immunostaining | Best Applications |
|---|---|---|---|---|---|
| NAFA Protocol | Excellent (epidermis and blastema fully intact) | High (equivalent to NAC) | Significant reduction | Excellent (no proteinase K) | Delicate regenerating tissues, combined FISH/IF experiments |
| NAC Protocol | Poor (epidermal damage, blastema shredding) | High | Moderate reduction | Poor (proteinase K digests epitopes) | High-abundance transcripts in mature tissues |
| Rompolas (NA) Protocol | Excellent | Weak/None | Significant reduction | Excellent | Immunofluorescence alone |
| Formamide Bleaching | Good | Enhanced | Dramatic reduction (copper sulfate quenching) | Good | Low-abundance transcripts, multiplex FISH |
The NAFA (Nitric Acid/Formic Acid) protocol demonstrates superior overall performance by achieving an optimal balance between tissue preservation and permeability. This method eliminates the need for proteinase K digestion, which is known to damage antigen epitopes and delicate regenerating tissues. Comparative studies show that NAFA preserves epidermal integrity significantly better than the established NAC protocol while maintaining equivalent FISH signal intensity for markers such as piwi-1 (neoblasts) and zpuf-6 (epidermal progenitors) [34].
For signal sensitivity enhancement, a short peroxide bleaching step in formamide dramatically improves probe penetration and subsequent signal intensity compared to overnight methanol-based bleaching. This modification reduces development time for both high- and low-abundance transcripts and improves signal consistency in densely-packed regions like the prepharyngeal area [33].
Detailed Experimental Protocols
NAFA Fixation Protocol for TSA-FISH
Reagents Required:
- Nitric acid (1%)
- Formic acid (2.5%)
- EGTA (5mM)
- Formaldehyde (4%)
- PBSTx (1X PBS with 0.3% Triton X-100)
- Methanol
Procedure:
- Mucus Removal: Starve planarians for at least 1 week before fixation. Treat with N-acetyl-cysteine (NAC) in planarian water for 5-7 minutes with gentle agitation [33].
- Fixation: Transfer planarians to freshly prepared NAFA fixative (1% nitric acid, 2.5% formic acid, 4% formaldehyde, 5mM EGTA in PBS). Incubate for 1.5-2 hours at room temperature with gentle rotation [34].
- Permeabilization: Wash samples 3×5 minutes with PBSTx. Incubate in 100% methanol for 10 minutes at room temperature [34].
- Rehydration: Gradually rehydrate through methanol/PBSTx series (75%, 50%, 25% methanol in PBSTx), 5 minutes each.
- Post-fixation: Post-fix with 4% formaldehyde in PBSTx for 10 minutes to maintain tissue integrity during subsequent hybridization steps.
- Storage: Store fixed samples in PBSTx at 4°C for immediate use or in methanol at -20°C for long-term preservation.
Critical Steps:
- Always prepare NAFA fixative fresh immediately before use
- Include EGTA to chelate calcium and inhibit RNases
- Adjust fixation time based on planarian size (reduce to 1 hour for fragments <3mm)
- Avoid proteinase K treatment to preserve antigenicity for combined immunofluorescence [34]
Enhanced Permeabilization and Bleaching for Sensitive Detection
Formamide Bleaching Solution:
- Formamide (100%)
- Hydrogen peroxide (3%)
- In 1X PBS
Procedure:
- After rehydration, incubate fixed planarians in formamide bleaching solution for 1-2 hours at room temperature [33].
- Wash 3×5 minutes with PBSTx.
- For autofluorescence quenching, treat with copper sulfate solution (10mM in 50mM ammonium acetate buffer, pH 5.0) for 1 hour [33].
- Wash thoroughly with PBSTx before proceeding to hybridization.
Optimized Blocking for TSA-FISH:
- Prepare blocking solution: 1X Roche Western Blocking Reagent in PBSTx with 0.3% Triton X-100 [33].
- Block samples for 2-4 hours at room temperature with gentle agitation.
- Proceed directly to hybridization with hapten-labeled probes.
Table 2: Research reagent solutions for planarian TSA-FISH
| Reagent | Composition/Type | Function in Protocol | Optimization Notes |
|---|---|---|---|
| NAFA Fixative | 1% nitric acid, 2.5% formic acid, 4% formaldehyde, 5mM EGTA | Tissue fixation and permeabilization | Preserves epitopes for IF; replaces proteinase K step |
| Formamide Bleaching Solution | 3% Hâ‚‚Oâ‚‚ in 100% formamide | Tissue permeabilization and autofluorescence reduction | Replaces overnight methanol bleaching; enhances signal |
| Roche Western Blocking Reagent | Solution in PBSTx with 0.3% Triton X-100 | Reduce non-specific antibody binding | Superior to casein or PEBR for anti-DIG and anti-FAM antibodies |
| Copper Sulfate Quenching Solution | 10mM in 50mM ammonium acetate buffer, pH 5.0 | Reduce tissue autofluorescence | Essential for low-abundance targets; improves signal-to-noise |
| OneSABER Probes | ssDNA concatemers with initiator sequences | Modular probe system for FISH | Enables multiple signal development methods from one probe set |
Workflow Integration and Visualization
The following workflow diagram illustrates the integrated sample preparation process for planarian TSA-FISH, highlighting critical decision points and quality check stages:
Technical Notes and Troubleshooting
Preserving Delicate Tissues: The regeneration blastema is particularly susceptible to damage from permeabilization treatments. The NAFA protocol eliminates this concern by avoiding proteinase K entirely, instead using controlled acid treatment to achieve permeabilization while maintaining tissue architecture. For regenerating samples <24 hours post-amputation, reduce formic acid concentration to 1.5% and incubation time to 1 hour [34].
Multiplex TSA-FISH Considerations: For sequential TSA development, effective peroxidase quenching between rounds is essential. Incubation with 2% sodium azide for 30 minutes completely inactivates HRP without damaging tissue morphology or antigenicity. Alternative quenching methods including Hâ‚‚Oâ‚‚ treatment or low pH buffers may compromise sample integrity and subsequent labeling rounds [33] [7].
Probe Penetration Optimization: Difficulties with probe penetration to internal tissues can be addressed by incorporating a heat-induced antigen retrieval step (65°C for 10 minutes in citrate buffer) after formamide bleaching. This treatment significantly improves hybridization efficiency for internal structures like the pharynx and reproductive organs without compromising tissue integrity [33].
The optimized fixation and permeabilization methods detailed in this application note address the specific challenges of planarian tissue preparation for TSA-FISH experiments. The NAFA protocol represents a significant advancement over traditional methods by enabling superior preservation of delicate regenerating tissues while maintaining compatibility with sensitive fluorescence detection. When integrated with formamide bleaching and copper sulfate autofluorescence quenching, researchers can achieve exceptional signal-to-noise ratios even for low-abundance transcripts. These standardized protocols provide the foundation for robust and reproducible TSA-FISH in planarian regeneration research, ultimately supporting more precise spatial gene expression analysis in this powerful model system.
In the pursuit of visualizing gene expression patterns in planarians through tyramide signal amplification fluorescence in situ hybridization (TSA-FISH), researchers confront two formidable anatomical barriers: a surface layer of mucus and pervasive body pigmentation. These obstacles impede probe penetration and generate high background autofluorescence, severely compromising the detection of low-abundance transcripts. The critical sample preparation steps of mucous removal and pigment bleaching are therefore paramount for success. Within planarian research, two primary bleaching methodologies have emerged: the traditional overnight methanol-based peroxide bleach and a more recent approach utilizing a short formamide-based peroxide bleach. This Application Note details both protocols, provides a quantitative comparison of their performance, and frames their application within the context of advanced planarian TSA-FISH research, offering researchers a clear guide to optimize their experimental outcomes.
The Scientist's Toolkit: Essential Reagents for Planarian TSA-FISH
The following table catalogues key reagents critical for implementing the bleaching and detection protocols discussed in this note.
Table 1: Key Research Reagent Solutions for Planarian TSA-FISH
| Reagent | Function/Application | Key Details |
|---|---|---|
| Formamide Bleaching Solution | Pigment bleaching & tissue permeabilization [33] | Hydrogen peroxide in formamide. Replaces overnight methanol bleaching. |
| Roche Western Blocking Reagent (RWBR) | Blocking non-specific antibody binding [33] | Dramatically reduces background in FISH, particularly for anti-DIG and anti-FAM antibodies. |
| Triton X-100 | Detergent in blocking & wash buffers [33] | Improves signal specificity, especially when used at 0.3% concentration. |
| Anti-hapten Antibodies (DIG, DNP, FAM) | Primary detection of hybridized probes [33] | Conjugated to peroxidase for TSA. Performance is enhanced with optimized blocking. |
| Tyramide Signal Amplification (TSA) Kits | Enzymatic signal amplification [24] | Provides fluorescent dye tyramides and HRP conjugates for high-sensitivity detection. |
| Sodium Azide | Peroxidase quenching for multiplex FISH [33] | Effectively quenches HRP activity between sequential TSA rounds, preventing false signal. |
| Copper Sulfate | Quenching endogenous autofluorescence [33] | Treating samples post-hybridization reduces planarian tissue autofluorescence. |
| For-DL-Met-DL-Phe-DL-Met-OH | For-DL-Met-DL-Phe-DL-Met-OH|High-Quality Research Peptide | For-DL-Met-DL-Phe-DL-Met-OH is a synthetic tripeptide for research use only (RUO). Explore its applications in peptide science. Not for human or veterinary diagnosis or therapy. |
| 2-(Methylthio)-9H-carbazole | 2-(Methylthio)-9H-carbazole, MF:C13H11NS, MW:213.30 g/mol | Chemical Reagent |
Quantitative Comparison: Formamide vs. Methanol Bleaching
The choice between formamide and methanol bleaching methods has significant implications for protocol duration, signal quality, and tissue integrity. The following table summarizes a direct comparison based on experimental data.
Table 2: Quantitative Comparison of Formamide and Methanol Bleaching Methods [33]
| Parameter | Formamide Bleach | Methanol Bleach |
|---|---|---|
| Standard Bleaching Duration | 1 - 2 hours | Overnight (≈16 hours) |
| Time to Maximum Signal Intensity | Dramatically reduced | Standard development time |
| Signal Intensity | Increased | Baseline |
| Tissue Permeability | Improved, more consistent labeling in dense regions (e.g., prepharyngeal) | Standard |
| Requirement for Reduction Step | Not required; step slightly diminishes signal | Typically required |
| Compatibility with Pre-bleaching | Benefit is lost if tissue is pre-bleached in methanol | N/A |
| Effect on mRNA Integrity | mRNA remains stable | mRNA remains stable, but benefits of formamide are masked |
Detailed Experimental Protocols
Protocol 1: Short Formamide Peroxide Bleaching for Enhanced Sensitivity
This protocol, adapted from King & Newmark (2013), is designed for maximum signal intensity and is ideal for detecting low-abundance mRNAs [33].
- Mucous Removal and Fixation: Begin by treating planarians with a mucous-removing agent such as N-acetyl-cysteine. Fix animals in formaldehyde-based fixative, as previously described [33].
- Formamide Peroxide Bleaching:
- Prepare the bleaching solution by diluting hydrogen peroxide in formamide (exact concentration can be optimized, e.g., 2-6%).
- Incubate fixed and permeabilized planarians in the bleaching solution for 1 to 2 hours at room temperature.
- Note: Bleaching for 30 minutes shows dramatic improvement, but 1-2 hours is optimal. Do not pre-bleach samples in methanol.
- Post-Bleaching Washes: Rinse the planarians thoroughly in phosphate-buffered saline (PBS) with Tween 20 (PBT) to remove all traces of formamide and peroxide.
- Hybridization and Detection: Proceed with standard FISH steps: hybridization with hapten-labeled riboprobes, antibody detection, and TSA development [33] [24].
Protocol 2: Traditional Overnight Methanol Peroxide Bleaching
This established protocol is effective for many applications but may be less sensitive for low-abundance targets [33].
- Mucous Removal and Fixation: As in Protocol 1.
- Methanol Peroxide Bleaching:
- Prepare a solution of hydrogen peroxide in 100% methanol.
- Incubate fixed planarians in the bleaching solution overnight at 4°C or room temperature.
- Post-Bleaching Washes and Reduction (Optional): Rehydrate samples and wash with PBT. A reduction step using sodium borohydride is often included after methanol bleaching to improve permeability [33].
- Hybridization and Detection: Continue with standard FISH and TSA steps.
Optimizing the Complete TSA-FISH Workflow for Planarians
Beyond bleaching, several key modifications to the standard FISH protocol are critical for achieving high signal-to-noise ratios in planarians [33].
- Enhanced Blocking and Washing: Supplement blocking buffers with Roche Western Blocking Reagent (RWBR) and replace Tween 20 with 0.3% Triton X-100 in wash and antibody buffers. This combination dramatically reduces non-specific background staining without compromising signal intensity [33].
- Quenching Autofluorescence: After hybridization and detection, treat samples with a solution of copper sulfate to quench broad-wavelength endogenous autofluorescence, significantly improving the signal-to-noise ratio [33].
- Multiplexing with Azide Quenching: For multicolor FISH requiring sequential TSA reactions, incubate samples with sodium azide between development rounds. Azide is the most effective method for quenching residual peroxidase activity, preventing false-positive signals in subsequent TSA rounds [33].
Integration with Broader Research Context
Mastering sample preparation is the foundation upon which advanced imaging and mechanistic studies are built. The ability to reliably detect low-abundance transcripts via optimized FISH is crucial for investigating the molecular machinery of planarian regeneration. For instance, highly sensitive FISH has been instrumental in elucidating the expression of key patterning genes like wnt1 and notum in planarian muscle cells following injury [35]. Furthermore, the ultrafast distant wound response, which is propagated through longitudinal muscle fibers and is essential for whole-body regeneration, can be characterized using these techniques [36]. Finally, the high-resolution, quantitative data generated from optimized FISH can be integrated with cutting-edge imaging technologies, such as expansion tiling light sheet microscopy (TLSM), to create detailed 3D reconstructions of gene expression within the context of entire neural-muscular networks [37]. Thus, the protocols detailed here are not isolated techniques but are integral components of a powerful research pipeline aimed at unraveling the complexities of regeneration.
In the field of planarian research, whole-mount in situ hybridization (WISH) and fluorescent in situ hybridization (FISH) are indispensable techniques for determining gene expression patterns, which is fundamental to understanding regenerative processes [33]. The ability to detect low-abundance transcripts is particularly critical for elucidating the molecular mechanisms governing stem cell dynamics and tissue regeneration [33] [13]. This application note details optimized protocols for probe hybridization, focusing on buffer composition and temperature parameters, specifically framed within the context of tyramide signal amplification (TSA)-based FISH methodologies for planarian research. These optimizations provide significant improvements in signal intensity and sensitivity, enabling researchers to overcome the challenges associated with detecting particularly low-abundance transcripts in planarians [33].
The Scientist's Toolkit: Research Reagent Solutions
The following table details essential reagents and materials used in optimized planarian FISH protocols, along with their specific functions:
Table 1: Key Research Reagents for Planarian FISH
| Reagent/Material | Function/Application | Optimization Notes |
|---|---|---|
| Formamide | Component of hybridization buffer; affects stringency and probe specificity. | Used in a short peroxide bleaching step to dramatically enhance tissue permeability and signal intensity [33] [13]. |
| Roche Western Blocking Reagent (RWBR) | Additive to blocking buffer to reduce non-specific background. | Dramatically reduces background staining for anti-DIG and anti-FAM antibodies in fluorescent TSA without sacrificing signal intensity [33]. |
| Triton X-100 | Detergent in wash and blocking buffers. | Using 0.3% concentration improves signal specificity, particularly with anti-DIG and anti-FAM antibodies [33]. |
| Sodium Azide (NaN₃) | Quenching agent for peroxidase activity between TSA rounds. | Most effective method for quenching peroxidase activity between sequential rounds of tyramide signal amplification in multiplex FISH, preserving tissue integrity [33]. |
| Copper Sulfate | Quenching agent for endogenous tissue autofluorescence. | Virtually eliminates planarian autofluorescence across a broad range of wavelengths, improving signal-to-noise ratio [33] [13]. |
| Glucose Oxidase | Enzyme for generating Hâ‚‚Oâ‚‚ in situ in TSA systems. | Used in fluorochromized tyramide-glucose oxidase (FT-GO) systems to stably supply Hâ‚‚Oâ‚‚, improving operational stability of the TSA reaction [7]. |
| Tyramide Conjugates | Substrates for peroxidase-catalyzed deposition in TSA. | Fluorochromized tyramide (FT) allows direct fluorescence detection without additional staining steps, simplifying multiplexing [7]. |
| Benzamide, 4-bromo-3-ethyl- | Benzamide, 4-bromo-3-ethyl-, CAS:1228826-63-8, MF:C9H10BrNO, MW:228.09 g/mol | Chemical Reagent |
| 5,6-Difluoroisoquinoline | 5,6-Difluoroisoquinoline|RUO |
Optimized Hybridization Parameters
Buffer Composition and Formamide Bleaching
A critical finding for enhancing planarian FISH sensitivity is the implementation of a peroxide bleaching step in formamide, which replaces the traditional overnight methanol-based bleach [33] [13]. This modification profoundly improves probe penetration and subsequent signal intensity.
Table 2: Optimized Hybridization Buffer and Bleaching Conditions
| Parameter | Standard Condition | Optimized Condition | Impact on Signal |
|---|---|---|---|
| Bleaching Solution | Methanol with Hâ‚‚Oâ‚‚ (overnight) | Formamide with Hâ‚‚Oâ‚‚ (1-2 hours) | Dramatically reduced development time and increased signal intensity for both chromogenic and fluorescent detection [33]. |
| Bleaching Duration | ~12-16 hours (overnight) | 1-2 hours | Signal intensity reaches maximum after 1-2 hours of formamide bleaching; longer incubations do not further enhance signal [33]. |
| Pre-bleaching | Not applicable | Avoid pre-bleaching in methanol | Pre-bleaching in methanol eliminates the benefit of formamide bleaching [33]. |
| Reduction Step | Often included | Can be omitted | The reduction step slightly diminishes signal intensity in formamide-bleached animals [33]. |
| Blocking Buffer | Standard protein-based blockers | Addition of Roche Western Blocking Reagent (RWBR) | Dramatically reduces background, particularly for anti-DIG and anti-FAM antibodies, without significant signal loss [33]. |
| Detergent in Washes | Tween 20 | Triton X-100 (0.3%) | Results in a slight but noticeable improvement in signal specificity [33]. |
The optimized bleaching solution is typically prepared with 5% formamide and 1.2% Hâ‚‚Oâ‚‚ in a 1:5 dilution of 20X SSC [33]. The mechanism is believed to be improved tissue permeability, as signal intensity increases with bleaching time up to a maximum at 1-2 hours.
Hybridization Temperature Optimization
While the specific optimal hybridization temperature can vary depending on the probe, the optimized protocol for planarians typically involves hybridization at 56°C [33]. This temperature, combined with the formamide concentration in the hybridization buffer (typically 50%), provides the right balance between hybridization specificity and signal preservation. Heat-induced antigen retrieval (HIAR) is also employed for FISH on regenerating planarians, providing a better balance between permeabilization of mature tissues and preservation of fragile regenerating tissues [33] [13].
Experimental Workflow for Optimized Planarian FISH
The following diagram illustrates the comprehensive workflow for performing optimized FISH on planarians, incorporating the critical steps for buffer and temperature optimization.
Detailed Protocol for Tyramide Signal Amplification FISH
Sample Preparation and Pre-hybridization
- Fixation and Mucus Removal: Fix planarians in formaldehyde-based fixatives. Remove mucus using N-acetyl-cysteine [33] [13].
- Formamide Peroxide Bleaching: Incubate planarians in bleaching solution containing 5% formamide and 1.2% Hâ‚‚Oâ‚‚ in a 1:5 dilution of 20X SSC for 1-2 hours at room temperature [33]. This step is critical for enhancing tissue permeability and signal intensity.
- Permeabilization: Treat with proteinase K to further permeabilize tissues. For regenerating tissues, a balance must be struck to allow probe penetration while preserving fragile tissue morphology [33].
Hybridization and Post-Hybridization Washes
- Pre-hybridization: Incubate samples in hybridization buffer for approximately 1 hour at the hybridization temperature.
- Probe Hybridization: Add gene-specific RNA probes diluted in hybridization buffer. Hybridize at 56°C overnight [33].
- Stringency Washes: Perform post-hybridization washes with SSC buffers containing 0.3% Triton X-100 to reduce background and improve signal specificity [33].
Signal Detection and Amplification
- Blocking: Incubate samples in blocking buffer supplemented with Roche Western Blocking Reagent (RWBR) to minimize non-specific antibody binding [33].
- Antibody Incubation: Incubate with peroxidase (POD)-conjugated anti-hapten antibodies (e.g., anti-DIG-POD, anti-FAM-POD).
- Tyramide Signal Amplification: Develop signal using fluorochromized tyramide (FT) and Hâ‚‚Oâ‚‚. For enhanced stability, the FT-GO system can be used, where Hâ‚‚Oâ‚‚ is generated in situ by the enzymatic reaction between glucose and glucose oxidase [7].
- Autofluorescence Quenching: For FISH, incubate samples in copper sulfate solution to virtually eliminate endogenous planarian autofluorescence [33] [13].
- Multiplexing: For detecting multiple targets, after each round of TSA, quench peroxidase activity by incubating with 2% sodium azide (NaN₃) before proceeding to the next antibody and TSA round [33].
Troubleshooting and Technical Notes
- Low Signal Intensity: Ensure the formamide peroxide bleaching step is performed for the full 1-2 hours and that no pre-bleaching in methanol has been conducted. Check the concentration and integrity of probes, particularly for low-abundance targets.
- High Background: Increase the concentration of RWBR in the blocking buffer. Ensure that stringency washes contain 0.3% Triton X-100 and are performed at the correct temperature. For TSA-based methods, optimize the concentration of fluorochromized tyramide and Hâ‚‚Oâ‚‚ (or glucose/glucose oxidase) to prevent over-development.
- Tissue Damage in Regenerates: For fragile regenerating tissues, consider adjusting the proteinase K concentration and time, and employ heat-induced antigen retrieval (HIAR) as a gentler alternative [33] [13].
- Signal Crossover in Multiplex FISH: Verify the efficiency of peroxidase quenching with sodium azide between TSA rounds. Ineffective quenching will lead to false positive signals in subsequent channels [33].
The optimized probe hybridization conditions detailed in this application note—specifically the implementation of formamide-based peroxide bleaching, refined buffer compositions with RWBR and Triton X-100, and precise temperature control—significantly enhance the sensitivity and specificity of FISH in planarian research. Integrated with the high signal amplification of TSA, these protocols provide a powerful methodological framework for visualizing low-abundance transcripts, thereby advancing our capacity to decipher the complex gene regulatory networks that underpin planarian regeneration and stem cell biology.
Tyramide Signal Amplification (TSA) is a catalytic reporter deposition technique that dramatically enhances the sensitivity of fluorescence in situ hybridization (FISH), enabling researchers to detect low-abundance mRNA transcripts that are critical for understanding planarian biology. In the study of planarian stem cells and regeneration mechanisms, researchers frequently encounter challenging, low-abundance transcripts that conventional detection methods cannot reliably visualize [33]. The TSA method leverages the potent activity of horseradish peroxidase (HRP) enzymes, which are conjugated to probes or antibodies. When HRP encounters hydrogen peroxide (Hâ‚‚Oâ‚‚), it activates tyramide substrates, converting them into highly reactive intermediates that rapidly form covalent bonds with electron-rich amino acids on nearby proteins, effectively depositing numerous fluorophore tags at the site of target recognition [38]. This signal amplification process, when properly optimized and executed, can improve detection sensitivity by orders of magnitude, making it indispensable for visualizing weakly expressed fate-specification transcripts, signaling molecules, and regulatory genes that govern planarian stem cell behavior and regenerative patterning.
The application of TSA-based FISH in planarian research has been particularly transformative for studying the complex dynamics of neoblast fate specification and the molecular cues that guide anterior-posterior axis regeneration [18]. For instance, critical polarity genes like notum, which is asymmetrically expressed in longitudinal muscle cells at anterior-facing wounds and determines head versus tail regeneration decisions, often require the heightened sensitivity afforded by TSA for clear visualization [39]. Similarly, elucidating the spatially intermingled distribution of fate-specified stem cells throughout the planarian body, as revealed through multiplexed error-robust FISH (MERFISH), relies on robust signal detection strategies [18]. This protocol details the optimized execution of the tyramide reaction specifically within the context of planarian FISH, incorporating key modifications that address the unique challenges of working with planarian tissues, including their inherent autofluorescence, dense parenchymal tissue, and the need to preserve delicate regenerating structures.
Technical Foundation of TSA
Mechanistic Principles of Tyramide Amplification
The exceptional sensitivity of the TSA reaction stems from its unique enzymatic mechanism that enables the localized deposition of numerous reporter molecules at target sites. The process begins when horseradish peroxidase (HRP), conjugated to a detection antibody or nucleic acid probe, encounters its substrate, hydrogen peroxide (H₂O₂) [38]. This interaction generates activated HRP intermediates that then catalyze the oxidation of phenol derivatives in tyramide reagents. The oxidation process creates highly reactive, short-lived tyramide radicals that possess an extremely brief diffusion radius of approximately 0.3 micrometers before reacting with electron-rich residues—primarily tyrosine, but also tryptophan and phenylalanine—on nearby proteins [40]. This restricted diffusion ensures that signal deposition remains tightly localized to the original HRP binding site, preserving spatial resolution while achieving substantial signal amplification.
The covalent bonding of fluorophore-labeled tyramides to tissue proteins creates a stable, permanent signal that withstands stringent washing procedures and even allows for multiple rounds of amplification in multiplexed experiments. Each HRP enzyme molecule can activate thousands of tyramide molecules per minute, resulting in an exponential increase in the number of fluorophores deposited at each target site compared to conventional direct or indirect immunofluorescence methods [38]. This amplification mechanism is particularly advantageous for detecting low-copy-number transcripts in planarians, such as those encoding fate-specifying transcription factors or early wound-induced genes, which may be present at only a few copies per cell but play decisive roles in regeneration and stem cell regulation [33].
Essential Reagent Solutions for TSA-FISH
The successful implementation of TSA in planarian research requires careful preparation and optimization of key reagent solutions. The composition of these solutions significantly impacts signal intensity, background staining, and overall signal-to-noise ratio.
Table 1: Essential Research Reagent Solutions for TSA-FISH in Planarians
| Reagent Solution | Key Components | Primary Function | Planarian-Specific Considerations |
|---|---|---|---|
| Tyramide Stock Solution | Tyramide-fluorophore conjugate, DMSO | Signal deposition substrate | Aliquot and store at -20°C protected from light; avoid freeze-thaw cycles |
| Amplification Buffer | Tris or Phosphate buffer, Hâ‚‚Oâ‚‚ | Provides optimal pH and oxidizing environment for HRP | Critical to optimize Hâ‚‚Oâ‚‚ concentration to balance signal and background |
| Blocking Buffer | Roche Western Blocking Reagent (RWBR), Triton X-100, maleic acid buffer | Reduces non-specific antibody binding | RWBR dramatically reduces background for anti-DIG and anti-FAM antibodies [33] |
| Post-Hybridization Wash Buffers | SSC buffer, Tween-20, Triton X-100 | Removes unbound probes while preserving tissue integrity | Addition of 0.3% Triton X-100 improves signal specificity [33] |
| Peroxidase Quenching Solution | Sodium azide (NaN₃) | Inactivates HRP between rounds of multiplexed FISH | Most effective method for preventing false signal in subsequent TSA rounds [33] |
The blocking step deserves particular attention in planarian FISH applications. Research has demonstrated that supplementing blocking buffers with Roche Western Blocking Reagent (RWBR), combined with the inclusion of Triton X-100 detergent, dramatically reduces background staining, particularly when using anti-digoxigenin (DIG) and anti-fluorescein (FAM) antibodies [33]. This modification is crucial for achieving the high signal-to-noise ratios necessary for detecting low-abundance transcripts in the dense, heterogeneous tissue environment of planarians.
Optimized TSA-FISH Protocol for Planarian Research
Pre-TSA Steps: Tissue Preparation and Hybridization
Proper tissue preparation is foundational to successful TSA amplification. For planarian studies, begin with a short bleaching step in formamide-based bleaching solution (4% H₂O₂ in 100% formamide) for 1-2 hours at room temperature to reduce background and improve tissue permeability [33]. This approach has been shown to significantly enhance signal intensity compared to traditional overnight methanol bleaching. Following fixation and bleaching, permeabilize tissues with proteinase K (8-12 μg/mL for 8-15 minutes depending on animal size) to facilitate probe access while preserving tissue morphology, particularly in fragile regenerating blastemas.
For hybridization, use standardized planarian FISH protocols with the following modifications to enhance TSA performance: increase hybridization temperature to 56°C, use optimized hybridization buffer containing 50% formamide, and extend hybridization time to 36-48 hours for low-abundance targets [33]. After hybridization, perform stringent washes with SSC buffers containing 0.3% Triton X-100 to reduce non-specific probe retention. Subsequently, incubate samples with peroxidase-conjugated anti-hapten antibodies (e.g., anti-DIG-POD, anti-DNP-POD) diluted in blocking buffer containing RWBR and Triton X-100 for 36-48 hours at 4°C to ensure adequate antibody penetration throughout the tissue.
Tyramide Reaction Execution and Optimization
The core TSA reaction requires precise optimization of multiple parameters to achieve maximum sensitivity while controlling background. The following workflow outlines the critical steps and decision points for executing the tyramide reaction in planarian samples:
Tyramide Working Solution Preparation
Prepare tyramide working solution immediately before use by diluting tyramide-fluorophore stock in amplification buffer. The optimal dilution factor must be determined empirically for each new tyramide batch and target transcript, but typically ranges from 1:50 to 1:1000 in amplification buffer containing 0.001-0.01% Hâ‚‚Oâ‚‚ [33]. For critical experiments targeting low-abundance planarian transcripts such as early wound-induced genes or fate-specification factors, prepare a dilution series to identify the optimal concentration that maximizes signal-to-noise ratio. The composition of the amplification buffer significantly impacts reaction efficiency; Tris-HCl (pH 7.5) or borate buffers (pH 8.5) at 50-100 mM concentration generally provide optimal enzymatic activity for HRP.
Reaction Execution and Monitoring
Apply the tyramide working solution to samples completely submerged in minimal volume to conserve reagent. Incubate at room temperature with gentle agitation, monitoring reaction progression periodically under a fluorescence microscope if possible. Reaction times typically range from 5 to 45 minutes, depending on target abundance and tyramide concentration [40]. For extremely low-abundance targets, longer incubation times may be necessary, but these increase the risk of background development. To stop the reaction, remove the tyramide solution and wash samples repeatedly in PBS or Tris buffer with 0.1% Tween-20 or Triton X-100. The covalent nature of tyramide deposition means the signal remains stable through subsequent washing and imaging steps.
Parameter Optimization for Maximum Sensitivity
Achieving maximum sensitivity requires systematic optimization of three key parameters that most significantly impact TSA performance:
Table 2: Quantitative Optimization Parameters for TSA in Planarian FISH
| Parameter | Typical Range | Effect on Signal | Effect on Background | Recommended Starting Point |
|---|---|---|---|---|
| Tyramide Concentration | 1:50 - 1:1000 dilution | Higher concentration increases signal intensity | Higher concentration increases background | 1:200 dilution for moderate abundance targets |
| Hâ‚‚Oâ‚‚ Concentration | 0.001% - 0.01% | Optimal at ~0.005%; higher levels may inactivate HRP | Increases significantly above 0.01% | 0.005% in amplification buffer |
| Reaction Time | 5 - 45 minutes | Longer time increases signal | Background accumulates with extended time | 15 minutes with monitoring |
| Detection Antibody Concentration | 1:500 - 1:2000 | Higher concentration increases potential signal | Increases non-specific binding | 1:1000 for peroxidase conjugates |
Recent studies have demonstrated that optimizing these parameters can achieve detection limits as low as 58 fg/mL in analogous applications, representing a 10-fold improvement in sensitivity over non-optimized methods [40]. In planarian research, this enhanced sensitivity enables reliable detection of critical low-abundance transcripts such as notum in longitudinal muscle cells after injury, which is essential for understanding the establishment of regeneration polarity [39].
Post-TSA Processing and Multiplexing
Following TSA development, thorough washing is critical to minimize background fluorescence. Perform 3-5 washes of 15-30 minutes each in PBSTx (PBS with 0.3% Triton X-100) at room temperature with gentle agitation. For multiplexed FISH applications requiring sequential detection of multiple targets, complete inactivation of HRP between TSA rounds is essential to prevent false-positive signal in subsequent channels. The most effective quenching method for planarian FISH involves incubation with 0.1% sodium azide (NaN₃) in PBSTx for 1-2 hours at room temperature, which efficiently inactivates residual peroxidase activity without damaging target mRNAs or compromising tissue integrity [33]. After azide quenching, confirm complete HRP inactivation by applying amplification buffer with H₂O₂ and tyramide substrate; no signal should develop if quenching was successful. Some protocols additionally incorporate a heat treatment step (65-70°C for 30-60 minutes) to denature antibodies while preserving RNA targets, particularly beneficial when working with regenerating planarians where tissue preservation is challenging [33].
Troubleshooting and Quality Control
Common Issues and Resolution Strategies
Even with optimized protocols, researchers may encounter challenges when implementing TSA for planarian FISH. The following table addresses common issues and provides evidence-based solutions:
Table 3: TSA Troubleshooting Guide for Planarian FISH
| Problem | Potential Causes | Solutions | Preventive Measures |
|---|---|---|---|
| High Background | Excessive tyramide concentration or reaction time | Reduce tyramide concentration by 2-5 fold; shorten reaction time | Include RWBR in blocking buffer; optimize detergent concentration |
| Weak or No Signal | HRP inactivation; insufficient tyramide | Use fresh Hâ‚‚Oâ‚‚; check antibody activity; increase tyramide concentration | Test reagents systematically; include positive controls |
| Non-Specific Signal | Incomplete peroxidase quenching (multiplexing) | Extend azide quenching time; include heat denaturation step | Validate quenching efficiency before subsequent rounds |
| Poor Tissue Morphology | Over-bleaching or over-permeabilization | Reduce formamide bleaching to 1 hour; optimize proteinase K concentration | Use gentler cell isolation procedures [39] |
| Inconsistent Staining | Variable reagent penetration | Extend antibody incubation times; include additional detergent | Ensure consistent tissue size and fixation across samples |
Controls and Validation
Rigorous experimental controls are essential for validating TSA-FISH results in planarian research. Include the following controls in every experiment: (1) No-primary-antibody control to assess non-specific tyramide deposition; (2) Sense probe or irrelevant probe control to evaluate probe specificity; (3) Biological positive controls with known expression patterns (e.g., smedwi-1 for neoblasts); and (4) For multiplex experiments, single-label controls to confirm spectral separation and absence of cross-talk between channels. Additionally, correlate TSA-FISH findings with complementary methodologies when possible, such as comparing expression patterns with single-cell RNA sequencing data or validating functional roles through RNA interference experiments [39] [18] [41].
Applications in Planarian Stem Cell and Regeneration Research
The exceptional sensitivity of optimized TSA has enabled critical advances in understanding planarian stem cell biology and regeneration mechanisms. In stem cell research, TSA-enhanced multiplexed FISH approaches like MERFISH have revealed that fate-specified neoblasts for different lineages (epidermal, muscle, intestinal, neural) are spatially intermingled throughout the planarian parenchyma without obvious clustering, suggesting that fate specification involves cell-intrinsic processes rather than precise positional cues [18]. This discovery was dependent on the ability to simultaneously detect multiple low-abundance fate-specifying transcription factors with single-cell resolution.
In regeneration studies, TSA has been indispensable for visualizing the asymmetric expression of polarity determinants such as notum, which is selectively activated in longitudinal muscle cells at anterior-facing wounds within hours of injury [39]. The sensitivity required to detect these early, spatially restricted expression patterns necessitates the signal amplification provided by TSA. Similarly, studying the complex regulatory networks that control anterior-posterior patterning, including genes like NR1I3 that modulate Wnt signaling to promote proper regeneration polarity, relies on sensitive detection methods capable of resolving subtle expression differences in intact and regenerating planarians [41].
The ongoing development of increasingly multiplexed FISH technologies, combined with optimized TSA protocols, continues to expand our understanding of planarian biology by enabling comprehensive mapping of gene expression patterns with cellular resolution. These technical advances provide the foundation for deciphering the complex regulatory networks that govern stem cell behavior and regenerative patterning in these remarkable organisms.
Within the field of planarian research, the precise spatial mapping of gene expression is fundamental to understanding the mechanisms underlying regeneration and stem cell biology. Whole-mount fluorescent in situ hybridization (FISH) enables the visualization of transcript distributions within the context of the entire organism. However, the detection of low-abundance mRNAs and the simultaneous colocalization of multiple genes present significant technical challenges. This application note details a robust protocol for multicolor FISH in planarians, utilizing sequential rounds of tyramide signal amplification (TSA) to achieve high-sensitivity, multiplexed detection of gene expression patterns. The modifications presented here, developed within the context of planarian research, provide solutions for enhanced signal intensity, reduced background, and effective sequential detection, thereby offering researchers a powerful tool for detailed molecular anatomical studies [33] [13].
Key Principles and Optimizations for Planarian FISH
Planarian tissues present unique challenges for FISH, including inherent autofluorescence, the presence of sticky mucous, and the delicate nature of regenerating tissues. The following optimizations are critical for success.
Enhanced Permeabilization and Signal Intensity
A short bleaching step in formamide dramatically enhances signal intensity for both colorimetric and fluorescent detection. This treatment improves tissue permeability, allowing for better probe penetration, particularly in densely packed regions like the prepharyngeal area. Replacing the traditional overnight methanol bleach with a 1-2 hour bleach in a formamide-based solution significantly reduces the development time for probes and improves the final signal-to-noise ratio, a crucial factor for detecting low-abundance transcripts [33].
Suppression of Background and Autofluorescence
Achieving a high signal-to-noise ratio is paramount for sensitive FISH, especially since the TSA reaction cannot be monitored and stopped in real-time. The use of Roche Western Blocking Reagent (RWBR) in the blocking buffer dramatically reduces non-specific background staining without compromising signal intensity. Furthermore, the addition of 0.3% Triton X-100 to wash and blocking buffers improves signal specificity. For fluorescent applications, a copper sulfate quenching step can be employed to virtually eliminate tissue autofluorescence, which is a common issue in planarians [33].
Effective Peroxidase Quenching for Sequential Rounds
Multicolor FISH with TSA relies on sequential rounds of hybridization, antibody binding, and tyramide development. A critical step between each round is the complete inactivation of the horseradish peroxidase (HRP) enzyme from the previous cycle. Residual HRP activity will lead to false-positive signal in subsequent detection rounds. Direct comparison of quenching methods has demonstrated that incubation with sodium azide (NaN₃) is the most effective method for quenching peroxidase activity without being detrimental to the specimen or subsequent antibody-antigen interactions [33] [42]. This allows for reliable visualization of multiple transcripts in the same sample.
Table 1: Key Optimizations for Planarian Multicolor FISH
| Challenge | Solution | Key Reagent/Modification | Effect |
|---|---|---|---|
| Low Signal Intensity | Formamide Bleaching | Hydrogen peroxide in formamide | Enhances tissue permeability and signal intensity [33] |
| High Background | Optimized Blocking | Roche Western Blocking Reagent (RWBR) | Dramatically reduces non-specific antibody binding [33] |
| Tissue Autofluorescence | Chemical Quenching | Copper sulfate solution | Virtually eliminates autofluorescence across a broad wavelength range [33] |
| Residual POD Activity | Peroxidase Quenching | Sodium azide (NaN₃) | Effectively inactivates HRP between TSA rounds for multiplexing [33] [42] |
| Low-Abundance Transcripts | Signal Amplification | Iterative tyramide signal amplification (TSA) | Enables detection of rare transcripts [33] |
Reagent and Material Solutions
The following table catalogues the essential reagents and materials required for implementing this optimized multicolor FISH protocol.
Table 2: Research Reagent Solutions for Multicolor FISH
| Item | Function/Description | Example/Critical Note |
|---|---|---|
| Hapten-Labeled Probes | Labeled antisense RNA probes for target mRNA hybridization. | Digoxigenin (DIG), fluorescein (FAM), and dinitrophenol (DNP) are common haptens [33] [43]. |
| Anti-Hapten-HRP Antibodies | Primary detection of hapten-labeled probes. | Anti-DIG-HRP, anti-FAM-HRP, anti-DNP-HRP. Specificity is critical for multiplexing [33] [24]. |
| Fluorochromized Tyramides | TSA substrate for HRP-catalyzed deposition. | Commercially available or bench-made (e.g., Cy3-, FITC-, Cy5-tyramide). Bench-made TAMRA-tyramide reported as highly sensitive [43]. |
| Formamide Bleaching Solution | Tissue permeabilization and signal enhancement. | Hydrogen peroxide in a formamide-based buffer [33]. |
| Roche Western Blocking Reagent | Blocking agent for reduced background. | Superior to other blocking reagents like casein for anti-hapten antibodies [33]. |
| Triton X-100 | Detergent in wash and blocking buffers. | Using 0.3% concentration improves signal specificity [33]. |
| Sodium Azide (NaN₃) | Peroxidase quenching agent. | Essential for inactivating HRP between sequential TSA rounds [33] [42]. |
Experimental Workflow for Sequential Multicolor FISH
The following diagram outlines the core iterative workflow for a two-color FISH experiment, which can be expanded to three colors by adding another cycle of antibody development, TSA, and quenching.
Detailed Protocol
Step 1: Sample Preparation and Hybridization Begin with fixed planarians. For enhanced signal, perform a short bleaching step by incubating samples in a hydrogen peroxide solution prepared in formamide for 1-2 hours at room temperature. This is followed by proteinase K treatment for tissue permeabilization. After pre-hybridization, simultaneously hybridize the planarians with all hapten-labeled antisense RNA probes (e.g., DIG-, FAM-, and DNP-labeled) in hybridization buffer. Incubate at 55°C for 20-24 hours [33] [24].
Step 2: First Round of TSA Detection After post-hybridization washes, block samples in a buffer containing RWBR and 0.3% Triton X-100 to minimize background. Apply the first anti-hapten primary antibody conjugated to HRP (e.g., anti-DIG-HRP). Following antibody incubation and washes, develop the signal by incubating with the first fluorochromized tyramide substrate (e.g., Cy3-tyramide) in the presence of hydrogen peroxide. The HRP enzyme catalyzes the deposition of the fluorescent tyramide, labeling the sites of the first transcript [33] [24].
Step 3: Peroxidase Quenching To inactivate the HRP from the first detection round, treat the samples with a solution of sodium azide. This critical step prevents the first HRP from catalyzing the TSA reaction in subsequent rounds, which would cause false colocalization signal. Incubate for a sufficient time to ensure complete quenching of peroxidase activity [33] [42].
Step 4: Second and Subsequent Rounds of Detection Apply the second anti-hapten antibody conjugated to HRP (e.g., anti-FAM-HRP). This antibody will bind to its respective hapten on the second probe. Develop the signal with a second, spectrally distinct fluorochromized tyramide (e.g., FITC-tyramide). For a three-color FISH, repeat the quenching step and then proceed with the third anti-hapten-HRP antibody and its corresponding tyramide substrate [33] [43].
Step 5: Imaging and Analysis After completing all detection rounds, mount the samples and image using a fluorescence or confocal microscope. The use of copper sulfate in the final washes can be incorporated to quench endogenous autofluorescence, further improving the signal-to-noise ratio. Analyze the images to determine the spatial expression and potential colocalization of the target transcripts [33].
Quantitative Data from Protocol Optimizations
The optimizations presented in this note are supported by empirical data demonstrating their effectiveness in enhancing the FISH procedure.
Table 3: Impact of Key Protocol Modifications on Signal Quality
| Parameter Optimized | Standard Condition | Optimized Condition | Observed Outcome |
|---|---|---|---|
| Bleaching Method [33] | Overnight in Methanol | 1-2 hours in Formamide | Dramatic reduction in development time; maximum signal intensity achieved. |
| Blocking Reagent [33] | Standard Blocking Buffer | RWBR in Buffer | Dramatically reduced background for anti-DIG and anti-FAM antibodies. |
| Detergent in Washes [33] | Tween-20 only | 0.3% Triton X-100 | Noticeable improvement in signal specificity, especially for anti-DIG and anti-FAM. |
| POD Accelerator [43] | None | 150-450 μg/mL 4-Iodophenol | Dose-dependent, significant increase in specific signal intensity. |
| Viscosity Agent in Hyb [43] | None | 5% Dextran Sulfate | Significant increase in signal intensity due to macromolecular crowding. |
Solving Common Problems and Enhancing Signal-to-Noise Ratio
In the field of planarian research, whole-mount fluorescent in situ hybridization (FISH) using tyramide signal amplification (TSA) is a critical technique for determining gene expression patterns, particularly for low-abundance transcripts [33] [13]. This exceptional sensitivity, however, presents significant technical challenges that must be addressed for accurate data interpretation. Planarian tissues exhibit substantial endogenous autofluorescence across a broad spectrum of wavelengths and contain endogenous peroxidase activity, both of which contribute to high background noise that can obscure specific signal detection [33]. Within the context of a broader thesis on TSA-FISH in planarian research, effective quenching of these endogenous sources becomes paramount for achieving the signal-to-noise ratio necessary to elucidate the expression patterns of key regulatory genes, such as fate-specifying transcription factors that guide neoblast differentiation [4] [18]. This application note provides detailed, optimized protocols for using copper sulfate to quench autofluorescence and azide to quench peroxidase activity, enabling clearer and more reliable detection of gene expression in planarian studies.
Background and Significance
Planarians have emerged as a powerful model organism for studying regeneration, stem cell biology, and germ cell development [33]. Their remarkable regenerative capabilities are driven by pluripotent stem cells called neoblasts, which can produce all adult cell types [4] [18]. Investigating the mechanisms underlying these processes often relies on visualizing spatial gene expression patterns through FISH. The TSA method enhances detection sensitivity for low-abundance transcripts but is susceptible to interference from endogenous planarian pigments and enzymes [33].
The challenges are twofold. First, planarian tissue autofluoresces across a broad wavelength range, creating a poor signal-to-noise ratio that is particularly problematic for detecting weakly expressed genes [33]. Second, multicolor FISH experiments using sequential rounds of TSA require complete inactivation of peroxidase activity between developments to prevent false signals in subsequent detection rounds [33]. Without effective quenching, background interference can compromise data quality and lead to inaccurate interpretation of gene expression patterns, ultimately hindering progress in understanding fundamental principles of stem cell regulation and regeneration in planarians.
The table below summarizes the key quantitative findings from the optimization of quenching treatments for planarian FISH.
Table 1: Quantitative Summary of Quenching Treatment Efficacy
| Treatment | Target | Optimal Concentration & Duration | Key Outcome Metrics | Comparison to Alternatives |
|---|---|---|---|---|
| Copper Sulfate | Autofluorescence | Not specified in detail [33] | "Virtually eliminates autofluorescence" [33] | More effective than other quenchers (e.g., TrueBlack, Sudan Black B) which can diminish imaging depth [44] |
| Sodium Azide | Peroxidase Activity | Not specified in detail [33] | "Most effectively quenches peroxidase activity" between TSA rounds [33] | Superior to hydrogen peroxide-based quenching; least detrimental to subsequent detection rounds [33] |
Experimental Protocols
Copper Sulfate Quenching of Autofluorescence
This protocol is designed to be integrated into a standard planarian FISH procedure following probe hybridization and antibody detection steps.
Materials Required:
- Copper sulfate solution (CuSOâ‚„)
- Ammonium acetate
- Ethanol
- Standard phosphate-buffered saline (PBS) or maleic acid buffer
Methodology:
- Following the final wash steps after tyramide development, prepare a quenching solution of copper sulfate in a buffer containing ammonium acetate in ethanol [33].
- Immerse the planarian samples completely in the copper sulfate quenching solution.
- Incubate for the prescribed duration. The specific concentration and time should be optimized empirically for your specific system, as the original publication does not specify exact details.
- After incubation, thoroughly rinse the samples with buffer (e.g., PBS or maleic acid buffer) to remove any residual copper sulfate.
- Proceed with mounting the samples for microscopy imaging. The reduction in background autofluorescence will result in a significantly improved signal-to-noise ratio.
Azide Quenching of Endogenous Peroxidase
This protocol is critical for multicolor FISH experiments involving sequential rounds of TSA with different haptens.
Materials Required:
- Sodium azide (NaN₃)
- Hydrogen peroxide (Hâ‚‚Oâ‚‚)
- Standard wash buffers (e.g., PBSTx - PBS with Triton X-100)
Methodology:
- After completing the first round of tyramide signal amplification and development, treat the planarian samples with a solution of sodium azide [33].
- The treatment should be performed for a duration sufficient to fully inactivate the peroxidase enzyme from the previous round. The exact concentration and incubation time were not specified in the source and may require optimization.
- Following azide treatment, wash the samples thoroughly with an appropriate buffer to remove the quenching agent.
- Proceed directly with the next round of antibody incubation and tyramide development for a different probe. The azide quenching step prevents residual peroxidase activity from generating false-positive signals in subsequent detection rounds.
Workflow and Signaling Pathways
The following diagram illustrates the logical workflow for integrating these quenching treatments into a multicolor FISH protocol, highlighting the critical decision points that ensure experimental fidelity.
Research Reagent Solutions
The table below lists key reagents essential for implementing the quenching protocols described in this note, along with their critical functions in the experimental workflow.
Table 2: Essential Reagents for Quenching Protocols in Planarian FISH
| Reagent | Function/Application | Technical Notes |
|---|---|---|
| Copper Sulfate (CuSOâ‚„) | Quenches broad-spectrum tissue autofluorescence by reducing background noise. | Use in a solution with ammonium acetate in ethanol [33]. |
| Sodium Azide (NaN₃) | Effectively quenches peroxidase activity between sequential rounds of TSA development. | Prevents false-positive signals in multicolor FISH; superior to H₂O₂ [33]. |
| Formamide | Component of hybridization buffer and bleaching solution. | A short peroxide bleach in formamide dramatically enhances WISH/FISH signal intensity [33]. |
| Triton X-100 | Non-ionic detergent used in wash and blocking buffers. | Using 0.3% Triton X-100 improves signal specificity, especially with anti-DIG and anti-FAM antibodies [33]. |
| Roche Western Blocking Reagent (RWBR) | Protein-based blocking agent. | Addition to blocking buffer dramatically reduces background for anti-hapten antibodies without significant signal loss [33]. |
| Tyramide Signal Amplification (TSA) Kits | Enzymatic amplification system for high-sensitivity fluorescence detection. | Critical for detecting low-abundance transcripts; requires peroxidase quenching for multi-round use [33]. |
The meticulous quenching of endogenous peroxidase activity with azide and autofluorescence with copper sulfate represents a critical methodological advancement for TSA-FISH in planarian research [33]. These protocols directly address the key technical challenges of high background and signal crossover, enabling the precise detection of low-abundance transcripts. By implementing these optimized methods, researchers can more reliably elucidate the complex spatial expression patterns of genes governing neoblast fate specification and regeneration [4] [18]. This enhanced capability strengthens planarians as a model system for uncovering fundamental principles of stem cell biology and regenerative medicine.
Fluorescence in situ hybridization (FISH), particularly when combined with Tyramide Signal Amplification (TSA), provides an exceptionally powerful tool for detecting low-abundance nucleic acid targets. However, this high sensitivity comes with a significant challenge: nonspecific background signal that can obscure true positive results. In planarian research, where the goal is often to detect rare RNA transcripts in heterogeneous tissues containing pluripotent stem cells (neoblasts) and their progeny, minimizing background is paramount for accurate interpretation [4] [21]. Background in TSA-FISH primarily arises from nonspecific probe binding, endogenous enzyme activity, and nonspecific antibody interactions, but can also stem from cellular autofluorescence or sample aggregation, especially in complex biological samples like planarian tissues [45].
The catalytic nature of TSA, which utilizes horseradish peroxidase (HRP) to generate high-density labeling of target sequences, can amplify these nonspecific signals alongside the specific signal, sometimes up to 100-fold compared to conventional methods [1]. Therefore, implementing advanced blocking strategies is not merely an optional optimization step but a fundamental requirement for generating reliable, interpretable data in sensitive applications such as studying gene expression during planarian stem cell differentiation and regeneration [4] [21].
Principles of Background Generation and Blocking
Understanding the sources of background is the first step in effectively suppressing it. The mechanism of TSA involves HRP catalyzing the conversion of tyramide derivatives into highly reactive radicals that covalently bind to tyrosine residues on proteins in the immediate vicinity of the probe binding site [1]. While this localized deposition is key to the high spatial resolution of TSA, any HRP activity that is not specifically bound to the target can lead to widespread, diffuse background staining.
The primary sources of background in TSA-FISH and their characteristics are summarized in the table below.
Table 1: Common Sources of Background in TSA-FISH and Their Characteristics
| Source of Background | Manifestation | Primary Cause |
|---|---|---|
| Nonspecific Probe Binding | Diffuse cytoplasmic or nuclear staining | Cross-hybridization to off-target RNA/DNA sequences, especially in low-complexity or repetitive genomic regions [46]. |
| Nonspecific Antibody Binding | Widespread fluorescent signal across the sample | Hydrophobic or ionic interactions between detection antibodies and tissue components [1]. |
| Endogenous Enzyme Activity | High background across entire tissue section | Endogenous peroxidases (e.g., in red blood cells) or phosphatases that remain active after fixation [1]. |
| Cellular Autofluorescence | Signal in negative controls, often in specific organelles | Natural fluorescence of molecules like lipofuscin in lysosomes or NADPH in the cytoplasm. |
| Sample Aggregation | Clustered, irregular signal spots | Incomplete tissue dispersion, leading to cell clumps that trap reagents and scatter light [45]. |
A critical, yet often overlooked, aspect of background reduction begins at the probe design stage. For RNA-FISH, probes with high specificity are essential. Software tools like TrueProbes, which integrate genome-wide BLAST-based binding analysis with thermodynamic modeling, can generate probe sets with enhanced target selectivity by minimizing the probability of off-target binding, thereby reducing the underlying cause of background [46].
Reagent Solutions for Effective Blocking
A robust TSA-FISH experiment relies on a toolkit of specific reagents, each chosen to address a particular step in the workflow and mitigate a specific source of background.
Table 2: Essential Research Reagent Solutions for TSA-FISH
| Reagent / Solution | Function & Rationale | Specific Use Case |
|---|---|---|
| TSA Blocking Reagent | A proprietary solution included in TSA kits to occupy nonspecific protein-binding sites before tyramide incubation, preventing nonspecific deposition of the tyramide conjugate [1]. | Essential for all TSA reactions to ensure clean, specific signal amplification. |
| Species-Matched Normal Serum | Serum from the same species as the secondary antibody is used to block nonspecific binding sites for immunoglobulins on the tissue sample. | Blocking for 30-60 minutes after the permeabilization step, before applying the primary or secondary antibody. |
| HRP-Conjugated Antibodies | Highly purified antibodies conjugated to HRP for specific detection of primary antibodies or hapten-labeled probes. | Used as the enzyme source for the TSA reaction. Antibody fragments (e.g., from Zenon Kits) can reduce background [1]. |
| Biotin-XX Tyramide | A hapten-labeled tyramide used for indirect TSA. Requires a subsequent detection step but allows for signal multiplication. | Detected with fluorescent streptavidin after the TSA reaction. Useful for extremely low-abundance targets [1]. |
| Hydrogen Peroxide (Hâ‚‚Oâ‚‚) | A component of the TSA reaction buffer that is essential for the HRP-mediated activation of tyramide [1]. | Added to the tyramide working solution immediately before application to the sample. |
| Alexa Fluor Tyramides | Fluorescently labeled tyramides that provide bright, photostable signals. Available in various colors for multiplexing. | Used for direct detection in the TSA reaction. Kits are available that combine these with HRP-secondaries and buffer [1]. |
Application-Optimized Protocols for Planarian Research
The following protocols are tailored for planarian research, where detecting the expression of fate-specifying transcription factors (FSTFs) in neoblasts or their progeny is crucial for understanding colony growth and regeneration [4] [21].
Comprehensive Protocol for TSA-FISH on Planarian Tissue Sections
This protocol assumes fixed planarian tissue sections on gelatin-coated slides [45].
Deparaffinization and Rehydration (if using paraffin sections):
- Xylene: 2 x 10 minutes
- 100% Ethanol: 2 x 5 minutes
- 95%, 70%, 50% Ethanol: 2 minutes each
- DEPC-treated PBS: 2 x 5 minutes
Permeabilization:
- Incubate slides with a permeabilization buffer (e.g., 0.5% Triton X-100 in PBS) for 20 minutes at room temperature.
- Rinse with PBS.
Pre-hybridization Blocking:
- Prepare a pre-hybridization buffer containing components to reduce nonspecific probe binding (e.g., formamide, SSC, Denhardt's solution, tRNA).
- Apply buffer to sections and incubate in a humidified chamber at the hybridization temperature for 1 hour.
Hybridization:
Post-Hybridization Washes:
- Carefully remove coverslips and wash slides with pre-warmed SSC buffers of decreasing salinity to remove unbound and nonspecifically bound probes.
- A final wash in PBS prepares the sample for immunodetection.
Immunological Blocking and Detection:
- Quench Endogenous Peroxidases: Incubate slides with 1% Hâ‚‚Oâ‚‚ in PBS for 30 minutes in the dark to inactivate endogenous peroxidases [1]. Rinse with PBS.
- Block Nonspecific Antibody Binding: Apply a blocking solution containing 1-5% species-matched normal serum and 1-3% BSA in PBS for 60 minutes at room temperature.
- Apply HRP-Conjugate: Incubate with HRP-conjugated anti-hapten antibody (e.g., anti-DIG-HRP) or streptavidin-HRP, diluted in blocking solution, for 60-90 minutes at room temperature.
- Wash: Rinse thoroughly with PBS to remove unbound conjugate.
Tyramide Signal Amplification:
- Prepare the tyramide working solution by diluting fluorescent tyramide (e.g., Alexa Fluor tyramide) in the provided amplification buffer and adding Hâ‚‚Oâ‚‚ to the recommended concentration immediately before use [1].
- Apply the solution to the slides and incubate for 5-10 minutes. The short incubation time is critical to prevent background buildup.
- Stop the reaction by washing slides extensively with PBS.
Counterstaining and Mounting:
Diagram 1: TSA-FISH Workflow for Planarians
Optimized Multiplexing and Validation Strategy
For multiplex TSA-FISH, where multiple targets are detected in the same sample, sequential rounds of amplification are required. A critical step between rounds is the inactivation of the HRP from the previous cycle by treating the slides with 1% Hâ‚‚Oâ‚‚ for 30 minutes after the first TSA reaction is complete and before applying the next HRP-conjugated antibody [1]. This prevents cross-talk and false-positive colocalization.
Validation is crucial. Controls must include:
- No-Probe Control: Hybridization without any probe to assess background from immunodetection and autofluorescence.
- Sense Probe or Scrambled Probe Control: To assess sequence-specificity of the signal.
- RNAi Knockdown Validation: For planarian research, using RNAi to knock down the target gene (e.g., as in
pou4-2(RNAi)orsoxB1-2(RNAi)studies) provides a biological negative control, where a significant reduction in signal confirms probe specificity [21].
Quantitative Analysis and Troubleshooting
Accurate quantification of FISH signals, especially in complex samples like planarian tissue that may contain autofluorescent structures, requires robust image analysis. Simple intensity thresholding can be inconsistent because fluorescence intensity can vary between experiments [45]. Advanced clustering methods, such as Fuzzy c-Means (FCM) clustering, can be applied to intensity data to automatically classify cells into target (positive) and non-target (negative) populations without relying on a fixed, manually set threshold, leading to more reliable and reproducible quantification [45].
Table 3: Troubleshooting Guide for High Background in TSA-FISH
| Problem | Potential Cause | Recommended Solution |
|---|---|---|
| High diffuse background across entire tissue | Incomplete quenching of endogenous peroxidases. | Increase Hâ‚‚Oâ‚‚ quenching incubation time or concentration. Include a positive control for peroxidase activity. |
| High diffuse background across entire tissue | Nonspecific binding of the HRP-conjugated antibody. | Increase the concentration of normal serum in the blocking solution. Titrate the HRP-antibody to use the lowest effective concentration. |
| High diffuse background across entire tissue | Tyramide reaction incubated for too long. | Shorten the tyramide incubation time (start with 5 minutes). |
| Punctate or speckled background in negative regions | Nonspecific probe hybridization. | Increase the stringency of post-hybridization washes (e.g., lower salt concentration, higher temperature). Use a computational tool like TrueProbes to redesign probes for higher specificity [46]. |
| High background in cell aggregates | Incomplete tissue dispersion trapping reagents [45]. | Optimize sample dispersion protocols, such as sonication duration for complex samples [45]. |
| Weak or No Specific Signal | Over-fixation or inadequate permeabilization. | Titrate fixation time and optimize permeabilization reagent and duration. |
| Weak or No Specific Signal | HRP conjugate inactivated by residual peroxide. | Ensure thorough washing after the endogenous peroxidase quenching step before applying the HRP conjugate. |
Mastering advanced blocking strategies is fundamental to exploiting the full power of TSA-FISH in demanding applications like planarian stem cell research. By understanding the biochemical origins of background and systematically applying the right reagents—from sophisticated probe design software and dedicated TSA blocking reagents to serum-based protein blocks and peroxidase quenching solutions—researchers can achieve exceptional signal-to-noise ratios. The protocols and troubleshooting guides outlined here provide a concrete pathway to obtaining clean, quantifiable, and reliable data, enabling the precise detection of low-abundance transcripts that drive fate decisions in neoblast colonies and illuminate the remarkable regenerative capabilities of planarians.
The efficacy of therapeutic and diagnostic antibodies is often limited by biological barriers presented by dense tissues. These barriers, characterized by dense extracellular matrix, high interstitial pressure, and complex cellular architecture, significantly impede antibody penetration and distribution. This challenge is acutely present in solid tumors, neural tissues, and dense organ systems, often resulting in subtherapeutic drug concentrations at the target site and compromised treatment outcomes.
Planarian research provides a unique model for studying these challenges due to its complex mesenchymal environment where neoblasts (planarian stem cells) are distributed throughout a dense parenchymal space. Recent spatial transcriptomic studies using multiplexed error-robust fluorescence in situ hybridization (MERFISH) have revealed that specialized neoblasts for different lineages (epidermal, muscle, intestinal, neural) exist in a highly intermingled spatial distribution without clear clustering, often located at significant distances from their target tissues [18]. This complex spatial organization presents a challenging environment for antibody penetration, mirroring the delivery challenges seen in mammalian dense tissues.
Key Optimization Strategies and Molecular Formats
Advanced Antibody Formats for Enhanced Tissue Penetration
Nanobodies and Small Fragment Engineering: Camelid-derived single-domain antibody fragments (VHHs), known as nanobodies, represent a breakthrough technology for dense tissue penetration. Their small size (approximately 15 kDa versus 150 kDa for conventional IgG) enables superior diffusion through dense tissues [47]. Their remarkable stability under extreme temperature and pH conditions, coupled with the ability to be produced cost-effectively in microbial systems, makes them ideal for diagnostic and therapeutic applications in challenging environments [47]. The long, finger-like complementarity-determining region (CDR3) of nanobodies allows them to bind unique, concave epitopes that are often inaccessible to conventional antibodies, potentially opening new targeting possibilities in dense tissues [47].
Bispecific and Multispecific Formats: Bispecific antibodies (bsAbs) represent a strategic shift from monospecific targeting, enabling engagement with two different antigens or epitopes simultaneously [47] [48]. This dual-targeting capability unlocks novel mechanisms of action, particularly relevant for complex tissues. In planarian research, where cell fate specification involves complex signaling pathways, bsAbs could be designed to simultaneously target multiple positional control genes (PCGs) or fate-specifying transcription factors (FSTFs). From a therapeutic perspective, T-cell engagers—a prominent class of bsAbs—can physically bridge T-cells to cancer cells, triggering a targeted immune attack even in poorly penetrated tissues [48]. The clinical success of agents like blinatumomab, mosunetuzumab, and tarlatamab in hematologic malignancies demonstrates the potency of this approach, though application in dense solid tumors remains challenging [48].
Antibody-Drug Conjugates (ADCs) and Novel Conjugates: ADCs combine the specificity of antibodies with the potency of cytotoxic payloads, creating "smart chemotherapy" that minimizes collateral damage to healthy tissues [47]. Next-generation ADC innovation focuses on three key areas: novel payloads (including immune-stimulating agents and protein degraders), advanced linker technology for stable circulation and precise payload release, and bispecific ADCs that recognize two different tumor antigens to address heterogeneity [47]. Beyond traditional ADCs, emerging formats like antibody-oligonucleotide conjugates (AOCs) combine the antigen-specific binding capability of an antibody with the gene-regulatory function of an oligonucleotide, enabling targeted therapeutic intervention across a range of diseases [49]. While promising, these complex conjugates present significant manufacturing and quality control challenges that must be addressed to ensure therapeutic efficacy and minimize off-target toxicity.
Table 1: Quantitative Comparison of Antibody Formats for Dense Tissue Applications
| Antibody Format | Molecular Weight (kDa) | Tissue Penetration Potential | Multitargeting Capability | Production Complexity |
|---|---|---|---|---|
| Full-size IgG | 150 | Low | No | High |
| Nanobody | 15 | High | No (monospecific) | Low (microbial systems) |
| Bispecific IgG | 150 | Low | Yes | Very High |
| scFv | 25-30 | Medium-High | No (monospecific) | Medium |
| ADC | 150-160 | Low | No (but delivers payload) | Very High |
| AOC | 155-165 | Low | No (but delivers oligonucleotide) | Very High |
Affinity and Valency Optimization
The relationship between antibody affinity and tissue penetration follows a paradoxical pattern where extremely high affinity (Kd < nM) can sometimes reduce overall efficacy by causing antibodies to bind immediately to the first encountered antigen in perivascular regions, preventing deeper tissue penetration—a phenomenon known as the "binding site barrier." Moderate affinity antibodies (Kd in nM range) may achieve more uniform tissue distribution by allowing for binding-site dissociation and rebinding events that facilitate deeper penetration.
Valency engineering offers additional control over antibody behavior. While multivalent binding increases functional affinity (avidity) and can improve target retention, it may also exacerbate penetration issues. Monovalent formats often achieve superior penetration but faster clearance. Innovative solutions include conditionally multivalent antibodies that remain monovalent until activated by proteases in the target tissue, providing both penetration and high avidity.
Computational and AI-Driven Design
Artificial intelligence is revolutionizing antibody optimization by predicting how antibody sequence modifications affect stability, affinity, and specificity. Machine learning models can predict antibody folding and binding characteristics in silico, dramatically reducing laboratory guesswork and development time [47]. Generative AI approaches can now design entirely novel antibody sequences or 3D structures tailored to bind specific targets with desired characteristics [47]. These computational approaches are particularly valuable for engineering antibodies targeting complex epitopes in dense tissues, such as those found in the planarian parenchymal environment where neighboring neoblasts frequently make divergent fate choices [18].
Experimental Protocols for Penetration and Specificity Assessment
Protocol 1: Tyramide Signal Amplification FISH for Low-Abundance Targets in Dense Tissues
Application Note: This protocol adapts standard FISH methodology with tyramide signal amplification (TSA) to enhance detection sensitivity for low-abundance mRNA targets in dense planarian tissues, particularly useful for visualizing fate-specifying transcription factors (FSTFs) expressed in neoblasts.
Reagents and Equipment:
- Planarian tissue sections (5-10 μm thickness)
- DIG- or FITC-labeled RNA probes targeting genes of interest
- Anti-DIG or Anti-FITC HRP-conjugated antibodies
- Tyramide-based fluorescent substrates (Alexa Fluor tyramides recommended)
- Hydrogen peroxide solution
- Proteinase K solution
- Hybridization buffer, wash buffers
- Fluorescence microscope with high-resolution imaging capabilities
Procedure:
- Tissue Preparation: Fix planarian tissue in 4% paraformaldehyde, embed in paraffin, and section at 5-10 μm thickness. Mount sections on charged slides.
- Deparaffinization and Rehydration: Deparaffinize slides in xylene, followed by ethanol series (100%, 95%, 70%) and finally PBS.
- Proteinase Digestion: Treat sections with Proteinase K (1-10 μg/mL in PBS) for 5-15 minutes at room temperature to permeabilize tissue. Optimize concentration and time to balance tissue preservation and probe accessibility.
- Hybridization: Apply DIG-labeled RNA probes (50-100 ng/μL) in hybridization buffer and incubate overnight at 55-65°C in a humidified chamber.
- Post-Hybridization Washes: Perform stringency washes with SSC buffers (2× SSC, 0.2× SSC, 0.1× SSC) at hybridization temperature to reduce non-specific binding.
- TSA Amplification: a. Block endogenous peroxidase activity with 0.3-1% Hâ‚‚Oâ‚‚ in PBS for 30 minutes. b. Apply anti-DIG-HRP antibody (1:500-1:1000 dilution) for 1 hour at room temperature. c. Wash thoroughly to remove unbound antibody. d. Apply fluorescent tyramide substrate (diluted 1:50-1:100 in amplification buffer) for 2-10 minutes, monitoring development.
- Counterstaining and Mounting: Counterstain with DAPI (1 μg/mL) for nuclear visualization, and mount with anti-fade mounting medium.
- Imaging and Analysis: Image using a fluorescence microscope with appropriate filter sets. For multiplexing, repeat the TSA process with different probes and tyramide substrates, incorporating HRP inactivation between rounds.
Troubleshooting Notes:
- Excessive background may indicate insufficient washing or excessive tyramide incubation time.
- Weak signal may require increased probe concentration, longer hybridization time, or extended tyramide development.
- For multiplexing, always proceed from least abundant to most abundant targets.
Protocol 2: Quantitative Assessment of Antibody Penetration in Dense Tissues
Application Note: This protocol describes a method for quantitatively evaluating antibody penetration efficiency in dense tissues using fluorescently labeled antibodies and spatial image analysis, applicable to both planarian and mammalian tissue models.
Reagents and Equipment:
- Test antibodies (fluorescently labeled)
- Tissue sections or whole-mount preparations
- Microfluidic delivery system (optional)
- Confocal or light-sheet fluorescence microscope
- Image analysis software (e.g., ImageJ, Imaris)
Procedure:
- Tense Tissue Preparation: Use 200-500 μm thick tissue sections or small whole-mount tissue fragments to preserve three-dimensional architecture.
- Antibody Application: Apply fluorescently labeled antibodies at varying concentrations (0.1-10 μM) in physiological buffer. Include control antibodies with known penetration characteristics.
- Distribution Period: Incubate tissues with antibodies for varying durations (1-48 hours) at physiological temperature with gentle agitation.
- Fixation and Clearing: Fix tissues with 4% PFA and optionally use tissue clearing techniques (e.g., CLARITY, CUBIC) for improved imaging depth.
- Image Acquisition: Acquire z-stack images using confocal or light-sheet microscopy at multiple random fields per sample.
- Quantitative Analysis: a. Measure fluorescence intensity as a function of distance from tissue surface or vasculature. b. Calculate penetration depth (distance where signal drops to half of maximum). c. Determine distribution uniformity (coefficient of variation across tissue regions).
- Data Interpretation: Compare penetration parameters across different antibody formats, sizes, and affinities.
Table 2: Key Parameters for Antibody Penetration Assessment
| Parameter | Measurement Method | Optimal Values for Dense Tissues |
|---|---|---|
| Penetration Depth | Distance where signal drops to 50% of maximum | >100 μm for fragments, >50 μm for full IgG |
| Distribution Uniformity | Coefficient of variation across tissue regions | <0.5 indicates uniform distribution |
| Binding Site Barrier Effect | Ratio of peripheral to deep tissue signal | <2.0 indicates minimal barrier effect |
| Absolute Concentration | Calibrated fluorescence intensity | Varies by target abundance and application |
Integration with Planarian Research and Signaling Pathways
Planarian Fate Specification as a Model System
Planarians offer a powerful model for studying antibody penetration challenges due to their complex tissue organization. Neoblasts (planarian stem cells) are distributed throughout the mesenchymal space in a highly intermingled manner, with neighboring neoblasts frequently making divergent fate choices [18]. Specialized neoblasts for different lineages (epidermal, muscle, intestinal, neural) exist in overlapping spatial domains without clear segregation, creating a challenging environment for targeted antibody delivery [18].
Research has shown that fate specification in neoblasts is regulated through expression of fate-specific transcription factors (FSTFs), with specification from naive to post-mitotic differentiating state potentially occurring within a single cell cycle [4]. This rapid specification process creates a dynamic target environment for molecular interventions. The spatial distribution of these fate choices is not random but follows positional information governed by conserved signaling pathways including Wnt, Notum, and other positional control genes (PCGs) [41].
Key Signaling Pathways in Planarian Patterning
The Wnt/β-catenin signaling pathway plays a crucial role in anteroposterior (AP) patterning in planarians. Multiple Wnt genes are expressed in a graded fashion along the AP axis to promote tail regeneration [41]. Inhibition of Wnt signaling components such as β-catenin-1, wnt1, teashirt, and wntless results in the formation of a head instead of a tail at posterior-facing wounds [41]. Conversely, knockdown of APC, a negative regulator of β-catenin, causes regeneration of tails in place of heads [41].
Notum, a secreted Wnt inhibitor, is required for head regeneration and displays asymmetric expression at anterior and posterior wounds, determining regeneration outcomes [41]. Recent research has identified NR1I3, a nuclear receptor family transcription factor, as an upstream regulator of Wnt signaling that mediates AP patterning through regulation of notum activation [41]. NR1I3 RNA interference (RNAi) during regeneration causes ectopic head formation in the posterior blastema, correlated with symmetric notum expression at wounds [41].
Diagram 1: Planarian AP Patterning Pathway
Antibody Validation in Planarian Research Context
Validating antibody specificity in planarian research presents unique challenges due to the evolutionary distance from commonly used model organisms and the presence of many planarian-specific genes. Proper validation is essential for reliable interpretation of experimental results, particularly when studying complex processes like fate specification and tissue patterning.
Key validation steps include:
- Knockdown Validation: Where possible, demonstrate reduced signal following RNAi-mediated gene knockdown.
- Expression Pattern Comparison: Compare staining patterns with known spatial expression data from in situ hybridization.
- Tagged Protein Expression: Express epitope-tagged versions of target proteins and demonstrate co-localization with antibody signal.
- Peptide Competition: Pre-incubate antibodies with immunizing peptides to demonstrate specific binding blockade.
- Multiple Epitope Targeting: Use multiple antibodies against different epitopes of the same protein to confirm staining patterns.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents for Antibody Applications in Dense Tissues
| Reagent Category | Specific Examples | Primary Function | Considerations for Dense Tissues |
|---|---|---|---|
| Tyramide Signal Amplification Reagents | Fluorescent tyramides (Alexa Fluor series), HRP-conjugated antibodies | Signal amplification for low-abundance targets in challenging tissues | Critical for detecting weakly expressed FSTFs in planarian tissues |
| Tissue Clearing Reagents | CUBIC, CLARITY, ScaleS | Render tissues transparent for improved antibody penetration and deep imaging | Essential for 3D reconstruction of antibody distribution patterns |
| Nanobody Reagents | Anti-GFP nanobodies, epitope-specific VHHs | Superior penetration in dense tissues due to small size | Can be engineered as bispecific formats for complex targeting |
| Bispecific Antibody Platforms | BiTE, DART, TandAb constructs | Engage multiple targets simultaneously for enhanced specificity and efficacy | T-cell engagers can activate immune response in poorly penetrated areas |
| Validation Tools | Recombinant planarian proteins, RNAi reagents | Confirm antibody specificity in evolutionarily distant systems | Crucial for planarian research due to limited commercial antibodies |
| Advanced Microscopy Reagents | Multiplexed FISH reagents, 3D imaging compatible mountants | Enable visualization of antibody distribution and target engagement in 3D space | Required for quantitative penetration assessment |
| 4-Pteridinamine, 7-phenyl- | 4-Pteridinamine, 7-phenyl-, CAS:73384-11-9, MF:C12H9N5, MW:223.23 g/mol | Chemical Reagent | Bench Chemicals |
| 5-Iodo-1-pentanol acetate | 5-Iodo-1-pentanol acetate, CAS:65921-65-5, MF:C7H15IO3, MW:274.10 g/mol | Chemical Reagent | Bench Chemicals |
Diagram 2: Antibody Validation Workflow
Antibody optimization for dense tissues requires a multifaceted approach combining advanced molecular formats, sophisticated delivery strategies, and rigorous validation methods. The integration of nanobodies, bispecific constructs, and signal amplification technologies represents the current state-of-the-art in overcoming penetration barriers. Planarian research provides a valuable model system for testing these optimization strategies due to its complex tissue organization and well-characterized patterning mechanisms.
Future developments will likely focus on conditionally active antibodies that become functional only in target tissues, computationally designed antibodies with optimized biophysical properties, and novel conjugation strategies that enhance tissue distribution while maintaining target specificity. As these technologies mature, they will undoubtedly enhance our ability to both study complex biological systems like planarian regeneration and develop more effective therapeutic antibodies for challenging disease targets in dense tissues.
Balancing Permeabilization and Tissue Integrity, Especially in Regenerating Blastemas
In the field of planarian research, whole-mount in situ hybridization (WISH) and its more sensitive variant, tyramide signal amplification FISH (TSA-FISH), are indispensable tools for visualizing gene expression patterns. These techniques are particularly valuable for studying the molecular underpinnings of regeneration, a process driven by neoblasts (pluripotent stem cells) and manifested in the formation of a regeneration blastema—the delicate, unpigmented tissue at the wound edge that gives rise to new structures [4] [50]. The central challenge in applying these techniques lies in achieving effective tissue permeabilization to allow probe penetration while preserving the structural integrity of the blastema and the surrounding tissue. Traditional protocols often rely on harsh treatments like proteinase K digestion and aggressive mucolytic agents, which can compromise delicate tissues, leading to damage or destruction of the very structures researchers aim to study [50]. This application note details the Nitric Acid/Formic Acid (NAFA) protocol, a robust method that successfully balances these competing demands, enabling high-quality gene expression analysis in homeostatic and regenerating planarians.
Comparative Analysis of Permeabilization Methods
We systematically evaluated the performance of three fixation and permeabilization protocols—the novel NAFA protocol, the established NAC protocol, and the NA (Rompolas) protocol—across key parameters critical for planarian research. The following table summarizes the quantitative and qualitative findings, with a focus on compatibility with TSA-FISH and the preservation of regenerating blastemas.
Table 1: Comparative Analysis of Planarian Permeabilization and Fixation Protocols
| Evaluation Parameter | NAFA Protocol | NAC Protocol | NA (Rompolas) Protocol |
|---|---|---|---|
| Epidermis Integrity | Well-preserved [50] | Noticeable breaches and damage [50] | Well-preserved [50] |
| Blastema Preservation | Excellent preservation of delicate blastema tissue [50] | Damage and shredding of regeneration blastema [50] | Not specified in search results |
| Chromogenic WISH Signal | Strong signal for internal (piwi-1) and external (zpuf-6) markers [50] | Strong signal, but with concurrent epidermal damage [50] | No detectable signal for piwi-1 or zpuf-6 [50] |
| Fluorescent Signal Intensity | Intense and clear [50] | Intense and clear [50] | Much weaker signal [50] |
| Immunostaining Compatibility | High; brighter signal for anti-H3P and preserved epitopes [50] | Weaker immunostaining signal; epitopes potentially disrupted [50] | Qualitative staining similar to NAFA for some antibodies [50] |
| Key Differentiating Step | Acid-based permeabilization without proteinase K [50] | Proteinase K digestion and aggressive NAC treatment [50] | Acid-based, but without formic acid [50] |
| Recommended for TSA-FISH on Blastemas? | Highly Recommended | Not Recommended | Not Recommended |
Detailed Experimental Protocols
The NAFA Fixation and Permeabilization Protocol for TSA-FISH
The following protocol is optimized for planarians, including those undergoing regeneration, and is compatible with subsequent TSA-FISH and immunofluorescence procedures [50].
Reagents:
- Fixative Solution: 4% Formaldehyde in PBS
- Nitric Acid Solution: 5% Nitric Acid (HNO₃)
- Permeabilization Solution: 5% Formic Acid in PBSTx (PBS with 0.3% Triton X-100)
- PBSTx Washing Buffer: PBS with 0.3% Triton X-100
- Glycine Solution: 2 mg/mL Glycine in PBSTx
- Optional: EGTA can be included to inhibit nucleases and preserve RNA integrity [50].
Procedure:
- Fixation: Fix planarians in 4% formaldehyde for 20-30 minutes at room temperature or overnight at 4°C.
- Wash: Rinse animals 3 x 5 minutes with PBSTx.
- Nitric Acid Treatment: Incubate planarians in 5% Nitric Acid for 5 minutes.
- Wash: Rinse thoroughly 3 x 5 minutes with PBSTx.
- Formic Acid Permeabilization: Incubate planarians in 5% Formic Acid for 10-12 minutes.
- Wash and Quench: Rinse 3 x 5 minutes with PBSTx, then incubate in Glycine solution for 10 minutes to quench residual aldehyde groups.
- Final Wash: Perform a final 3 x 5 minute wash with PBSTx.
- Proceed to Hybridization: The samples are now ready for the standard TSA-FISH procedure.
Workflow for Evaluating Permeabilization Efficacy and Tissue Integrity
The following diagram illustrates the key steps for implementing and validating the NAFA protocol in a research setting.
Evaluating Permeabilization Protocols
The Scientist's Toolkit: Essential Research Reagents
Successful application of the NAFA protocol and subsequent TSA-FISH relies on a set of key reagents. The following table outlines these essential materials and their functions.
Table 2: Key Research Reagent Solutions for Planarian TSA-FISH
| Reagent/Material | Function in the Protocol | Specific Example / Note |
|---|---|---|
| Formaldehyde | Crosslinking fixative that preserves tissue architecture and immobilizes biomolecules. | Typically used at 4% in PBS. |
| Nitric Acid (HNO₃) | Acid treatment that initiates permeabilization of the planarian epidermis. | Used at 5% concentration for ~5 minutes [50]. |
| Formic Acid | Carboxylic acid that completes permeabilization, allowing probe penetration without protease use. | 5% solution in PBSTx; can be substituted with acetic or lactic acid [50]. |
| Triton X-100 | Non-ionic detergent that solubilizes membranes, further enhancing permeability. | Standard component (0.1-0.3%) of washing buffers (PBSTx). |
| Proteinase K | Protease that digests proteins to permeabilize tissue. | Avoided in the NAFA protocol to preserve tissue integrity and antigen epitopes [50]. |
| N-Acetyl Cysteine (NAC) | Mucolytic agent that breaks down mucus. | Considered aggressive in established protocols; can damage epidermis and blastema [50]. |
| Anti-acetylated Tubulin Antibody | Immunostaining marker for cilia; used to assess epidermal integrity. | Integrity is well-preserved with NAFA and NA protocols, but damaged with NAC [50]. |
| Anti-H3P (Phospho-Histone H3) Antibody | Immunostaining marker for mitotic cells. | Signal is brighter with the NAFA protocol compared to NAC and NA protocols [50]. |
| DAPI (4',6-diamidino-2-phenylindole) | Fluorescent nuclear counterstain. | Standard dye for visualizing all nuclei in fluorescence imaging [51]. |
| 2,6-Dichloro-4-ethylphenol | 2,6-Dichloro-4-ethylphenol, CAS:7495-69-4, MF:C8H8Cl2O, MW:191.05 g/mol | Chemical Reagent |
| 8-(Benzylsulfanyl)quinoline | 8-(Benzylsulfanyl)quinoline|Research Compound | 8-(Benzylsulfanyl)quinoline is a quinoline derivative for research use only (RUO). Explore its potential applications in medicinal chemistry and chemical biology. |
The NAFA protocol represents a significant methodological advancement for gene expression studies in planarian regeneration. By replacing destructive proteinase K digestion with a controlled acid-based permeabilization step, it successfully resolves the core conflict between probe access and morphological preservation. This enables researchers to obtain clear, robust TSA-FISH signals while maintaining the integrity of critical structures like the regeneration blastema, thereby providing more reliable and high-fidelity data for understanding the molecular mechanisms of stem cell biology and regeneration.
Tyramide signal amplification fluorescence in situ hybridization (TSA-FISH) has become an indispensable technique in planarian research, enabling highly sensitive detection of gene expression patterns essential for studying regeneration and stem cell biology. The technique is particularly valuable for visualizing low-abundance transcripts in these experimentally flatworms, where precise gene expression patterns govern the remarkable regenerative capabilities of neoblasts (pluripotent stem cells). However, researchers often face challenges with weak signals or high background that can compromise data interpretation. This application note provides a comprehensive troubleshooting framework for optimizing TSA-FISH in planarians, incorporating both established and novel methodological improvements to achieve robust, publication-quality results.
Understanding TSA-FISH in Planarian Context
TSA-FISH leverages the catalytic activity of peroxidase enzymes to deposit fluorochromized tyramine molecules at sites of probe hybridization, resulting in substantial signal amplification through the generation of highly reactive tyramide radicals that covalently bind to electron-rich amino acids near the target [7]. This exceptional sensitivity makes it particularly suitable for detecting low-abundance transcripts in planarians, such as fate-specifying transcription factors that determine neoblast differentiation into over 125 distinct cell types [18].
The planarian experimental system presents unique challenges for TSA-FISH, including high mucus secretion that impedes probe penetration, endogenous peroxidase activity, broad tissue autofluorescence, and delicate regenerating tissues that are easily damaged by standard permeabilization methods [33] [34]. The following sections address these challenges through optimized protocols and systematic troubleshooting approaches.
Optimized Experimental Workflows
Sample Preparation and Fixation
Proper sample preparation is critical for successful TSA-FISH in planarians. The recently developed Nitric Acid/Formic Acid (NAFA) protocol offers significant advantages over traditional methods by better preserving delicate tissues like the regeneration blastema and epidermis while maintaining excellent RNA integrity [34].
NAFA Fixation Protocol:
- Mucus Removal: Incubate planarians in 5% N-acetyl-cysteine (NAC) for 5-10 minutes with gentle agitation [34]
- Fixation: Transfer to NAFA fixative (containing nitric acid, formic acid, and EGTA in PBS) for 2-4 hours at room temperature [34]
- Washing: Rinse 3× with PBSTx (PBS with 0.3% Triton X-100)
- Dehydration: Gradual methanol series (25%, 50%, 75% in PBSTx) followed by 100% methanol
- Storage: Store at -20°C in 100% methanol until use
Comparative studies demonstrate that the NAFA protocol preserves epidermal integrity significantly better than traditional NAC-based methods while maintaining comparable probe penetration for internal markers like piwi-1 (neoblasts) and zpuf-6 (epidermal progenitors) [34].
Hybridization and Signal Detection
Enhanced Permeabilization and Bleaching: Traditional methanol bleaching can be replaced with a short formamide-based bleaching step that dramatically improves signal intensity. Incubate fixed samples in formamide bleaching solution (containing hydrogen peroxide) for 1-2 hours, which enhances tissue permeability while reducing autofluorescence [33].
Optimized Hybridization Conditions:
- Pre-hybridization: Rehydrate through methanol series and pre-hybridize in hybridization buffer for 2-4 hours at 56°C
- Probe Hybridization: Incubate with hapten-labeled riboprobes (DIG, DNP, or FITC) at 56°C for 16-40 hours
- Post-hybridization Washes: Perform stringent washes with SSC buffers containing 0.3% Triton X-100 to reduce non-specific binding [33]
TSA Signal Amplification:
- Blocking: Incubate with modified blocking buffer containing Roche Western Blocking Reagent and 0.3% Triton X-100 for 4-16 hours [33]
- Peroxidase Conjugation: Incubate with anti-hapten peroxidase conjugates (anti-DIG, anti-DNP, or anti-FITC) for 24-48 hours at 4°C
- Tyramide Development: Develop signal with fluorochromized tyramide (e.g., FITC-tyramide, Cy3-tyramide) in reaction buffer containing glucose oxidase for stable hydrogen peroxide production [7]
- Peroxidase Quenching: For multiplex experiments, incubate with 2% sodium azide between development rounds to completely inactivate peroxidase [33] [7]
Troubleshooting Common Problems
Weak or Absent Signal
Weak hybridization signals represent one of the most frequent challenges in planarian TSA-FISH. The table below summarizes the primary causes and evidence-based solutions:
Table 1: Troubleshooting Weak Signal in Planarian TSA-FISH
| Cause | Solution | Rationale |
|---|---|---|
| Insufficient permeabilization | Replace methanol bleaching with 1-2 hour formamide bleach [33] | Formamide dramatically improves tissue permeability without damaging mRNA integrity |
| Inefficient probe penetration | Include 0.3% Triton X-100 in wash and blocking buffers [33] | Enhanced detergent action improves probe access to internal tissues |
| Low-abundance target | Use glucose oxidase-based TSA system for stable Hâ‚‚Oâ‚‚ production [7] | Enzymatic Hâ‚‚Oâ‚‚ generation maintains consistent reaction conditions |
| Suboptimal fixation | Implement NAFA protocol instead of proteinase K digestion [34] | Acid-based fixation preserves RNA and epitopes while allowing adequate penetration |
| Excessive development time | Standardize development time using positive control probes | Prevents reaction exhaustion; TSA proceeds rapidly to completion [33] |
The fluorochromized tyramide-glucose oxidase (FT-GO) system provides particularly robust signal amplification, demonstrating 9.7 to 34.1-fold greater sensitivity compared to conventional indirect immunofluorescence detection methods [7].
High Background Fluorescence
Excessive background staining can obscure specific signals and complicate data interpretation. The following approaches effectively reduce background in planarian TSA-FISH:
Table 2: Addressing High Background in Planarian TSA-FISH
| Background Type | Solution | Rationale |
|---|---|---|
| Non-specific antibody binding | Use Roche Western Blocking Reagent in blocking buffer [33] | Specifically reduces background from anti-hapten antibodies |
| Tissue autofluorescence | Quench with 0.25% copper sulfate in 50mM ammonium acetate buffer [33] | Effectively reduces planarian autofluorescence across multiple wavelengths |
| Endogenous peroxidase | Include 1-2% sodium azide in incubation buffers [33] | Inhibits endogenous peroxidase activity without damaging tissues |
| Non-specific tyramide deposition | Optimize tracer concentration and include competitive inhibitors [7] | Reduces hydrophobic interactions with planarian tissues |
| Incomplete washing | Extend wash times and include 0.3% Triton X-100 [33] | Enhanced detergent action improves removal of unbound reagents |
The modified blocking buffer with Roche Western Blocking Reagent and Triton X-100 dramatically reduces background, particularly for anti-DIG and anti-FAM antibodies commonly used in TSA-FISH [33].
Multiplexing Challenges
Multiplex TSA-FISH enables simultaneous detection of multiple transcripts, which is particularly valuable for studying complex gene regulatory networks in planarian stem cells and regeneration. Effective multiplexing requires complete peroxidase inactivation between development rounds. Sodium azide (2% in PBS) provides the most effective quenching while preserving tissue integrity and antigenicity for subsequent rounds [33] [7].
Sequential Multiplexing Protocol:
- Complete first TSA development as described above
- Quench peroxidase activity with 2% sodium azide for 1-2 hours at room temperature
- Wash thoroughly with PBSTx (3× 15 minutes)
- Proceed with next round of peroxidase conjugation and TSA development
- Repeat quenching between subsequent rounds
This approach has been successfully used to simultaneously visualize multiple specialized neoblast populations, revealing their spatially intermingled organization within the planarian parenchyma [18].
Research Reagent Solutions
Table 3: Essential Reagents for Planarian TSA-FISH
| Reagent | Function | Application Notes |
|---|---|---|
| Roche Western Blocking Reagent | Reduces non-specific antibody binding | Critical for anti-DIG and anti-FAM antibodies; dramatically improves signal-to-noise [33] |
| Glucose oxidase | Enzymatic Hâ‚‚Oâ‚‚ generation for TSA | Provides stable peroxide supply; improves operational stability [7] |
| Fluorochromized tyramide | TSA substrate | Direct detection without secondary reagents; reduces background [7] |
| Formamide bleaching solution | Tissue permeabilization and bleaching | Superior to methanol bleaching; significantly enhances signal intensity [33] |
| Sodium azide | Peroxidase quenching | Essential for multiplex FISH; most effective quenching method [33] [7] |
| Triton X-100 | Detergent for enhanced permeability | Critical at 0.3% concentration in wash and blocking buffers [33] |
Workflow Visualization
Figure 1: Optimized workflow for planarian TSA-FISH showing critical steps for achieving high signal-to-noise ratios. Yellow boxes indicate sample preparation stages, green represents hybridization and blocking, blue shows detection steps, and red indicates final analysis. The loopback arrow illustrates the iterative process for multiplex experiments.
Figure 2: Logical troubleshooting framework for common TSA-FISH problems in planarians, connecting specific issues (red) with their underlying causes (yellow) and evidence-based solutions (green).
Applications in Planarian Research
The optimized TSA-FISH protocols described herein have enabled significant advances in planarian research. For example, multiplexed error-robust FISH (MERFISH) combined with TSA principles has revealed that fate-specified neoblasts for different lineages are spatially intermingled throughout the planarian mesenchyme, with neighboring neoblasts frequently making divergent fate choices [18]. This spatial organization challenges traditional models of stem cell niche organization and highlights the importance of high-resolution visualization techniques for understanding regenerative mechanisms.
Similarly, enhanced TSA-FISH methods have facilitated the study of rare cell populations and low-abundance transcripts during planarian regeneration, providing insights into the molecular mechanisms governing polarity restoration and tissue patterning [10]. The ability to reliably detect weakly expressed genes such as signaling ligands and transcription factors has been particularly valuable for reconstructing the regulatory networks that coordinate whole-body regeneration.
Robust TSA-FISH is achievable in planarians through systematic optimization of sample preparation, hybridization conditions, and signal detection. The protocols and troubleshooting guidelines presented here address the unique challenges posed by planarian tissues while leveraging the exceptional sensitivity of tyramide-based amplification. By implementing these evidence-based improvements—particularly the NAFA fixation protocol, formamide bleaching, enhanced blocking conditions, and the FT-GO detection system—researchers can overcome common problems with weak signals or high background. These advances support increasingly sophisticated experimental approaches, including multiplex gene expression analysis and single-cell resolution mapping, that continue to drive discoveries in planarian regeneration and stem cell biology.
Ensuring Specificity and Exploring Next-Generation FISH Technologies
Tyramide Signal Amplification (TSA) dramatically enhances the sensitivity of Fluorescence In Situ Hybridization (FISH), enabling the detection of low-abundance targets. However, this increased sensitivity also raises the risk of non-specific background signal. For researchers studying planarians—model organisms prized for regeneration and stem cell biology—ensuring signal specificity is paramount for accurate interpretation of gene expression patterns. This application note details the essential validation controls required to confirm the specificity of your TSA-FISH signals within the context of planarian research.
Core Principles of TSA-FISH Specificity
TSA, also known as Catalyzed Reporter Deposition (CARD), utilizes the catalytic activity of horseradish peroxidase (HRP) to generate high-density labeling of a target sequence in situ [1]. The process involves a probe binding to the target, which is typically detected by an HRP-labeled conjugate. The HRP enzyme then catalyzes the activation of multiple labeled tyramide molecules, which form covalent bonds with electron-rich amino acid residues (e.g., tyrosine) in proteins immediately surrounding the target site [1] [7]. This deposition results in signal amplification up to 100-fold compared to conventional methods [22].
The very power of this amplification means that any non-specific probe binding or residual enzyme activity can lead to false positive signals. Therefore, a rigorous set of controls is necessary to distinguish true signal from artifact.
Essential Validation Controls for Planarian TSA-FISH
The following controls are critical for validating TSA-FISH experiments. They should be incorporated into every experimental run to ensure the reliability of your data.
Probe Specificity Controls
The foundation of a specific FISH signal is a specific probe.
- No-Probe Control: Omit the specific probe from the hybridization mixture. This controls for non-specific binding of the detection reagents (e.g., anti-digoxigenin-HRP) and the tyramide itself. Any signal indicates a need for better blocking or purification of reagents.
- Competition Assay with Unlabeled Probe: Co-hybridize the labeled probe with a large excess of the same, unlabeled probe. The unlabeled probe should compete for binding sites, significantly reducing or eliminating the specific fluorescent signal. This confirms that signal is due to sequence-specific hybridization [52].
- Target-Specific Mismatch Control: Use a probe with one or several central base pair mismatches to the target sequence. This destabilizing mismatch should abolish specific hybridization, and the absence of signal confirms that the wild-type probe's binding is specific [52].
- BLAST Analysis: In silico analysis of the probe sequence against the planarian genome (e.g., Schmidtea mediterranea) is a mandatory first step to predict potential off-target binding sites and ensure specificity [52] [53].
Enzymatic and Amplification Controls
These controls address the TSA amplification step, a common source of non-specific background.
- No-HRP Control: Omit the HRP-conjugated detection antibody or streptavidin. This verifies that the tyramide reagent is not depositing signal non-specifically without enzymatic activation.
- HRP Inactivation Control for Multiplexing: For multi-round TSA-FISH, the HRP from the previous round must be completely inactivated before initiating the next. Incubation with sodium azide (NaN₃) is an effective method for quenching peroxidase activity without damaging the sample or its antigenicity [7]. A control round where NaN₃ is applied prior to the next tyramide should show no carry-over signal.
- Fluorochromized Tyramide-Glucose Oxidase (FT-GO) System: Consider adopting systems that generate hydrogen peroxide enzymatically in situ (e.g., via glucose oxidase) rather than adding it directly. This provides a more stable and controlled supply of Hâ‚‚Oâ‚‚, improving the signal-to-noise ratio and operational stability of the TSA reaction [7].
Biological Controls
These controls use the biological material itself to assess specificity.
- Sense Probe / Scrambled Sequence Control: Hybridize with a sense strand probe or a probe with a scrambled sequence. This is a standard negative control to assess background from non-specific nucleic acid interactions.
- Knockdown/Knockout Validation: Where possible, use RNA interference (RNAi) to knock down the target mRNA in planarians. A significant reduction in FISH signal in knockdown animals compared to controls provides the strongest possible biological evidence for probe specificity.
- Tissue Specificity Control: Know the expected expression pattern for your target gene. If a gene is known to be expressed only in a specific planarian cell type (e.g., neoblasts in the parenchyma), signal appearing in other tissues (e.g., the pharynx or nerve cords) may indicate off-target binding.
Quantitative Assessment of Controls
The data from your validation controls should be quantified wherever possible to establish objective pass/fail criteria. The following table summarizes key metrics.
Table 1: Quantitative Metrics for TSA-FISH Validation Controls
| Control Type | Measurement | Acceptance Criterion | Tool/Method |
|---|---|---|---|
| No-Probe Control | Mean fluorescence intensity (MFI) in target region | MFI ≤ 2x background MFI | ImageJ / FIJI |
| Mismatch Control | Signal-to-Background Ratio | ≥ 90% signal reduction vs. specific probe | ImageJ / FIJI |
| FT-GO vs. Direct Hâ‚‚Oâ‚‚ [7] | Signal-to-Noise Ratio (SNR) | SNR significantly higher with enzymatic Hâ‚‚Oâ‚‚ generation | Custom analysis or software |
| Post-Knockdown | Number of detectable signal spots | ≥ 70% reduction in spot count | Automated spot detection (e.g., U-FISH [54]) |
Experimental Protocol: Validating a New Probe in Planarians
This is a sample workflow for validating a new oligonucleotide probe targeting a planarian mRNA.
Day 1: Sample Preparation and Hybridization
- Fixation: Fix planarians in 4% formaldehyde in 1x PBS.
- Permeabilization: Treat with proteinase K (concentration and duration optimized for planarian tissue).
- Pre-hybridization: Pre-hybridize in a suitable buffer for 1-2 hours at the hybridization temperature.
- Hybridization: Divide samples and hybridize overnight with:
- Test Sample: Specific, labeled probe (e.g., 10 ng/μL).
- No-Probe Control: Hybridization buffer only.
- Mismatch Control: Labeled mismatch probe (e.g., 10 ng/μL).
Day 2: Signal Amplification and Detection
- Stringency Washes: Perform a series of washes with SSC buffers to remove unbound probe.
- Blocking: Incubate in a blocking buffer (e.g., with 1% BSA) to minimize non-specific antibody binding.
- HRP Conjugate: Incubate with HRP-conjugated anti-label antibody (e.g., anti-DIG-HRP).
- TSA Reaction: Incubate with fluorophore-labeled tyramide substrate in amplification buffer. For the No-HRP control, omit the HRP conjugate in this step.
- Counterstaining and Mounting: Counterstain nuclei with DAPI and mount for imaging.
Day 3: Imaging and Analysis
- Image Acquisition: Acquire images of all control and test samples using identical microscope settings (exposure time, laser power, gain).
- Quantitative Analysis: Measure fluorescence intensities and signal-to-background ratios for each sample as outlined in Table 1.
- AI-Assisted Spot Detection (Optional): For high-throughput or single-molecule analysis, use tools like U-FISH, a deep learning method that provides universal, rapid, and precise detection of FISH spots across diverse image types, improving accuracy and objectivity [54].
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for TSA-FISH Validation
| Reagent / Kit | Function in Validation | Key Features |
|---|---|---|
| TyraMax Dyes [22] | Fluorochromized tyramides for direct detection | Wide selection of bright, photostable dyes from blue to near-IR; stable in buffer for automated workflows. |
| TSA Kits [1] | Provide all critical reagents (tyramide, HRP-conjugates, buffers) for a standardized workflow. | Ensures consistency and includes blocking reagent to help minimize background. |
| Zenon HRP Antibody Labeling Kits [1] | Label primary antibodies directly with HRP, bypassing secondary detection. | Useful for multiplexing and for validating signals when using antibodies from the same host species. |
| Sodium Azide (NaN₃) [7] | Quenches HRP activity between rounds of multiplex TSA-FISH. | Effective inactivation method that is less deleterious to tissue and antigens than heat or low-pH treatments. |
| U-FISH Software [54] | AI-powered spot detection for objective, quantitative analysis of FISH signals. | Enhances image analysis; provides consistent spot detection across diverse datasets without manual parameter tuning. |
| Tigerfish Software [53] | Designs oligonucleotide probes targeting repetitive DNA intervals. | Crucial for checking probe specificity in complex genomes and avoiding off-target binding to repetitive elements. |
Schematic Workflow of TSA-FISH and Key Validation Points
The following diagram illustrates the core TSA-FISH protocol and highlights the critical steps where validation controls must be applied.
Rigorous validation is not an optional extra but a fundamental component of robust TSA-FISH experiments in planarian research. By systematically implementing the probe, enzymatic, and biological controls outlined here, researchers can confidently assign specific gene expression patterns to the remarkable stem cells and tissues of planarians. This rigorous approach ensures that the powerful sensitivity of TSA is matched by the specificity required for meaningful biological discovery.
Fluorescence in situ hybridization (FISH) serves as a cornerstone technique for visualizing nucleic acid sequences within their native cellular and tissue contexts. While conventional FISH provides essential spatial information, its utility is often constrained by limited sensitivity and resolution, particularly when detecting low-abundance targets. This challenge is acutely present in planarian research, where the need to visualize weakly expressed fate-specification genes in neoblasts is paramount for understanding regenerative mechanisms. Tyramide Signal Amplification FISH (TSA-FISH) has emerged as a powerful alternative that significantly enhances these parameters. This application note provides a direct comparison between these two methodologies, contextualized within planarian stem cell research, to guide researchers in selecting and implementing the optimal approach for their experimental goals.
Principles and Mechanisms
Conventional FISH relies on fluorescently labeled nucleic acid probes that hybridize directly to complementary target sequences within fixed samples. The sensitivity of this method is fundamentally limited by the number of fluorophores that can be attached to each probe without compromising hybridization efficiency.
TSA-FISH (also known as Catalyzed Reporter Deposition, CARD) employs an enzymatic amplification step that dramatically increases detection capability. The technique utilizes horseradish peroxidase (HRP)-conjugated reagents (typically secondary antibodies or streptavidin) that bind to the hapten-labeled probe. Upon addition of hydrogen peroxide, the HRP enzyme catalyzes the conversion of fluorescently labeled tyramide substrates into highly reactive radicals that covalently bind to electron-rich tyrosine residues on proteins in the immediate vicinity of the hybridization site. This reaction results in the deposition of numerous fluorophores per target, enabling substantial signal amplification [1] [30].
Table 1: Fundamental Principles of Conventional FISH vs. TSA-FISH
| Characteristic | Conventional FISH | TSA-FISH |
|---|---|---|
| Signal Generation | Direct fluorescence from labeled probes | Enzyme-catalyzed deposition of fluorescent tyramides |
| Amplification Mechanism | None | Horseradish peroxidase (HRP)-mediated activation |
| Key Components | Fluorophore-conjugated probes | Hapten-labeled probes, HRP-conjugates, tyramide substrates |
| Spatial Resolution | Limited by signal intensity, especially for low-abundance targets | Enhanced localization due to confined deposition zone |
| Theoretical Signal Multiplier | 1x (no amplification) | Up to 100x or more [1] |
Molecular Workflows of FISH Techniques
Performance Comparison: Sensitivity and Resolution
Quantitative assessments demonstrate the superior performance characteristics of TSA-FISH across multiple parameters critical for planarian research, particularly when studying weakly expressed fate-specification genes in neoblasts or rare neuronal subtypes.
Table 2: Quantitative Performance Comparison Between Conventional FISH and TSA-FISH
| Performance Metric | Conventional FISH | TSA-FISH | Experimental Evidence |
|---|---|---|---|
| Detection Sensitivity | Limited to moderate abundance targets | Up to 10x higher signal amplification [25] | Flow cytometry detection of low-abundance estrogen receptors [1] |
| Signal-to-Noise Ratio | Variable, lower for rare transcripts | ~2x improvement in SNR [25] | Immunofluorescence in cleared zebrafish brain [25] |
| Spatial Resolution | Diffusion-limited, ~200-500 nm | Excellent, ~50-100 nm due to confined deposition [1] | Subcellular localization in HeLa cells [1] |
| Target Abundance Threshold | Moderate to high copy number | Single-copy genes, low-abundance mRNAs [55] | Detection of single mRNA molecules [55] |
| Multiplexing Capacity | Limited by fluorophore availability | Enhanced via sequential staining with antibody stripping [25] | Multiple rounds of labeling in volumetric imaging [25] |
The enhanced sensitivity of TSA-FISH is particularly valuable in planarian research applications such as:
- Visualizing low-expression transcription factors (e.g.,
pou4-2,soxB1-2) in small neoblast subpopulations [21] - Detecting weakly expressed neuronal genes in sparse mechanosensory neurons [21]
- Resolving expression patterns of fate-specific transcription factors in spatially intermingled neoblast populations [18]
Experimental Protocols
Conventional FISH Protocol for Planarians
Sample Preparation:
- Fix planarians in 4% paraformaldehyde with 1% DMSO for 1 hour at room temperature.
- Dehydrate through ethanol series (50%, 70%, 85%, 95%, 100%) and store at -20°C indefinitely.
- Rehydrate through descending ethanol series and permeabilize with proteinase K (50 μg/mL) for 1 hour.
- Post-fix in 4% paraformaldehyde for 30 minutes [56].
Hybridization and Detection:
- Prehybridize in hybridization buffer for 1-4 hours at 45-57°C.
- Hybridize with fluorophore-labeled probes (15-30 base pairs) overnight at 45-57°C.
- Wash stringently to remove non-specifically bound probes.
- Mount and image immediately using fluorescence microscopy [56] [57].
TSA-FISH Protocol for Enhanced Sensitivity
Sample Preparation and Hybridization:
- Follow conventional FISH protocol steps 1-3 for sample preparation.
- For combined protein-RNA detection (IF/FISH), perform immunofluorescence first, followed by post-fixation.
- Hybridize with hapten-labeled (digoxigenin or biotin) probes overnight at optimal temperature [56] [25].
Signal Amplification:
- Block endogenous peroxidases with 0.3% Hâ‚‚Oâ‚‚ in methanol for 30 minutes.
- Incubate with HRP-conjugated anti-hapten antibody (e.g., anti-DIG-HRP) at 1:500-1:1000 dilution for 1 hour.
- Prepare tyramide working solution: Fluorescent tyramide (1:100) in amplification buffer with 0.0015% Hâ‚‚Oâ‚‚.
- Incubate with tyramide working solution for 1-2 hours, protected from light.
- Wash thoroughly with PBST to stop reaction [1] [25] [30].
Critical Considerations for Planarian Tissue:
- For optimal probe penetration in thick tissues, use alternative permeabilization methods (xylenes/ethanol) instead of proteinase K when preserving protein epitopes [56].
- Adjust hybridization temperature based on probe GC content and length.
- Optimize tyramide incubation time to balance signal intensity with background.
TSA-FISH Experimental Workflow
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagent Solutions for TSA-FISH in Planarian Research
| Reagent/Category | Specific Examples | Function in Protocol | Application Notes |
|---|---|---|---|
| Fixation Agents | 4% Paraformaldehyde with 1% DMSO | Preserves tissue morphology and RNA localization | Critical for planarian ovary and whole-mount samples [56] |
| Permeabilization Methods | Proteinase K (50 μg/mL), Xylenes, RIPA detergent | Enables probe access to intracellular targets | Use proteinase K for RNA only; xylenes/detergents for IF/FISH [56] |
| Probe Types | Digoxigenin-labeled RNA probes, ssDNA SABER concatemers | Target-specific recognition | RNA probes offer higher sensitivity; SABER allows modular design [56] [12] |
| HRP Conjugates | Anti-DIG-HRP, Streptavidin-HRP | Links hapten-labeled probes to amplification system | Zenon antibody labeling kits useful for direct HRP conjugation [1] |
| Tyramide Reagents | Alexa Fluor tyramides, Cyanine tyramides | Signal amplification substrates | Different fluorophores enable multiplexing; low non-specific binding variants preferred [1] [30] |
| Blocking Reagents | Normal goat serum, BSA, TSA blocking reagent | Reduces non-specific background | Essential for minimizing off-target tyramide deposition [1] [25] |
Applications in Planarian Research
The enhanced sensitivity of TSA-FISH enables critical applications in planarian stem cell biology that are challenging with conventional FISH:
Neoblast Fate Specification Studies: TSA-FISH allows visualization of low-abundance fate-specific transcription factors (FSTFs) in individual neoblasts, revealing that fate choices are spatially intermingled rather than clustered by lineage. This has been instrumental in demonstrating that neighboring neoblasts frequently make divergent fate choices for tissues of different locations and functions [18].
Mechanosensory Neuron Regeneration: Research on Smed-pou4-2, a regulator of mechanosensory neurons in Schmidtea mediterranea, benefits from TSA-FISH sensitivity for detecting expression in sparse neuronal subtypes. The technique enables precise mapping of pou4-2 expression in dorsal ciliated stripes and regeneration blastemas [21].
Multiplexed Spatial Transcriptomics: Advanced FISH methodologies like MERFISH (multiplexed error-robust FISH) build upon TSA principles to simultaneously map hundreds of RNA species with single-cell resolution in planarian tissues. These approaches have revealed the spatial organization of distinct cell types and their progenitor populations [18].
TSA-FISH represents a significant advancement over conventional FISH for applications requiring high sensitivity and resolution, particularly in the context of planarian stem cell research. The enzymatic amplification provided by the TSA system enables detection of low-abundance transcripts that are often undetectable with conventional methods, while maintaining excellent spatial resolution. For researchers investigating gene expression patterns in neoblast subpopulations, rare cell types, or weakly expressed regulatory genes, TSA-FISH provides the necessary sensitivity to answer previously intractable questions about planarian regeneration and stem cell biology. The protocol modifications for planarian tissues, particularly regarding permeabilization and fixation, ensure optimal results for these complex whole-mount samples.
In the field of molecular biology, detecting small nucleic acid targets has historically presented a significant challenge due to the limited signal intensity of traditional fluorescence in situ hybridization (FISH). This is particularly relevant in advanced research models like planarians, where understanding the spatial organization of gene expression is crucial for unraveling the mechanisms of regeneration and stem cell (neoblast) biology [18]. Tyramide Signal Amplification (TSA)-FISH overcomes this barrier by enabling the robust mapping of probes less than 1 kilobase (kb) in size, providing researchers with a powerful tool to investigate gene expression with high sensitivity and spatial resolution. This application note details the methodology and quantitative performance of TSA-FISH for small targets, framing it within the context of contemporary planarian research.
Quantitative Performance of TSA-FISH
The enhanced sensitivity of TSA-FISH is quantitatively demonstrated by its successful application to small probes that are undetectable via conventional FISH methods. The following table summarizes key experimental data from a foundational study:
Table 1: Performance of TSA-FISH with Small DNA Probes
| Probe Name | Species | Probe Size (base pairs) | Mapped Chromosomal Location | Detection with Conventional FISH |
|---|---|---|---|---|
| EST883227 (AA243820) | Human | 319 | 7p21 | No signal [58] |
| EST990006 (AA348546) | Human | 608 | 17q25 | No signal [58] |
| cmyc (exon 2) | Mouse | 855 | 15D2 | No signal [58] |
As the data shows, TSA-FISH readily produced signals for all three probes in both interphase and metaphase cells, whereas conventional FISH experiments yielded no detectable signal [58]. This establishes TSA as a critical method for targets where probe size is a limiting factor.
Detailed Experimental Protocol
TSA-FISH Protocol for Small Probes
This protocol is adapted for mapping small PCR-generated probes in planarian tissue sections or whole-mount specimens, integral for studying fate-specification in neoblasts [18].
I. Probe Generation and Labeling
- Probe Synthesis: Generate probe DNA via PCR, using genomic DNA or a plasmid clone as a template.
- Non-radioactive Labeling: During PCR amplification, incorporate hapten-labeled nucleotides directly into the product. Common haptens include:
- Biotin-11-dUTP or Biotin-16-dUTP
- Digoxigenin-11-dUTP
- Purification: Purify the labeled PCR product using a spin column or ethanol precipitation to remove unincorporated nucleotides.
II. Fluorescence In Situ Hybridization
- Sample Preparation: Prepare planarian tissue samples (e.g., cryosections or regenerating fragments) on charged glass slides. Fix with appropriate fixatives (e.g., 4% PFA).
- Permeabilization: Treat samples with a permeabilization agent (e.g., Proteinase K, detergent) to facilitate probe access.
- Denaturation: Co-denature the target DNA in the sample and the labeled probe together in a formamide-containing hybridization buffer at high temperature (e.g., 80°C for 10 minutes).
- Hybridization: Incubate the slides at the optimal hybridization temperature (typically 37°C) in a humidified chamber for a minimum of 4 hours, or overnight.
III. Tyramide Signal Amplification
- Blocking: After post-hybridization washes to remove unbound probe, incubate the slides in a blocking buffer (e.g., containing BSA and serum) to minimize non-specific binding.
- Enzyme Conjugation: Incubate the slides with a horseradish peroxidase (HRP) enzyme conjugated to a reporter molecule that binds to the hapten on your probe. For a biotinylated probe, use Streptavidin-HRP. For a digoxigenin-labeled probe, use an anti-digoxigenin-HRP antibody.
- Tyramide Reaction: Wash the slides to remove unbound HRP conjugate. Then, incubate with the fluorescently labeled tyramide substrate. The HRP enzyme catalyzes the deposition of numerous tyramide molecules at the site of probe hybridization, resulting in a massive signal amplification.
- Counterstaining and Mounting: Counterstain the nuclei with DAPI and mount the slides with an anti-fade mounting medium.
Workflow Visualization
The following diagram illustrates the logical sequence and key components of the TSA-FISH protocol:
The Scientist's Toolkit: Essential Research Reagents
Successful implementation of TSA-FISH for small targets requires a specific set of reagents. The following table details key materials and their functions within the protocol.
Table 2: Essential Reagents for TSA-FISH with Small Probes
| Reagent / Material | Function / Role in the Experiment |
|---|---|
| PCR-Generated Probe | A small (<1 kb), specific DNA sequence used to locate the target gene of interest. Its small size necessitates signal amplification [58]. |
| Biotin- or Digoxigenin-dUTP | Hapten-labeled nucleotide incorporated during PCR to tag the probe, enabling subsequent detection [58]. |
| Streptavidin-HRP or Anti-Digoxigenin-HRP | Enzyme conjugate that binds specifically to the hapten on the probe. HRP is the key enzyme that drives the amplification reaction [58]. |
| Fluorophore-Labeled Tyramide | The substrate for HRP. Upon activation, it precipitates and binds covalently to nearby tyrosine residues, depositing numerous fluorophores at the target site [58]. |
| Hybridization Buffer (with Formamide) | Creates the chemical environment for specific DNA annealing between the probe and the cellular target, while suppressing non-specific binding. |
| Mounting Medium with DAPI | Preserves the sample and provides a fluorescent counterstain for cell nuclei, allowing for the visualization of tissue architecture and subcellular localization. |
Application in Planarian Research
The power of highly sensitive in situ hybridization techniques like TSA-FISH is exemplified in modern planarian studies. For instance, research utilizing multiplexed error-robust FISH (MERFISH) has revealed that neoblasts expressing different fate-specific transcription factors (FSTFs) are spatially intermingled throughout the planarian body [18]. This means that a neoblast committed to becoming a neuron can be found immediately adjacent to one committed to becoming a muscle or epidermal cell, with no apparent local clustering of like fates [18].
The ability to map the expression of multiple, sometimes rare, transcripts simultaneously is fundamental to such discoveries. TSA-FISH provides a robust methodology to validate and extend these findings, especially for genes with low expression levels or when using particularly small probes, thereby deepening our understanding of stem cell fate specification in regeneration [4] [18].
Integrating MERFISH and Other Multiplexing Approaches with TSA
The freshwater planarian Schmidtea mediterranea has emerged as a powerful model organism for studying adult stem cell biology, regeneration, and tissue renewal [18] [59]. Its remarkable regenerative capabilities are driven by neoblasts—pluripotent adult stem cells that produce over 125 distinct cell types [18]. Understanding fate specification in these cells requires precise spatial mapping of gene expression patterns within the complex tissue architecture of the organism. Within this research context, Tyramide Signal Amplification (TSA) serves as a critical enhancement for fluorescence in situ hybridization (FISH), dramatically improving the detection of low-abundance nucleic acid targets [60]. When integrated with advanced multiplexing techniques like Multiplexed Error-Robust FISH (MERFISH), TSA enables researchers to achieve unprecedented spatial resolution in mapping gene expression and chromatin organization, providing new insights into the molecular mechanisms governing planarian regeneration [61] [18].
Tyramide Signal Amplification (TSA): Principles and Workflow
Tyramide Signal Amplification is a catalytic enhancement technique that utilizes the enzymatic activity of horseradish peroxidase (HRP) to deposit numerous fluorescent or chromogenic reporter molecules directly at the site of target recognition [60]. The fundamental workflow involves several key steps, as illustrated in the diagram below:
Diagram 1: TSA Signal Amplification Workflow
The TSA process provides significant advantages for planarian research. It enhances detection sensitivity by 10-5000 times compared to standard FISH protocols, allowing researchers to use substantially lower concentrations of primary antibodies or probes [60]. This is particularly valuable for detecting low-abundance transcripts in planarian stem cells and their progeny. Furthermore, because the activated tyramide derivatives covalently bind to tyrosine residues near the enzyme site, the signal remains localized with excellent spatial resolution, enabling precise subcellular localization of target molecules [60]. The covalent binding also allows for subsequent antibody stripping and multiple rounds of staining, making TSA ideal for multiplexed applications.
Advanced Multiplexed FISH Technologies
Recent advances in multiplexed FISH technologies have revolutionized spatial transcriptomics by enabling simultaneous measurement of hundreds to thousands of genes while preserving their spatial contexts. MERFISH (Multiplexed Error-Robust Fluorescence In Situ Hybridization) represents a particularly powerful approach that combines sequential hybridization with error-robust barcoding schemes to accurately identify numerous RNA species in their native tissue environment [61] [18]. In planarian research, MERFISH has been employed to characterize the spatial organization of distinct cell types and map fate specification across stem cells, successfully labeling all major tissue classes including intestine, epidermis, muscle, protonephridia, and various neuronal cell types [18].
The latest innovations continue to enhance these capabilities. MERFISH+ integrates chemical probe anchoring in protective hydrogels with high-throughput microfluidics and microscopy, supporting robust and repeated hybridization cycles across entire centimeter-scale tissue samples [61]. This enhanced version allows simultaneous quantification of over 1,800 genes while resolving the 3D organization of chromatin loci and their associated epigenomic marks, enabling the reconstruction of comprehensive spatially-resolved multi-omic atlases at subcellular resolution [61].
Integrated Protocols: Combining TSA with Multiplexed FISH
TSA-FISH Protocol for Planarian Tissue Sections
The following protocol outlines the key steps for performing TSA-FISH on planarian tissue sections, based on established methodologies with modifications for planarian applications [62]:
Sample Preparation:
- Fix planarians in appropriate fixative (e.g., 4% formaldehyde) and embed in optimal cutting temperature (OCT) compound for cryosectioning
- Prepare frozen sections (5-10 μm thickness) and transfer to charged slides
- Store slides at -80°C until use
Pre-hybridization Treatments:
- Permeabilization: Treat slides with 10 mM HCl for 10 minutes at 37°C to remove cytoplasm [62]
- Endogenous Peroxidase Quenching: Incubate sections with 1% hydrogen peroxide for 30 minutes at room temperature to quench endogenous peroxidase activity [62]
- RNase Protection: Digest with 100 μg/mL RNase A for 1 hour at 37°C to reduce background [62]
- Blocking: Apply 5× Denhardt's solution for 30 minutes at 37°C to reduce non-specific binding [62]
Hybridization and Detection:
- Probe Preparation: Prepare probe mixture containing 10-30 ng of labeled specific probe in 50% deionized formamide and 10% dextran sulfate in 2× SSC [62]
- Denaturation: Denature probe and chromosomes simultaneously for 5 minutes at 70°C [62]
- Hybridization: Apply probe mixture to slides and hybridize overnight at 37°C in a humidified chamber [62]
- TSA Signal Amplification:
- Counterstaining and Mounting: Counterstain with DAPI (0.5 μg/mL) and mount in antifade mounting medium [62]
MERFISH with TSA Enhancement for Planarian Studies
The integration of TSA with MERFISH provides enhanced sensitivity for detecting low-abundance transcripts in planarian tissues. The following protocol outlines the key modifications to standard MERFISH for planarian applications:
Probe Design and Validation:
- Design MERFISH encoding probes targeting planarian transcripts of interest (e.g., neoblast markers: smedwi-1, vasa-1; differentiation markers: tissue-specific transcription factors)
- Include additional TSA-compatible labels in probe design for signal amplification
- Validate probe specificity using known planarian tissue markers (e.g., intestinal mat, epidermal PRSS12, muscle colF-2) [18]
Multiplexed Hybridization with TSA Enhancement:
- Sample Preparation: Fix and permeabilize planarian tissue sections using optimized protocols for planarian tissues
- Primary Hybridization: Hybridize with MERFISH encoding probe library following standard MERFISH procedures
- Readout Hybridization with TSA: For each round of readout hybridization:
- Apply readout probes conjugated with HRP rather than direct fluorophores
- Develop with appropriate tyramide-fluorophore conjugate (e.g., Alexa Fluor Tyramide) for 2-10 minutes [60]
- Apply stop solution to halt HRP activity
- Image samples using appropriate fluorescence microscopy
- Strip probes and repeat for subsequent hybridization rounds
Image Processing and Analysis:
- Process images using MERFISH decoding algorithms to assign cellular barcodes
- Identify cell boundaries using nuclear stains (DAPI) and membrane markers
- Classify cell types based on pooled expression of multiple cell-type enriched markers [18]
- Spatial analysis of cell types and transcriptional states within the planarian body plan
Comparative Analysis of Multiplexing Approaches
Performance Metrics of FISH Technologies
Table 1: Comparison of FISH Technologies for Planarian Research
| Technology | Multiplexing Capacity | Sensitivity | Spatial Resolution | Key Applications in Planarian Research |
|---|---|---|---|---|
| Traditional FISH | 1-3 targets | Limited for low-abundance targets | Subcellular | Mapping single gene expression patterns |
| TSA-FISH | 1-5 targets with sequential staining | 10-5000x enhancement over conventional FISH [60] | Subcellular (excellent signal localization) | Detection of low-abundance transcripts, small RNAs |
| MERFISH | 100-10,000+ targets [61] [18] | Limited for low-expression genes | Subcellular (single-molecule resolution) | Comprehensive cell type mapping, fate specification analysis |
| MERFISH+ with TSA | 1,000+ targets with enhanced detection [61] | Enhanced for low-abundance targets | Subcellular in 3D across cm-scale tissues [61] | Whole-organ 3D mapping, multi-omic integration |
Research Reagent Solutions for Planarian Spatial Transcriptomics
Table 2: Essential Research Reagents for TSA-Enhanced Multiplexed FISH
| Reagent Category | Specific Examples | Function | Application Notes for Planarian Research |
|---|---|---|---|
| TSA Reagents | Alexa Fluor Tyramide SuperBoost Kits [60] | Signal amplification via HRP-catalyzed deposition | Ideal for low-abundance neoblast markers; use 10-5000x less primary antibody |
| Probe Systems | MERFISH encoding probes, species-specific hybridization probes [62] [18] | Target-specific nucleic acid detection | Design degenerate primers for planarian gene families; verify specificity by sequencing |
| HRP Conjugates | Anti-fluorescein-HRP, Poly-HRP secondary antibodies [62] [60] | Enzyme conjugation for TSA reaction | Poly-HRP provides enhanced sensitivity for critical low-expression targets |
| Fluorophores | Alexa Fluor dyes (488, 546, 555, 568, 594, 647) [60] | Signal detection across multiple channels | Compatible with standard filter sets and planarian autofluorescence considerations |
| Mounting Media | Antifade with DABCO [62] | Preservation of fluorescence signal | Include DAPI for nuclear counterstaining (0.5 μg/mL) |
Data Integration and Analytical Frameworks
Computational Integration of Multimodal Spatial Data
The integration of MERFISH and TSA-FISH data with other omics technologies requires sophisticated computational frameworks. The Spateo-VI generative integration framework has been developed specifically to harmonize spatial transcriptomic and chromatin data to reconstruct 3D spatially-resolved multi-omic atlases [61]. This approach has enabled the characterization of 3D cellular neighborhoods and transcriptional gradients in complex tissues.
For planarian research, population-based genome structure modeling provides another valuable approach for integrating spatial transcriptomics data with other genomic information. This method uses maximum likelihood estimation to generate populations of 3D genome structures statistically consistent with experimental data, allowing characterization of the nuclear microenvironment of genomic regions [63]. The relationship between these integrated analytical approaches is illustrated below:
Diagram 2: Computational Integration of Multimodal Spatial Data
Applications to Planarian Stem Cell and Regeneration Research
The integration of MERFISH with TSA enhancement has revealed fundamental insights into planarian biology. Studies using these technologies have demonstrated that fate specification in neoblasts occurs in a highly intermingled manner, with neighboring stem cells frequently making divergent fate choices for tissues of different locations and functions [18]. This spatial intermingling of fate specification challenges traditional models of regeneration that предполага strict lineage restriction or positional guidance.
Specifically, MERFISH analysis has shown that specialized neoblasts for different lineages (epidermal, muscle, neural, intestinal) are distributed throughout the mesenchymal space without overt spatial organization [18]. These specialized neoblasts are often located closer to non-target tissues than to their target tissues, indicating that fate choice involves stem-cell intrinsic processes rather than direct proximity to destination tissues [18]. This discovery fundamentally changes our understanding of pattern formation in regeneration, suggesting it is driven primarily by the migratory assortment of progenitors from mixed and spatially distributed fate-specified stem cells.
Troubleshooting and Optimization Guidelines
Common Challenges and Solutions in TSA-Enhanced Multiplexed FISH
High Background Signal:
- Cause: Incomplete quenching of endogenous peroxidase activity or excessive tyramide development time
- Solution: Optimize hydrogen peroxide concentration and incubation time; titrate tyramide development time (typically 5-15 minutes for different targets) [62]
- Planarian-specific consideration: Planarian tissues may have higher endogenous peroxidase activity; consider increasing quenching step duration
Insufficient Signal:
- Cause: Inadequate permeabilization, low probe concentration, or insufficient tyramide amplification
- Solution: Optimize permeabilization conditions (HCl concentration and duration); increase probe concentration (10-30 ng per reaction) [62]; use poly-HRP secondary antibodies for enhanced amplification [60]
Tissue Damage or Morphology Loss:
- Cause: Over-digestion during permeabilization or excessive heating during denaturation
- Solution: Reduce permeabilization time; optimize denaturation conditions (typically 5 minutes at 70°C) [62]
- Planarian-specific consideration: Planarian tissues may be more delicate than mammalian tissues; consider reduced permeabilization times
Poor Multiplexing Performance:
- Cause: Incomplete probe stripping between rounds or fluorophore bleaching
- Solution: Optimize stripping conditions (e.g., microwaving in citrate buffer pH6 until boiling for ~2 minutes, then 15 minutes at 20% power) [60]; use antifade mounting media and store slides in dark
Validation and Quality Control Measures
Specificity Controls:
- Include sense probes or scrambled sequence probes as negative controls
- Use known tissue-specific markers as positive controls (e.g., intestinal mat, epidermal PRSS12, muscle colF-2) [18]
- Validate probe specificity by sequencing cloned fragments when using degenerate primers [62]
Quantitation and Reproducibility:
- Establish standardized imaging conditions (exposure times, laser power)
- Include internal reference standards for signal quantification
- Perform technical replicates to assess reproducibility
- Use cell type classification based on pooled expression of multiple markers rather than single genes [18]
Spatial Accuracy Validation:
- Compare spatial patterns with known anatomical landmarks in planarians
- Validate using whole-mount FISH for selected targets [18]
- Correlate with single-cell RNA-seq data when available
The study of planarian biology, particularly the molecular mechanisms governing their remarkable regenerative capabilities, requires sophisticated tools to visualize and quantify complex cellular processes. Within the context of tyramide signal amplification (TSA)-based fluorescence in situ hybridization (FISH) research in planarians, the integration of advanced multiplexed imaging technologies opens new avenues for discovery. Mass cytometry (CyTOF) and Multiplexed Ion Beam Imaging (MIBI) represent next-generation platforms that transcend the limitations of conventional fluorescence microscopy, offering highly multiplexed detection of biomarkers with single-cell resolution. These technologies, when coupled with novel TSA reagents, provide unprecedented capability to elucidate the spatial organization of planarian stem cells (neoblasts) and their differentiation trajectories during regeneration.
The fundamental challenge in planarian research lies in comprehensively mapping the fate specification of neoblasts, which are responsible for producing over 125 distinct adult cell types [18]. Traditional FISH methods, while invaluable, are constrained by spectral overlap when attempting to visualize multiple targets simultaneously. The modifications to whole-mount FISH protocols for planarians, including formamide bleaching, optimized blocking buffers, and copper sulfate quenching of autofluorescence, have significantly improved signal sensitivity [33]. Building upon these foundations, mass cytometry and MIBI introduce a paradigm shift by utilizing metal-labeled tags instead of fluorophores, effectively eliminating spectral overlap and enabling simultaneous measurement of dozens of parameters [64] [65]. This application note details how these technologies, enhanced with novel TSA reagents, can be deployed to address fundamental questions in planarian biology.
Mass cytometry (cytometry by time-of-flight, or CyTOF) and Multiplexed Ion Beam Imaging by Time of Flight (MIBI-TOF) are grounded in mass spectrometry detection, which provides a fundamentally different readout mechanism compared to optical microscopy. In mass cytometry, cells in suspension are stained with antibodies tagged with stable heavy-metal isotopes, nebulized, and then analyzed by inductively coupled plasma time-of-flight mass spectrometry (ICP-TOF-MS) [64]. This approach allows for multiparametric analysis of up to 50 biomarkers simultaneously on single cells, with current theoretical limits extending to 130 parameters [64]. The key advantage is the virtual absence of spectral spillover, as mass spectrometry easily distinguishes isotopes with minimal overlap.
MIBI-TOF operates on a similar principle but is designed for in situ imaging of tissue sections. In MIBI-TOF, tissue sections stained with metal-tagged antibodies are interrogated by a primary ion beam (typically oxygen) that ablates a small spot, generating secondary ions that are analyzed by time-of-flight mass spectrometry [65] [66]. The result is a highly multiplexed image depicting sub-cellular expression and localization for dozens of distinct proteins, preserving spatial context at single-cell resolution. This capability is particularly valuable for planarian research, where the spatial relationships between neoblasts and their differentiated progeny are critical for understanding patterning during regeneration.
Table 1: Comparative Analysis of Multiplexed Imaging Technologies
| Feature | Mass Cytometry (CyTOF) | Multiplexed Ion Beam Imaging (MIBI) | Conventional Fluorescence Microscopy |
|---|---|---|---|
| Detection Principle | ICP-TOF-MS | Secondary Ion MS (SIMS-TOF) | Photon detection with optical filters |
| Multiplexing Capacity | High (theoretically up to 130 parameters) [64] | High (dozens of parameters) [65] | Limited (typically 3-5 colors due to spectral overlap) |
| Spatial Context | No (single-cell suspension) | Yes (preserves tissue architecture) | Yes (preserves tissue architecture) |
| Resolution | Single-cell | Subcellular (~1 μm) [65] | Diffraction-limited (~200 nm) |
| Key Applications | Deep immunophenotyping, signaling analysis | Spatial proteomics, tumor microenvironments | Standard protein and RNA localization |
The recent development of novel TSA reagents that incorporate heavy-metal tags rather than fluorophores bridges the sensitivity of enzymatic amplification with the multiplexing capability of mass spectrometry [67]. These reagents enable the detection of low-abundance biomarkers—such as fate-specific transcription factors in rare neoblast populations—by leveraging the catalytic activity of peroxidase-conjugated antibodies to deposit numerous metal-tagged tyramide molecules at the target site. This signal amplification is critical for detecting weakly expressed genes that regulate planarian stem cell fate decisions.
Experimental Protocols for Planarian Profiling
Sample Preparation and Staining for MIBI
The transition from standard planarian FISH protocols to MIBI-compatible staining requires careful adaptation to preserve tissue morphology and antigen integrity while incorporating metal-labeled reagents.
Protocol: MIBI Sample Preparation and Staining for Planarian Sections
Tissue Fixation and Sectioning:
- Fix planarians in 4% formaldehyde in PBS. The optimization of fixation is critical, as various steps during sample processing profoundly influence antibody-antigen interactions [68].
- Embed fixed samples in optimal cutting temperature (OCT) compound and cryosection at 5-10 μm thickness.
- Mount sections on conductive glass slides (e.g., indium tin oxide-coated slides).
Mucus Removal and Bleaching:
- Treat sections with N-acetyl-cysteine to remove mucus, a planarian-specific challenge that can impede probe penetration [33] [68].
- For pigmented planarians, a short bleaching step in formamide-based peroxide solution dramatically enhances signal intensity by reducing background and improving permeability, a significant improvement over methanol-based bleaching [33].
Antigen Retrieval and Permeabilization:
- Perform heat-induced antigen retrieval (HIAR) using a citrate-based buffer. This step is particularly crucial for regenerating tissues, as it provides a better balance between permeabilization of mature tissues and preservation of fragile regenerating tissue morphology [33].
- Permeabilize with 0.5% Triton X-100 in PBS.
Multiplexed Immunostaining with Metal-Labeled Antibodies:
- Block non-specific binding with a modified blocking buffer containing Roche Western Blocking Reagent (RWBR) and 0.3% Triton X-100, which dramatically reduces background for various anti-hapten antibodies [33].
- Incubate with a cocktail of primary antibodies against target planarian antigens (e.g., neoblast markers like SMEDWI-1, muscle fibers, neuronal markers) [18] [68].
- Incubate with corresponding secondary antibodies conjugated to metal isotopes (e.g., lanthanides) using polymer-based systems to maximize metal loading.
- For low-abundance targets, use metal-tagged TSA reagents. Apply peroxidase-conjugated antibodies specific to haptens on RNA probes or primary antibodies, followed by development with heavy-metal-conjugated tyramide [67].
Post-staining Processing:
- Counterstain with a DNA intercalator (e.g., Ir-191/193) to identify individual nuclei.
- Fix with 2% glutaraldehyde to crosslink antibodies to the tissue and enhance stability during MIBI analysis.
Data Acquisition and Analysis on MIBI-TOF
Instrument Calibration:
- Tune the MIBI-TOF instrument using a uniform metal slide to calibrate mass detectors and optimize primary ion beam focus.
Image Acquisition:
- Define regions of interest (ROIs) within the planarian section, ensuring coverage of key anatomical landmarks.
- Acquire images with a pixel size of 0.5-1 μm and a dwell time of 10-50 ms/pixel, adjusting based on metal abundance to ensure sufficient counts for low-abundance targets.
Data Preprocessing and Cell Segmentation:
- Preprocess the data to correct for dead time and detector noise.
- Use the nuclear signal from the DNA intercalator to perform cell segmentation, identifying individual cell boundaries.
- Extract single-cell expression data for all metal tags within each segmented cell.
Spatial Analysis and Visualization:
- Analyze the spatial distribution of different cell types. For instance, apply this to test the finding that neoblasts with different fate specifications are spatially intermingled rather than clustered [18].
- Calculate cell-cell proximity metrics and perform neighborhood analysis to identify recurrent cellular communities within the planarian mesenchyme.
Diagram: MIBI workflow for planarian tissue analysis, highlighting the metal-TSA step.
The Scientist's Toolkit: Key Reagent Solutions
The successful implementation of mass cytometry and MIBI in planarian research depends on a specialized set of reagents and tools. The table below catalogues essential solutions for designing and executing these multiplexed experiments.
Table 2: Research Reagent Solutions for Advanced Multiplexed Imaging
| Reagent Category | Specific Examples | Function and Application |
|---|---|---|
| Metal-Labeled Antibodies | MaxPar Antibodies [69] | Pre-conjugated antibodies against common targets for use in CyTOF and MIBI. |
| Metal Conjugation Kits | MaxPar X8 Antibody Labeling Kits [69] | Enable custom labeling of antibodies with lanthanide metals for novel targets. |
| TSA Reagents | Novel Metal-Tagged Tyramides [67] | Amplify signals for low-abundance targets (e.g., fate-specific transcription factors). |
| Sample Barcoding | Palladium Barcoding Reagents [69] | Allow multiplexing of multiple samples, improving staining consistency and reducing reagent consumption. |
| Cell Identification | DNA Intercalators (Ir-191/193) | Distinguish single cells in both suspension (CyTOF) and tissue (MIBI) analysis. |
| Panel Design Tools | Online Browser-Based Panel Design [69] | Assist in building CyTOF panels from large antibody catalogs without spectral overlap concerns. |
| Planarian-Specific Antibodies | Anti-SMEDWI-1, Anti-Neoblast Markers [68] | Key reagents specifically generated for identifying and characterizing planarian stem cells. |
Data Analysis and Interpretation in Planarian Systems
The raw data generated from CyTOF and MIBI experiments require specialized computational approaches to extract biologically meaningful insights into planarian stem cell biology. The high-dimensional, single-cell data from CyTOF can be analyzed using dimensionality reduction techniques such as t-Distributed Stochastic Neighbor Embedding (t-SNE) or Uniform Manifold Approximation and Projection (UMAP) to visualize the continuum of neoblast states and identify putative transitional populations during fate specification. For instance, this approach could reveal the relationship between naive neoblasts and those expressing fate-specific transcription factors for epidermis, muscle, or intestine [18] [66].
The analysis of MIBI data adds the critical dimension of spatial context. After cell segmentation based on nuclear and membrane markers, the expression matrix can be used to classify cells into known planarian cell types. Spatial analysis can then test fundamental hypotheses about neoblast organization, such as whether specialized neoblasts of the same fate cluster together or are distributed in an intermingled fashion [18]. Quantitative spatial metrics—including nearest-neighbor distances, Ripley's K-function, and cell-cell proximity scores—can be calculated to statistically compare the distributions of different specialized neoblast classes relative to each other and to their target tissues.
Diagram: Data analysis workflow for multiplexed imaging, from raw data to biological interpretation.
The integration of these high-dimensional data types with planarian functional genomics is particularly powerful. For example, the spatial expression patterns of key regulatory genes identified through FISH or single-cell RNA sequencing can be validated and extended through MIBI using antibodies against the corresponding proteins. This multi-modal approach provides a more comprehensive understanding of how gene expression networks are translated into cellular organization and tissue patterning during planarian regeneration.
Future Outlook and Concluding Remarks
The convergence of planarian biology with advanced multiplexed technologies represents a significant frontier in regenerative biology. The ongoing development of reagents for mass cytometry continues to expand the scope of information that can be obtained, particularly through new small molecule reagents that enable monitoring of active biochemistry at the cellular level [64]. For planarian research, this could translate to tools for simultaneously measuring cell signaling activity, metabolic state, and lineage markers in the same cell populations, providing unprecedented insight into the regulation of neoblast behavior.
The automation of data analysis through machine learning approaches will be crucial for handling the vast datasets generated by these technologies [66]. As these tools become more accessible, the synergy between multiplexed imaging and automated image analysis promises to drive a new era in fundamental research and comparative pathology. For the planarian research community specifically, the adoption of mass cytometry and MIBI, enhanced with novel TSA reagents, offers a path to systematically decode the complex logic of fate specification in adult stem cells, with potential implications for understanding the fundamental principles of tissue repair and regeneration across the animal kingdom.
Conclusion
Tyramide Signal Amplification FISH represents a transformative methodology for planarian research, providing the necessary sensitivity to visualize low-abundance mRNAs that are crucial for understanding stem cell dynamics and regeneration. The optimized protocols discussed—from formamide bleaching and copper sulfate quenching to advanced blocking strategies—directly address the unique challenges posed by planarian tissues. As the field advances, the integration of TSA with emerging multiplexing technologies like MERFISH and mass cytometry promises to unlock unprecedented capabilities for mapping complex gene expression networks in a spatial context. The continued refinement of these techniques will not only deepen our understanding of planarian biology but also provide broadly applicable insights into stem cell regulation, tissue homeostasis, and regenerative medicine, with potential implications for clinical diagnostics and therapeutic development.
References
- 1. TSA and Other Peroxidase-Based Signal Amplification ... [https://www.thermofisher.com/us/en/home/references/molecular-probes-the-handbook/ultrasensitive-detection-technology/tyramide-signal-amplification-technology.html]
- 2. Tyramide Signal Amplification (TSA) Methodology [https://www.news-medical.net/life-sciences/Tyramide-Signal-Amplification-(TSA)-Methodology.aspx]
- 3. Tyramide signal amplification-based detection system [https://www.sciencedirect.com/science/article/abs/pii/S1386142525012053]
- 4. Stochastic cell-intrinsic stem cell decisions control colony ... [https://elifesciences.org/articles/100885]
- 5. Pluripotent Stem Cell Plasticity is Sculpted by a Slit ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12080947/]
- 6. Research progress and perspectives on the application of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11804208/]
- 7. Fluorochromized tyramide-glucose oxidase as a multiplex ... [https://www.nature.com/articles/s41598-022-19085-9]
- 8. Tyramide Signal Amplification (TSA) | Fluorescence Imaging [https://www.tocris.com/product-type/tyramide-signaling-amplification-tsa]
- 9. Nanozyme-enhanced tyramine signal amplification probe ... [https://www.sciencedirect.com/science/article/abs/pii/S1385894724095846]
- 10. A comparative analysis of planarian regeneration ... [https://elifesciences.org/reviewed-preprints/105707]
- 11. Systemic identification and characterization of the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12518275/]
- 12. One probe fits all: a highly customizable modular RNA in situ ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12148028/]
- 13. In situ hybridization protocol for enhanced detection ... [https://experts.illinois.edu/en/publications/in-situ-hybridization-protocol-for-enhanced-detection-of-gene-exp]
- 14. A comparative analysis of planarian genomes reveals ... [https://www.nature.com/articles/s41467-024-52380-9]
- 15. Allometry of cell types in planarians by single-cell ... - PMC - NIH [https://pmc.ncbi.nlm.nih.gov/articles/PMC12057665/]
- 16. Smed-pou4-2 regulates mechanosensory neuron ... [https://elifesciences.org/reviewed-preprints/107718]
- 17. Planarian RNAi knockdown: feeding once might just be ... [https://www.frontiersin.org/journals/neuroscience/articles/10.3389/fnins.2025.1546196/full]
- 18. Fate specification is spatially intermingled across planarian ... [https://www.nature.com/articles/s41467-023-43267-2]
- 19. CMN Weekly (23 May 2025) - Your ... [https://crisprmedicinenews.com/news/cmn-weekly-23-may-2025-your-weekly-crispr-medicine-news/]
- 20. Sensitive Multiplexed Fluorescent In Situ Hybridization ... [https://pubmed.ncbi.nlm.nih.gov/31552667/]
- 21. Smed-pou4-2 regulates mechanosensory neuron ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12626421/]
- 22. Tyramide Signal Amplification [https://biotium.com/technology/primary-secondary-antibody-conjugates/tyramide-signal-amplification/]
- 23. TSA and Other Peroxidase-Based Signal Amplification ... [https://www.thermofisher.com/uk/en/home/references/molecular-probes-the-handbook/ultrasensitive-detection-technology/tyramide-signal-amplification-technology.html]
- 24. Fluorescence In Situ Hybridization using TSA [https://www.thermofisher.com/us/en/home/references/protocols/cell-and-tissue-analysis/microscopy-protocol/fluorescent-in-situ-hybridization-using-tsa.html]
- 25. TSA-PACT: a method for tissue clearing ... - Cell & Bioscience [https://cellandbioscience.biomedcentral.com/articles/10.1186/s13578-023-01043-1]
- 26. Signal Amplification for Fluorescent Staining of Single ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12508258/]
- 27. Evolutionary dynamics of whole-body regeneration across ... [https://www.nature.com/articles/s41559-023-02221-7]
- 28. In situ hybridization protocol for enhanced detection of gene expression... [https://bmcdevbiol.biomedcentral.com/articles/10.1186/1471-213X-13-8]
- 29. In situ hybridization protocol for enhanced detection of gene expression... [https://pubmed.ncbi.nlm.nih.gov/23497040/]
- 30. (TSA) Applications Tyramide Signal Amplification [https://www.news-medical.net/life-sciences/Tyramide-Signal-Amplification-(TSA)-Applications.aspx]
- 31. Reduced adult stem cell fate specification led to eye reduction ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11696554/]
- 32. A comparative analysis of planarian regeneration ... [https://elifesciences.org/reviewed-preprints/105707/reviews]
- 33. In situ hybridization protocol for enhanced detection of gene ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3610298/]
- 34. A powerful and versatile new fixation protocol for ... [https://bmcbiol.biomedcentral.com/articles/10.1186/s12915-024-02052-3]
- 35. Planarian microtubules form a network within muscle and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12579930/]
- 36. Ultrafast distant wound response is essential for whole ... [https://www.sciencedirect.com/science/article/pii/S0092867423006955]
- 37. 3D reconstruction of neuronal allometry and ... [https://elifesciences.org/articles/101103]
- 38. A technical review and guide to RNA fluorescence in situ ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7085896/]
- 39. egal-1 and microtubules promote regeneration polarity in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12579938/]
- 40. Tyramide signal amplification for a highly sensitive ... [https://pubs.rsc.org/en/content/articlelanding/2025/an/d5an00078e]
- 41. NR1I3 modulates Wnt signaling to promote anterior–posterior ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12312329/]
- 42. A three dimensional immunolabeling method with ... [https://www.nature.com/articles/s42003-025-08317-z]
- 43. Multicolor fluorescent in situ hybridization to define abutting ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3088888/]
- 44. Immersionâ€Based Clearing and Autofluorescence ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12589860/]
- 45. Automated Image Analysis for Quantitative Fluorescence In ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC1892889/]
- 46. TrueProbes: Quantitative Single-Molecule RNA-FISH ... [https://elifesciences.org/reviewed-preprints/108599]
- 47. Key Trends Shaping Therapeutic Antibodies in 2025 and ... [https://www.diatheva.com/key-trends-shaping-therapeutic-antibodies-in-2025-and-beyond/]
- 48. Bispecific immunotherapy based on antibodies, T-cell ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2025.1679092/full]
- 49. Advances in the pharmaceutical development of antibody ... [https://www.sciencedirect.com/science/article/pii/S0928098725002908]
- 50. A powerful and versatile new fixation protocol for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11533299/]
- 51. An annotated fluorescence image dataset for training ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7419523/]
- 52. Specificity Re-evaluation of Oligonucleotide Probes for the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5446981/]
- 53. Tigerfish designs oligonucleotide-based in situ ... [https://www.nature.com/articles/s41467-024-45385-x]
- 54. U-FISH: a fluorescent spot detector for imaging-based spatial ... [https://genomebiology.biomedcentral.com/articles/10.1186/s13059-025-03736-x]
- 55. Fluorescence in Situ Hybridization - an overview [https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/fluorescence-in-situ-hybridization]
- 56. Optimized RNA ISH, RNA FISH and protein-RNA double ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4126239/]
- 57. Fluorescence in situ hybridization (FISH) in food pathogen ... [https://medcraveonline.com/IJMBOA/fluorescence-in-situ-hybridization-fish-in-food-pathogen-detection.html]
- 58. FISH applied to mapping PCR-labeled probes less than 1 kb ... [https://pubmed.ncbi.nlm.nih.gov/10489619/]
- 59. Single-cell transcriptomics in planaria: new tools allow new ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9704525/]
- 60. Tyramide Signal Amplification with SuperBoost Kits [https://www.thermofisher.com/us/en/home/life-science/cell-analysis/cellular-imaging/immunofluorescence/tyramide-signal-amplification-tsa.html]
- 61. MERFISH+, a large-scale, multi-omics spatial technology ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12637541/]
- 62. FISH with tyramide signal amplification (TSA-FISH) [https://bio-protocol.org/exchange/minidetail?id=10064090&type=30]
- 63. Evaluating the role of the nuclear microenvironment in gene ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10442234/]
- 64. Reagents for Mass Cytometry [https://pubmed.ncbi.nlm.nih.gov/36696538/]
- 65. High-Dimensional Tissue Profiling by Multiplexed Ion ... [https://pubmed.ncbi.nlm.nih.gov/34766270/]
- 66. Multiplexed Ion Beam Imaging: Insights into Pathobiology [https://pubmed.ncbi.nlm.nih.gov/34752710/]
- 67. New tyramide signal amplification (TSA) reagents ... - Stanford [https://techfinder.stanford.edu/technology/new-tyramide-signal-amplification-tsa-reagents-and-methods-expand-applications]
- 68. Generation of cell type-specific monoclonal antibodies for the ... [https://bmcdevbiol.biomedcentral.com/articles/10.1186/s12861-014-0045-6]
- 69. Mass Cytometry Kits, Reagents And Accessories [https://www.standardbio.com/products/kits-reagents-and-accessories/cytometry]