Revolutionizing Zebrafish Research: A Comprehensive Guide to ZEG Early Selection Protocol for Enhanced CRISPR Efficiency
This article provides a comprehensive examination of the Zebrafish Embryo Genotyper (ZEG) protocol, an automated non-invasive system for rapid genotyping of live zebrafish embryos.
Revolutionizing Zebrafish Research: A Comprehensive Guide to ZEG Early Selection Protocol for Enhanced CRISPR Efficiency
Abstract
This article provides a comprehensive examination of the Zebrafish Embryo Genotyper (ZEG) protocol, an automated non-invasive system for rapid genotyping of live zebrafish embryos. Targeting researchers, scientists, and drug development professionals, we explore ZEG's foundational technology that enables genetic material extraction with >90% survival rates, detailed methodological applications in CRISPR-Cas9 workflows where it demonstrates 17-fold increases in somatic editing efficiency, practical troubleshooting for DNA quality optimization, and rigorous validation through morphological, cellular, and behavioral analyses. The protocol represents a significant advancement over traditional fin clipping, offering substantial ethical and resource savings while accelerating genetic research timelines.
Understanding ZEG Technology: Principles and Advantages Over Traditional Genotyping
The zebrafish (Danio rerio) has emerged as a premier model organism in biomedical research due to its genetic tractability, high fecundity, and physiological similarities to humans. With 84% of human disease-associated genes having zebrafish orthologues, this model system plays a crucial role in functional genomics and disease modeling [1]. However, traditional genotyping methods have created a significant bottleneck in research workflows, impeding the full exploitation of this versatile model organism. The conventional approach of sacrificial genotyping or fin clipping in later developmental stages presents substantial ethical, practical, and scientific challenges that limit research efficiency and compromise animal welfare.
The fundamental issue stems from the standard practice of raising zebrafish to juvenile or adult stages (typically exceeding two months) before performing fin clipping for genotyping [1]. This approach generates substantial numbers of "surplus" animals that do not possess the desired genotypes but must be maintained until genotyping is complete [2]. From an ethical standpoint, this practice conflicts with the 3Rs principles (Replacement, Reduction, and Refinement) that govern humane animal research [2] [3]. The logistical consequences are equally significant – substantial resources are expended on raising, housing, and caring for animals that will ultimately not be used in experiments, creating inefficiencies that delay research advancement and increase costs.
Limitations of Traditional Genotyping Methods
Scientific and Technical Constraints
Traditional genotyping methods impose significant limitations on experimental design and scientific rigor. The inability to link genotype to phenotype at early developmental stages is particularly problematic when studying embryonic lethal mutations or early-onset phenotypes [1]. When researchers can only genotype after phenotypes manifest, they lose the ability to track the progression of these phenotypes from their initiation. Furthermore, mosaic founders (F0) generated through CRISPR-Cas9 editing exhibit varying degrees of editing across different cells, making early genotyping essential for understanding genotype-phenotype relationships in these critical animals [4].
The technical limitations of conventional fin clipping are equally constraining. The process requires anaesthesia or euthanisation of the animal, particularly challenging for small fish [3]. While catch-and-release fin clipping presents a non-lethal alternative, the released animals face risks of infection, diminished growth, and reduced survival due to the missing fin tissue [3]. Additionally, the DNA yield and quality from traditional methods can be variable, potentially affecting downstream applications like whole genome sequencing [3].
Welfare Considerations
The welfare implications of traditional genotyping methods extend beyond the immediate procedural stress. The current practice generates substantial numbers of animals that are raised to later developmental stages despite not being needed for research, creating an ethical dilemma regarding their ultimate disposition [2]. Some countries' ethics legislation insists these "surplus" animals should be maintained and left to die of natural causes, creating additional welfare challenges and resource burdens for research facilities [2].
Table 1: Comparison of Traditional Zebrafish Genotyping Methods
| Method | Developmental Stage | Key Limitations | Welfare Impact | Data Quality Concerns |
|---|---|---|---|---|
| Sacrificial Genotyping | Any stage, typically larval to adult | Complete loss of animal; prevents longitudinal studies | High - animal is euthanized | Precludes tracking individual animal development over time |
| Adult Fin Clipping | >2 months | Delayed genotyping; requires anesthesia | Moderate - invasive procedure with recovery required | Cannot link early phenotypes to genotype |
| Larval Fin Clipping | 3-5 dpf | Technically challenging; variable DNA yield | Low-moderate - tissue regeneration occurs | PCR efficiency can be variable [1] |
| Post-mortem Genotyping | After phenotype observation | Cannot preselect animals for experiments | High - animal cannot be recovered for breeding | Genotype-phenotype links are correlative, not predictive |
Advanced Genotyping Solutions
The Zebrafish Embryo Genotyper (ZEG) Device
The Zebrafish Embryo Genotyper (ZEG) represents a transformative approach to overcoming the genotyping bottleneck. This automated microfluidic system enables extraction of genetic material from live zebrafish embryos as early as 72 hours post-fertilization (hpf) in a manner that does not destroy the embryo [5] [4]. The technology utilizes microfluidic harmonic oscillation of an animal on an abrasive surface, which generates sufficient genetic material for analysis from 24 individual embryos in just 10 minutes with minimal handling [4].
The key advantage of the ZEG device is its non-destructive nature, allowing researchers to identify genotypes early in development and then raise only the desired embryos to adulthood. This approach has demonstrated remarkable success in improving the efficiency of CRISPR-Cas9 knock-in experiments, with studies reporting an almost 17-fold increase in somatic editing efficiency when combining ZEG with early genotyping and next-generation sequencing [5]. The benefit was particularly evident for alleles with lower somatic editing efficiencies, enabling researchers to selectively raise embryos with the highest rates of correctly edited cells.
Alternative Non-Invasive Methods
Beyond the ZEG device, researchers have developed additional non-invasive genotyping methods that address the limitations of traditional approaches. A shaking-based assay enables genotyping of live, early developmental stage zebrafish embryos by using low-frequency shaking to induce the detachment of a limited number of cells [2]. These cells are then analyzed using PCR-based genotyping approaches, providing reliable medium-throughput method for identification of genotypes at ethically acceptable developmental stages.
Another innovative approach involves mucus swabbing as a non-lethal alternative to fin clipping [3]. This method involves stroking a specialized swab along the fish's skin or gills to collect mucus containing DNA. While skin swabs treated with Proteinase K during extraction can match fin clips in whole genome sequencing performance, the DNA yield is generally lower than from fin clips [3]. This approach represents a viable non-invasive DNA sampling alternative, particularly for studies where preserving animal integrity is paramount.
Table 2: Advanced Non-Invasive Genotyping Methods for Zebrafish Research
| Method | Principle | Optimal Stage | Throughput | Key Advantages |
|---|---|---|---|---|
| ZEG Device | Microfluidic harmonic oscillation with abrasive surface | 72 hpf | High (24 embryos in 10 min) | Non-destructive; enables longitudinal studies; high PCR efficiency |
| Shaking-Based Assay | Low-frequency shaking induces cell detachment | Early developmental stages | Medium | Non-invasive; ethically acceptable developmental stage |
| Mucus Swabbing | DNA collection from skin or gill mucus | Juvenile to adult | Medium | Completely non-destructive; no tissue damage |
| Microscopic Tail Biopsy | Minute tissue collection with micro-scalpel | 3-5 dpf | Medium-high | High regeneration potential; lower mortality than standard fin clip |
Experimental Protocols and Workflows
ZEG Protocol for Early Embryo Genotyping
The ZEG protocol represents a standardized approach for early embryo genotyping that aligns with 3R principles. The following workflow details the critical steps for successful implementation:
Preparation: Position 72 hpf zebrafish larvae in the ZEG device. Ensure the microfluidic chambers are clean and the abrasive surfaces are in optimal condition for cell dissociation [4].
DNA Extraction:
- Load individual embryos into the ZEG chambers
- Activate the harmonic oscillation system for precisely calibrated intervals
- Collect the extraction medium containing genomic DNA
- The typical DNA concentration range from this process is 7-81 ng/μL, with an average of 34.0 ng/μL [4]
DNA Amplification and Analysis:
- Use 1.5 μL of the DNA supernatant per PCR reaction
- For optimal results with potentially fragmented DNA, employ Restorase DNA Polymerase, which significantly increases PCR amplification of damaged DNA templates [4]
- Target amplification of fragments smaller than 260 bp for most consistent results across all developmental stages [4]
Embryo Recovery and Rearing:
- Return successfully genotyped embryos to fresh embryo media
- Monitor for normal development and absence of morphological abnormalities
- Studies confirm that embryos subjected to ZEG extraction show no obvious behavioral or phenotypic abnormalities compared to control embryos [4]
Larval Fin Clipping Protocol
For laboratories without access to the ZEG device, larval fin clipping at 3-5 days post-fertilization provides a viable alternative with high survival rates and rapid tissue regeneration:
Anesthesia and Preparation:
- Anesthetize 3-5 dpf zebrafish larvae in ˜1.5 mM Tricaine in 1x E3 embryo media
- Prepare a dissection surface by taping a 9 cm Petri dish lid with autoclave tape across its interior surface [1]
Fin Clipping Procedure:
- Using a micro scalpel, section the caudal fin distal to the limit of blood circulation
- Apply steady downward pressure within the pigment gap site of the caudal fin to avoid damaging the notochord [1]
- Transfer the sectioned fin tissue to a small piece of filter paper for visualization
DNA Extraction and Genotyping:
- Transfer the filter paper with fin tissue to a 96-well PCR plate containing 25 μL of 50 mM NaOH solution
- Heat samples at 95°C for 5 minutes followed by cooling to 4°C for 10 minutes for tissue lysis
- Add 6 μL of 500 mM Tris-HCl, pH 8.0 to each sample to neutralize the solution [1]
- Use 1.5 μL of the DNA supernatant per PCR reaction
Larval Recovery:
- Return larvae to fresh embryo media in a 96-well tissue culture plate
- Observe fin regeneration over the following 48 hours
- Resume normal feeding and maintenance of successfully genotyped larvae
The following workflow diagram illustrates the key decision points in selecting an appropriate genotyping method:
Genotyping Method Selection Workflow
Research Reagent Solutions
Successful implementation of advanced genotyping protocols requires specific reagents and materials optimized for each method. The following table details essential research reagent solutions for zebrafish genotyping:
Table 3: Essential Research Reagents for Advanced Zebrafish Genotyping Methods
| Reagent/Material | Application | Function | Specifications | Alternative Options |
|---|---|---|---|---|
| ZEG Device | Early non-invasive genotyping | Microfluidic harmonic oscillation for cell dissociation | Processes 24 embryos in 10 minutes | Not easily replaceable; core facility resource |
| Restorase DNA Polymerase | PCR amplification from minimal DNA | Enhanced amplification of damaged/degraded DNA | Critical for ZEG-extracted DNA | Standard Taq polymerase (less efficient) |
| Chelating Resin | DNA extraction from fin clips | Tissue lysis and DNA binding | 5% styrene-divinylbenzene copolymer | NaOH-based extraction [1] |
| Proteinase K | DNA extraction from swabs | Protein degradation for improved DNA yield | Essential for skin swab DNA concentration | Alternative proteases (less effective) [3] |
| Micro-scalpels | Larval fin clipping | Precise tissue sectioning | Fine tip for microscopic procedures | Conventional scalpels (less precise) |
| Copan 4N6FLOQSwabs | Mucus swabbing | DNA collection from skin/gills | Regular tip size for genetic sampling | Standard cotton swabs (lower DNA yield) [3] |
| ATL Buffer | Sample storage | Tissue preservation before DNA extraction | Maintains DNA integrity for swabs | Direct freezing (less effective) [3] |
Integration with CRISPR-Cas9 Gene Editing
The combination of early genotyping methods with CRISPR-Cas9 gene editing has revolutionized the generation of zebrafish disease models. Research demonstrates that optimal CRISPR-Cas9 components significantly improve knock-in efficiency, with Cas9 protein outperforming mRNA and non-target asymmetric PAM-distal (NAD) ssODN conformations achieving the highest editing rates [5]. At two different cacna1c gene loci, the NAD conformation with Cas9 protein significantly outperformed all other conditions, with average somatic editing efficiencies of 5.14% ± 0.71 and 2.83% ± 0.75 respectively [5].
The integration of early genotyping enables researchers to selectively raise embryos with the highest rates of correctly edited cells, particularly valuable for studying human pathogenic variants such as those associated with Long QT syndrome (c.2570C>G or p.(Pro857Arg)) and Brugada syndrome (c.989C>T or p.(Thr330Met)) [5]. The ZEG selection procedure has demonstrated successful germline transmission events in pre-selected embryos, confirming the practical utility of this approach for establishing stable genetic lines [5].
The following diagram illustrates the integrated workflow combining CRISPR-Cas9 gene editing with early genotyping selection:
Integrated CRISPR-Cas9 with Early Genotyping Workflow
The genotyping bottleneck in zebrafish research represents a significant challenge that has impeded research progress and compromised animal welfare. Traditional methods relying on sacrificial approaches or fin clipping in later developmental stages create ethical dilemmas and practical limitations that slow research advancement. The development and implementation of early non-invasive genotyping techniques, particularly the Zebrafish Embryo Genotyper (ZEG) device, provide transformative solutions that align with 3R principles while enhancing research efficiency.
The integration of these advanced genotyping methods with state-of-the-art gene editing technologies creates powerful workflows for generating and studying zebrafish disease models. By enabling early selection of efficiently edited embryos, researchers can achieve dramatic improvements in somatic editing efficiency – up to 17-fold increases – while reducing the number of surplus animals [5]. This approach is particularly valuable for modeling human disease variants and establishing stable genetic lines with confirmed germline transmission.
As zebrafish continue to play an increasingly important role in biomedical research, adopting these advanced genotyping methodologies will be essential for maximizing research output while maintaining the highest standards of animal welfare. The future of zebrafish research lies in the widespread implementation of these refined approaches, enabling more efficient, ethical, and scientifically rigorous studies that fully leverage the unique advantages of this valuable model organism.
The Zebrafish Embryo Genotyper (ZEG) represents a significant technological advancement in biomedical research, addressing a critical bottleneck in large-scale genetic screens using zebrafish. This automated system is designed to rapidly obtain genetic material from live zebrafish embryos and larvae for genotyping, while maintaining a high survival rate exceeding 90% [6] [7]. The core technology leverages microfluidic harmonic oscillation on a roughened glass surface to achieve this, enabling researchers to perform non-lethal genotyping at stages as early as 24–72 hours post-fertilization (hpf) [6] [8]. The development of ZEG is particularly transformative for research involving CRISPR-Cas9 mutagenesis and knock-in (KI) experiments, as it facilitates the early selection of genetically edited embryos, thereby dramatically improving the efficiency of generating desired mutant lines [5]. By integrating ZEG-based early genotyping with next-generation sequencing (NGS), researchers have achieved an almost 17-fold increase in somatic editing efficiency in CRISPR KI experiments, underscoring its vital role in modern genetic research [5].
Core Technological Principle & Mechanism
The operational principle of the ZEG device centers on using controlled mechanical vibration to gently dislodge cells from live embryos. The system consists of two main components: a disposable, roughened-glass "chip" that holds the embryos, and a base unit that generates precise oscillatory motion [6] [9].
- Microfluidic Harmonic Oscillation: The base unit houses a platform, suspended on springs, which is agitated by a small vibration motor. This motor generates harmonic oscillations at a specific frequency and voltage (typically 1.4 volts). This motion is transferred to the chip, causing the embryos to oscillate rapidly within their individual chambers [6].
- Surface Interaction for Cellular Extraction: Each chamber of the disposable chip features a circular roughened glass area, created via laser etching or a raster engraving technique [9]. As the embryos are harmonically oscillated over this abrasive surface, the gentle friction leads to the detachment of a small number of cells from the embryo's surface [6] [2]. This process is designed to be minimally invasive, ensuring the embryo remains viable for subsequent development and experimentation.
- Genetic Material Collection: The dislodged cells and DNA remain suspended in the surrounding embryo medium (E3). After a brief period of oscillation (typically 7 to 10 minutes for 24 embryos), a standard pipette is used to collect approximately 10 µL of fluid from each chamber. This fluid contains sufficient genetic material for downstream molecular analyses like PCR, sequencing, or high-resolution melt analysis (HRMA) [6].
The entire workflow, from loading to collection, is visualized in the following diagram:
Key Performance Data & Optimization
Extensive testing has been conducted to quantify the performance and optimize the operation of the ZEG device. The data below summarizes its key efficacy and safety metrics.
Table 1: Key Performance Metrics of the ZEG Device
| Performance Parameter | Reported Outcome | Experimental Context & Notes |
|---|---|---|
| Genotyping Sensitivity | > 90% [6] | Achieved with High-Resolution Melt Analysis (HRMA). Optimized protocol later achieved >80% sensitivity with gel electrophoresis [9]. |
| Embryo Survival Rate | > 90% [6] [7] | Observed at 72 hours post-fertilization (hpf) after ZEG processing. |
| Throughput | 24 embryos simultaneously in < 10 minutes [6] | Device also constructed in 16 and 48 chamber formats [6]. |
| Cell & DNA Yield | Sufficient for PCR, HRMA, and sequencing [6] | 5 µL of the 10 µL collected fluid used in an 11 µL PCR reaction [6]. |
| Impact on Development/Morphology | No apparent effects observed [6] | Gross morphology examination post-processing [6]. |
| Impact on Motor Behavior | No significant differences detected [6] | Analysis of spontaneous swimming, light-evoked responses, and tap startle at 7 dpf [6]. |
| Cellular/Molecular Stress | Low-level acute stress response, no long-lasting effects [10] [11] | Gene expression and tissue integrity analysis post-ZEG protocol [10]. |
Optimization efforts identified that the surface texture profile of the glass chip was the most critical factor influencing performance consistency. A shift to a raster engraving fabrication technique not only improved the consistency of PCR amplification and survivability but also reduced chip manufacturing time by 90% [9].
Detailed Experimental Protocol
This section provides a step-by-step protocol for using the ZEG device, from preparation to genotyping.
Materials and Reagents
Table 2: The Scientist's Toolkit: Essential Reagents and Materials for ZEG
| Item | Function / Application | Specifications / Notes |
|---|---|---|
| ZEG Base Unit & Chips | Core device for cellular extraction. | Chips with roughened glass chambers [6]. |
| Zebrafish Embryos/Larvae | Biological subject for genotyping. | Typically 24–72 hpf [6]. |
| E3 Embryo Medium | Standard medium for raising and handling embryos. | Used to suspend embryos during loading and processing [6]. |
| Standard Pipette & Tips | For manual loading and unloading of embryos and fluid collection. | Tips may be cut off for easier handling of embryos [6]. |
| PCR Master Mix | For amplification of extracted DNA. | e.g., LightScanner Master Mix or Power Up SYBR Green Master Mix [6]. |
| Primers | For locus-specific amplification during genotyping. | Designed for target allele (e.g., zc90 allele primers) [6]. |
| Trypan Blue & DAPI | (Optional) For cell counting and viability assessment. | Used with a hemocytometer post-extraction [6]. |
Step-by-Step Workflow
- Preparation: Raise zebrafish embryos at 28.5°C in E3 embryo medium and stage them by time and morphology [6]. Ensure the ZEG base unit and a clean chip are ready.
- Loading: Using a standard pipette with cut-off tips, manually transfer individual embryos or larvae into the chambers of the ZEG chip. Each chamber should contain the embryo in a volume of 10–14 µL of E3 medium. Loading 24 embryos typically takes about two minutes [6].
- Extraction:
- Carefully place the loaded chip onto the platform of the ZEG base unit.
- Cover the chip with the evaporation-limiting cover.
- Power the vibration motor at 1.4 volts and run for 7 to 10 minutes [6]. During this time, harmonic oscillation will cause cellular material to be dislodged into the medium.
- Collection: After the oscillation cycle is complete, use a standard pipette to carefully collect 10 µL of fluid from each chamber. This fluid now contains the genetic material for genotyping [6].
- Genotyping: Use 5 µL of the collected fluid directly in an 11 µL PCR reaction for genotyping [6]. The extracted DNA is compatible with various downstream analyses, including:
- Post-Processing Animal Care: Return the genotyped, live embryos to fresh E3 medium. They can now be selectively raised based on their genotype for downstream applications, testing, or to establish adult lines [6] [5].
The following diagram illustrates the critical path from extraction to final analysis, highlighting the key decision points for researchers.
Applications in CRISPR-Cas9 Knock-In and Early Selection
The integration of ZEG into CRISPR-Cas9 workflows has proven to be a powerful strategy for improving the efficiency of generating knock-in (KI) models, which are crucial for modeling human diseases caused by single base-pair substitutions [5].
- Enhanced Somatic Editing Efficiency: In studies aimed at modeling human CACNA1C mutations, the combination of early genotyping with ZEG and NGS-based detection resulted in an almost 17-fold increase in somatic editing efficiency compared to standard methods. This pre-selection allowed researchers to identify and raise only those embryos with the highest rates of correct editing [5].
- Optimization of CRISPR Components: Research utilizing ZEG has helped identify optimal CRISPR conditions. For instance, the use of Cas9 protein (as opposed to mRNA) with a non-target asymmetric PAM-distal (NAD) single-stranded deoxynucleotide (ssODN) repair template was found to significantly outperform other configurations, leading to higher KI efficiencies at multiple genetic loci [5].
- Reduction in Animal Use and Costs: By enabling genotyping at 72 hpf, ZEG allows researchers to cull non-edited or incorrectly edited embryos at a very early, pre-sentient stage. This aligns with the 3Rs principles (Replacement, Reduction, and Refinement) in animal research by significantly reducing the number of animals raised to adulthood, thereby saving time, space, and resources [5] [2].
Validation of Minimal Physiological Impact
A critical aspect of the ZEG protocol's validation is the comprehensive assessment of its impact on the processed embryos. Studies have confirmed that the procedure is minimally invasive and does not compromise the animal's utility for downstream assays.
- Gross Morphology and Survival: As noted in Table 1, survival rates consistently exceed 90%, with no apparent effects on body morphology or development observed under a dissecting microscope [6].
- Behavioral Analysis: Motor behavior tests conducted at 7 days post-fertilization (dpf) showed no significant differences between ZEG-processed larvae and controls. Tests included analysis of spontaneous swimming, light-evoked responses, and tap startle response [6].
- Cellular and Molecular Characterization: A dedicated study examining the cellular and molecular effects of the ZEG protocol found that although it induces a low-level acute stress response, there are no long-lasting effects on tissue integrity or stress-related gene expression. This confirms that the ZEG protocol is suitable for a wide variety of sensitive downstream physiological and molecular assays [10] [11].
The ZEG device, with its core technology of microfluidic harmonic oscillation on a roughened glass surface, addresses a fundamental challenge in zebrafish research. It provides a reliable, rapid, and efficient method for the non-lethal genotyping of early-stage embryos, with demonstrated survival and sensitivity rates exceeding 90%. Its integration into CRISPR-Cas9 workflows, particularly when combined with NGS, dramatically improves the efficiency of generating precise disease models. Furthermore, rigorous validation confirms its minimal physiological impact, ensuring that genotyped animals remain viable and healthy for further experimental use. The ZEG protocol therefore stands as an essential tool for enhancing the scale, efficiency, and ethical standards of zebrafish-based genetic research.
Within the context of establishing a robust Zebrafish Embryo Genotyper (ZEG) early selection protocol, the extraction of genetic material is a foundational step. Traditional methods often rely on the sacrifice of the embryo or invasive biopsies that compromise its viability, thereby preventing longitudinal studies and increasing experimental animal burden. Non-invasive extraction techniques address this critical limitation by enabling early genotype-phenotype correlation and facilitating the reduction of "surplus" animals in mutant line generation [12]. This document details the mechanics of two primary non-invasive methods—fin scratching for embryos and skin swabbing for adult zebrafish—providing application notes and standardized protocols to enhance research reproducibility and align with the 3R principles (Replace, Reduce, Refine) [12] [13].
Method Comparison and Selection
Selecting the appropriate non-invasive method depends on the developmental stage of the zebrafish and the specific research requirements. The following table summarizes the key characteristics of the two primary techniques to guide researchers.
Table 1: Comparison of Non-Invasive Genetic Material Collection Methods in Zebrafish
| Method | Target Stage | Key Advantage | Typical gDNA Concentration & Purity | Sample Processing Time | Impact on Animal |
|---|---|---|---|---|---|
| Fin Scratching (FS) [12] | Embryos (as early as 2 dpf) | Allows early selection and strategic culturing; compatible with subsequent phenotypic analysis. | Sufficient for at least two rounds of PCR-based genotyping. | Protocol is rapid, minimizes embryo handling time. | Minimally invasive; compatible with normal development and early in vivo analyses. |
| Skin Swabbing [13] | Adult zebrafish (≥ 2 mpf, ~20 mm length) | A true non-invasive alternative to fin clipping; induces less stress axis activation. | 31.68 ± 3.64 ng/µL (A260/A280 = 1.25 ± 0.09) | ~10 minutes to complete the entire protocol. | Non-invasive; no tissue damage, minimal stress. |
Protocol 1: Fin Scratching for Embryo Genotyping
This protocol enables the isolation of sufficient genomic DNA (gDNA) for genotyping from single zebrafish embryos as early as 2 days post-fertilization (dpf) through a minimally invasive tail fin scratch [12].
Experimental Workflow
The fin scratching method integrates directly with a Zebrafish Embryo Genotyper early selection pipeline, allowing researchers to select embryos with desired genotypes for downstream culture or experiments.
Reagents and Equipment
Table 2: Research Reagent Solutions for Fin Scratching Protocol
| Item | Function/Application | Specifications/Notes |
|---|---|---|
| Tricaine (Ethyl 3-aminobenzoate methanesulfonate) [12] [13] | Anesthesia | Used at 1.5 mg/mL in E3 medium to immobilize embryos for precise manipulation. |
| E3 Embryo Medium [12] [13] | Embryo rearing and recovery | Standard medium for maintaining zebrafish embryos post-procedure. |
| Proteinase K [13] | gDNA extraction | Digests proteins to release genomic DNA; heat-inactivated post-incubation. |
| DNA Collection Buffer [13] | DNA stabilization and extraction | Contains Tris-HCl and Proteinase K for efficient DNA release. |
| Fine Needle/Micromanipulator [12] | Fin scratching | Tool for minimally invasive tissue collection from the caudal fin. |
| Stereomicroscope [12] | Visualization | Essential for precise positioning and scratching of the embryo tail fin. |
| Thermal Cycler | Incubation | For heating samples to 98°C to inactivate Proteinase K and prepare gDNA for PCR. |
Detailed Methodology
- Anesthesia and Positioning: Transfer a 2 dpf embryo to a solution of 1.5 mg/mL tricaine in E3 medium. Under a stereomicroscope, position the embryo to allow clear access to the caudal fin tip [12] [13].
- Fin Scratching: Using a fine needle or a micromanipulator, gently scratch the tip of the tail fin to collect a minimal amount of tissue. The goal is to obtain bioptic material without compromising the embryo's viability or normal development [12].
- Recovery and DNA Extraction: Immediately transfer the embryo to fresh E3 medium for recovery and continued development. Place the minute tissue fragment collected on the needle into a microfuge tube containing 400 µL of DNA collection buffer (e.g., 30 mM Tris-HCl, 1 µg/µL Proteinase K) [13].
- gDNA Isolation: Vortex the tube for 10 seconds and incubate at 98°C for 5 minutes. Centrifuge the tube briefly at 16,000× g for 10 minutes. The resulting supernatant contains PCR-ready gDNA suitable for subsequent genotyping [13].
Protocol 2: Skin Swabbing for Adult Zebrafish Genotyping
This protocol describes a rapid, non-invasive method to obtain PCR-ready gDNA from adult zebrafish using skin mucus collection with a cotton swab [13].
Experimental Workflow
The skin swabbing method provides a true non-invasive alternative to fin clipping for adult zebrafish genotyping, minimizing animal stress and allowing for repeated sampling if necessary.
Reagents and Equipment
Table 3: Research Reagent Solutions for Skin Swabbing Protocol
| Item | Function/Application | Specifications/Notes |
|---|---|---|
| Cotton Swabs (Q-tips) [13] | Mucus collection | Sterile, non-abrasive swabs for collecting skin mucus from the caudal fin. |
| Proteinase K [13] | gDNA extraction | Digests proteins in mucus to release cellular DNA for PCR. |
| DNA Collection Buffer [13] | DNA stabilization and extraction | Tris-HCl based buffer with Proteinase K for efficient DNA release from swab. |
| Tricaine [13] | Anesthesia | Anesthetizes fish for safe and stress-free handling during swabbing. |
Detailed Methodology
- Anesthesia and Swabbing: Anesthetize an adult zebrafish (≥ 2 months post-fertilization) in 1.5 mg/mL tricaine. Blot the fish dry on paper towels to remove excess water. Using a clean cotton swab, firmly but gently swab the caudal fin region several times until the tip of the swab becomes yellowish from the mucus [13].
- Recovery: Immediately return the fish to system water for full recovery. Skin swabbing has been shown to induce less stress axis activation compared to fin clipping [13].
- gDNA Extraction: Cut the cotton tip (~1 cm of the wad) into a microfuge tube containing 400 µL of DNA collection buffer with Proteinase K. Vortex the tube for 10 seconds and incubate at 98°C for 5 minutes. Remove the cotton wad, and the resulting solution can be used directly as a template for PCR [13]. The typical gDNA concentration obtained is 31.68 ± 3.64 ng/µL, which is sufficient for robust PCR amplification [13].
The integration of non-invasive genetic material extraction methods, such as fin scratching for embryos and skin swabbing for adults, is instrumental in advancing the Zebrafish Embryo Genotyper (ZEG) early selection framework. These protocols enable researchers to determine genotype before phenotypic manifestation, strategically plan experiments, and significantly reduce the number of animals required for generating and maintaining mutant lines. By providing detailed, reproducible methodologies and quantitative performance data, this application note empowers the research community to adopt these refined practices, thereby enhancing the ethical standards, efficiency, and translational impact of zebrafish-based biomedical research.
Performance Metrics of the ZEG Device
The table below summarizes the key quantitative performance metrics of the Zebrafish Embryo Genotyper (ZEG) device, demonstrating its efficiency and reliability for live embryo genotyping.
| Metric | Performance Value | Experimental Details & Conditions |
|---|---|---|
| Survival Rate | >90% [6] | Assessed on n > 25 animals per experiment across >10 separate experimental replicates. Animals were examined post-ZEG procedure under a dissecting microscope by an examiner blinded to the experimental status [6]. |
| Genotyping Sensitivity | >90% [6] | The genetic material collected was successfully amplified by PCR and used for subsequent analysis, including sequencing, gel electrophoresis, or high-resolution melt analysis [6]. |
| Throughput | 24 embryos or larvae simultaneously [6] | The device is capable of processing 24 samples in a single run. Chips with 16, 24, or 48 chambers were constructed [6]. |
| Processing Time | <10 minutes for 24 samples [6] | This time includes the vibration motor operation period of 7 to 10 minutes for genetic material extraction [6]. |
| DNA Extraction Volume | 10 μL collected from a total chamber volume of 11-14 μL [6] | Embryos or larvae are manually loaded into chip wells in 10–14 μL of E3 medium or water. A standard pipette is used to collect 10 μL of fluid from each chamber post-processing [6]. |
| Somatic Editing Efficiency (with Early Selection) | ~17-fold increase reported in a CRISPR-KI study [14] | Achieved by combining ZEG-based early genotyping with next-generation sequencing for pre-selection of embryos with the highest rates of correctly edited cells [14]. |
Experimental Protocol: ZEG Operation and Genotyping
This section provides a detailed methodology for using the ZEG device for rapid, non-lethal genotyping of zebrafish embryos and larvae.
Zebrafish Embryo Genotyper (ZEG) Operation Protocol
Principle: Using microfluidic harmonic oscillation of the animal on a roughened glass surface, the ZEG obtains genetic material (cells and DNA) for genotyping while keeping the animal alive [6].
Materials:
- Biological Material: Zebrafish embryos or larvae (24–72 hours post-fertilization) [6].
- Equipment: ZEG base unit and disposable extraction chips [6].
- Reagents: E3 embryo medium [6].
Procedure:
- Loading: Manually load individual embryos or larvae into each chamber of the ZEG chip using a standard pipette with cut-off tips. Each chamber should contain the animal in 10–14 μL of E3 medium. Loading 24 embryos typically takes approximately two minutes [6].
- Extraction: Place the loaded chip onto the platform of the base unit and cover it with the evaporation-limiting cover. Power the vibration motor at 1.4 volts for 7 to 10 minutes. This agitation generates an abrasive environment that liberates cells and DNA into the surrounding medium [6].
- Collection: After the vibration cycle, use a standard pipette to collect 10 μL of fluid from each chamber. This sample contains the genetic material for downstream genotyping [6].
- Recovery: Return the live embryos or larvae to fresh E3 medium. They can subsequently be raised for downstream applications, testing, or to adulthood [6].
Downstream PCR-Based Genotyping Analysis
Principle: The genetic material obtained from the ZEG device is amplified by PCR and can be analyzed by various methods, including sequencing, gel electrophoresis, or high-resolution melt analysis (HRMA) [6].
Materials:
- Reagents:
- Embryo lysis buffer (e.g., 1 mL 10x PCR Gold Buffer, 30 μL NP-40, 30 μL Tween 20, up to 10 mL MilliQ H₂O) [15].
- Proteinase K [15].
- PCR Master Mix (e.g., containing 10x PCR gold buffer, dNTP mix, MgClâ‚‚, and deionized water) [15].
- Primers specific to the target genomic region [6].
- Restriction enzymes (if digestion is required) [15].
- Agarose gel electrophoresis reagents or HRMA master mix [6].
Procedure:
- DNA Preparation: Use 5 μL of the 10 μL fluid collected from the ZEG directly in an 11 μL volume PCR reaction [6]. Alternatively, for other protocols, embryos can be lysed in embryo lysis buffer supplemented with Proteinase K [15].
- PCR Amplification: Perform PCR amplification using primers and conditions optimized for the target allele [6] [15].
- Genotype Analysis:
- High-Resolution Melt Analysis (HRMA): Analyze PCR products using HRMA with an appropriate master mix (e.g., LightScanner Master Mix) [6].
- Gel Electrophoresis: For protocols requiring restriction fragment length polymorphism (RFLP) analysis, digest PCR products with the appropriate restriction enzyme and separate the fragments on an agarose gel to distinguish wild-type, heterozygous, and mutant genotypes [15].
- Next-Generation Sequencing (NGS): For precise quantification of editing efficiency, especially in CRISPR applications, use NGS on the ZEG-derived DNA. This allows for the identification and selective raising of embryos with the highest rates of somatic editing [14].
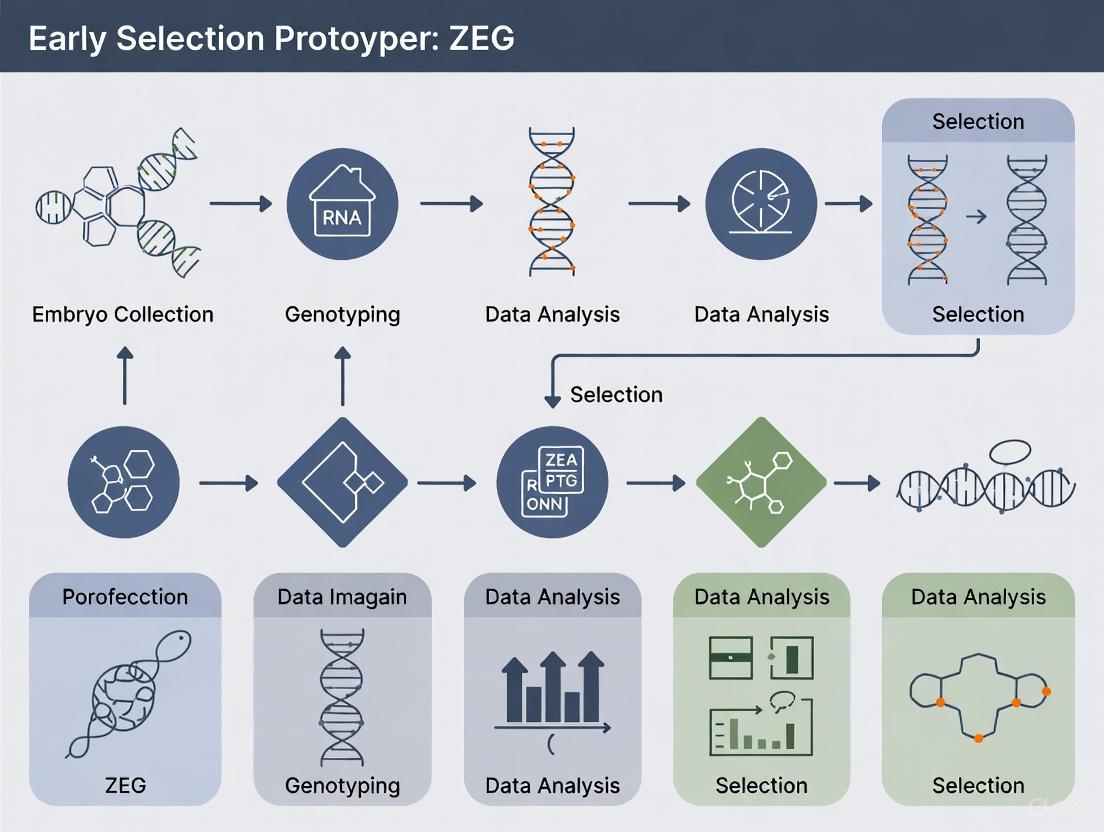
Workflow Diagram: ZEG Early Selection Protocol
The following diagram illustrates the integrated workflow for the early selection of genetically edited zebrafish using the ZEG device.
The Scientist's Toolkit: Research Reagent Solutions
The table below lists key reagents and materials essential for implementing the ZEG early selection protocol.
| Item | Function/Application | Example Details / Notes |
|---|---|---|
| ZEG Device & Chips | Automated extraction of genetic material from live zebrafish embryos/larvae. | The base unit agitates a disposable chip with 24 roughened glass chambers. Chips are made from standard glass slides and polyimide tape [6]. |
| E3 Embryo Medium | Standard medium for raising and maintaining zebrafish embryos. | Contains 5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl₂·2H₂O, 0.33 mM MgSO₄·7H₂O, and 0.002% methylene blue [15]. |
| Cas9 Protein | CRISPR-Cas9 genome editing component. | Using Cas9 protein (vs. mRNA) with a non-target asymmetric PAM-distal (NAD) ssODN conformation significantly increased KI efficiency in a cacna1c gene study [14]. |
| ssODN Repair Template | Homology-directed repair (HDR) template for precise knock-in. | A 120-nucleotide non-target asymmetric PAM-distal (NAD) conformation was optimal for introducing point mutations in the cacna1c gene [14]. |
| Embryo Lysis Buffer | Lysing embryos for DNA release in non-ZEG genotyping or post-ZEG processing. | Typically contains non-ionic detergents like NP-40 (0.3%) and Tween 20 (0.3%) in a PCR-compatible buffer [15]. |
| PCR Master Mix | Amplification of the target genomic region from extracted DNA. | A standard mix containing buffer, dNTPs, MgCl₂, and a thermostable DNA polymerase (e.g., AmpliTaq Gold) [15]. Can be prepared in bulk and stored at -20°C [15]. |
| High-Resolution Melt (HRMA) Master Mix | Post-PCR genotyping to identify genetic variants based on melt curve profiles. | Enables closed-tube genotyping without gel electrophoresis (e.g., LightScanner Master Mix) [6]. |
| PFM03 | PFM03, MF:C14H15NO2S2, MW:293.4 g/mol | Chemical Reagent |
| YE6144 | YE6144, MF:C21H27ClFN7O, MW:447.9 g/mol | Chemical Reagent |
The zebrafish (Danio rerio) is a cornerstone model organism in biomedical research, prized for its rapid development, genetic tractability, and high fecundity. However, a significant bottleneck has long constrained its full potential for large-scale genetic and drug screens: the absence of automated, non-lethal genotyping methods for early developmental stages. Traditionally, researchers have been forced to raise zebrafish to adulthood—a process consuming two to three months—before performing invasive fin clipping for genotyping. This approach is time-consuming, expensive, and requires maintaining excessive numbers of animals [6].
Framed within the broader thesis on ZEG early selection protocol research, this Application Note details a transformative solution: the Zebrafish Embryo Genotyper (ZEG). This automated system enables the rapid extraction of PCR-amplifiable genetic material from 24 embryos or larvae simultaneously in under 10 minutes, with greater than 90% genotyping sensitivity and animal survival rates [6]. This protocol revolutionizes research workflows by allowing for the selection of genetically desired individuals at 48-72 hours post-fertilization (hpf), making them immediately available for downstream experiments and drastically reducing the space and resources needed to raise animals to adulthood.
Key Advantages and Quantitative Workflow Transformation
The transition from traditional fin clipping to the ZEG protocol represents a paradigm shift in zebrafish colony management and experimental design. The following table summarizes the profound advantages of this new workflow.
Table 1: Comparative Analysis: ZEG vs. Traditional Fin Clipping
| Parameter | Zebrafish Embryo Genotyper (ZEG) | Traditional Fin Clipping |
|---|---|---|
| Animal Age | 48 - 72 hpf (embryos/larvae) [6] | ~ 3 months (adults) [6] |
| Processing Time | ~10 minutes for 24 samples [6] | 4-6 hours for 96 samples [6] |
| Genotyping Sensitivity | >90% [6] | High (established method) |
| Animal Survival Rate | >90% [6] | High, but invasive |
| Impact on Animal | Minimal invasion; animals recover and are available for experimentation [6] | Invasive; permanent fin clip |
| Downstream Utility | Animals available for phenotypic screening, drug testing, or raising [6] | Limited after procedure |
| Facility Space & Costs | Drastically reduced; only genotypically desired animals are raised | Significant; excess animals must be raised to ensure correct genotypes |
Detailed Experimental Protocol for ZEG Operation
Principle
The ZEG device operates on the principle of microfluidic harmonic oscillation. Live zebrafish embryos or larvae are placed in individual chambers with a roughened glass surface and subjected to controlled vibration. This motion creates an abrasive environment that gently dislodges cells and releases DNA from the animal into the surrounding medium, without compromising its viability [6].
Materials and Equipment
- ZEG Base Unit: Housing with a spring-suspended platform and a vibration motor (e.g., Precision Microdrive 312–108) [6].
- Disposable Extraction Chip: A glass slide with mechanically etched, roughened areas, assembled with a polyimide tape layer to create 24 shallow chambers [6].
- Pipettes and cut-off pipette tips.
- E3 Embryo Medium.
- PCR Reagents: Master mix (e.g., LightScanner Master Mix), primers, nuclease-free water [6].
- Materials for Downstream Analysis: Gel electrophoresis equipment, sequencer, or High-Resolution Melt Analysis (HRMA) instrument.
Step-by-Step Procedure
Table 2: Reagent Solutions and Materials for ZEG Genotyping
| Item | Function/Description |
|---|---|
| ZEG Disposable Chip | Provides the roughened-surface chamber for the abrasive extraction of genetic material from live embryos [6]. |
| E3 Embryo Medium | Standard medium for maintaining zebrafish embryos and larvae during the procedure [6]. |
| PCR Master Mix | Pre-mixed solution containing DNA polymerase, dNTPs, and buffer for the amplification of extracted DNA [6]. |
| High-Resolution Melt Analysis (HRMA) Dyes | Fluorescent dyes that allow for genotyping based on post-PCR dissociation curves, enabling precise variant identification [6]. |
| Trypan Blue & DAPI | Stains used for post-extraction cell counting and viability assessment on a hemocytometer [6]. |
Step 1: Animal Preparation
- Raise zebrafish embryos to the desired stage (48-72 hpf) at 28.5°C in E3 embryo medium [6].
- Manually dechorionate the embryos if necessary, depending on the experimental requirements and stage.
Step 2: Device Setup
- Ensure the ZEG base unit is on a stable, level surface.
- Place a new, disposable extraction chip onto the platform of the base unit.
Step 3: Sample Loading
- Using a standard pipette with a cut-off tip, manually transfer individual embryos or larvae into the chambers of the chip.
- Each chamber should be filled with 10–14 µL of E3 medium containing one animal. Loading 24 embryos typically takes approximately two minutes [6].
Step 4: Genetic Material Extraction
- Place the evaporation-limiting cover over the chip to prevent sample dehydration.
- Power the vibration motor at 1.4 volts for 7 to 10 minutes. During this time, harmonic oscillation agitates the animals against the roughened surface, dislodging cells and releasing DNA into the medium [6].
Step 5: Sample Collection
- Carefully remove the cover.
- Using a standard pipette, collect 10 µL of fluid from each chamber. This fluid contains the genetic material for genotyping.
- Immediately return the live animals to fresh E3 medium. Observations indicate no apparent effects on body morphology, development, or motor behavior tests post-procedure [6].
Step 6: Genotypic Analysis
- Use 5 µL of the collected fluid as the template in an 11 µL PCR reaction [6].
- Perform PCR amplification using conditions optimized for your target of interest.
- Analyze the PCR products via your preferred method:
Workflow Visualization
The following diagram illustrates the streamlined ZEG protocol alongside the traditional, cumbersome workflow, highlighting the dramatic reduction in time and resources.
Data Presentation and Analysis
The quantitative performance of the ZEG system has been rigorously validated. The following table consolidates key experimental data from validation studies, providing a clear summary of the system's efficiency and reliability.
Table 3: Quantitative Performance Metrics of ZEG Genotyping
| Metric | Result | Experimental Detail / Method of Analysis |
|---|---|---|
| Genotyping Sensitivity | >90% | PCR followed by sequencing, gel electrophoresis, or HRMA [6]. |
| Animal Survival Rate | >90% | Survival and morphology examined under a dissecting microscope by a blinded examiner [6]. |
| DNA Yield | Sufficient for PCR | 5 µL of 11 µL total collection volume used directly in PCR [6]. |
| Cell Count per Sample | Confirmed Presence | Samples incubated with Trypan Blue and DAPI, counted via hemocytometer [6]. |
| Behavioral Impact | No Apparent Effects | Motor behavior tests on 7 dpf larvae using video analysis (Noldus EthoVision) [6]. |
The ZEG protocol represents a fundamental workflow transformation in zebrafish-based research. By enabling rapid, non-lethal genotyping at the embryonic or larval stage, it effectively decouples genetic selection from the lengthy and resource-intensive process of raising animals to adulthood. This innovation directly addresses the critical bottleneck that has limited the scale and efficiency of zebrafish screens [6].
The implications for research are substantial. First, it dramatically increases genetic efficiency and throughput. Second, it empowers new experimental designs, particularly for embryonic or larval phenotypic analysis, where the genotype can now be known a priori. Third, it aligns with the 3Rs principle (Replacement, Reduction, and Refinement) in animal research by minimizing animal numbers and refining procedures to reduce suffering [6] [17].
Integrating the ZEG early selection protocol into a broader research thesis facilitates more ambitious projects, such as large-scale CRISPR mutagenesis screens, precise modeling of human genetic diseases, and high-throughput drug discovery. As one researcher from the University of Utah attested, "In the olden days, I would have had to sacrifice each embryo to genotype them... Now instead I have genotyped offspring at 48 hpf ready for experimentation, or to grow up, saving me valuable space in our zebrafish facility!" [17]. The ZEG system transforms the zebrafish model into a more powerful and efficient tool for modern biomedical discovery.
Implementing ZEG Protocol: Step-by-Step Workflow and CRISPR Integration
The Zebrafish Embryo Genotyper (ZEG) represents a transformative technological advancement in zebrafish research, enabling rapid, automated cellular extraction from live embryos and larvae for genotyping while maintaining high survival rates. This protocol details the comprehensive configuration of the ZEG device and preparation of its specialized microfluidic chips, which collectively facilitate high-throughput genetic screening by obtaining genetic material from 24 embryos simultaneously in less than 10 minutes with greater than 90% survival rates. Developed to address critical bottlenecks in large-scale mutagenesis and drug screening studies, the ZEG system eliminates the previous necessity to raise animals to adulthood for fin clipping, thereby significantly reducing labor, time, and resource requirements while enabling direct correlation of embryonic phenotypes with genotypes. This application note provides researchers, scientists, and drug development professionals with detailed methodologies for implementing this technology within early selection protocols for CRISPR-edited zebrafish models, substantially improving germline transmission efficiency by enabling pre-selection of embryos with the highest editing rates.
Device Specifications and Components
Base Unit Construction
The ZEG base unit operates the disposable extraction chips through precisely controlled mechanical agitation. The housing is constructed via 3-D printing (LulzBot TAZ6) and contains several integrated components: a power supply (Uctronics U5168), a chip-holding platform with a mounted 12mm (3V) vibration motor (Precision Microdrive 312–108) on its undersurface, and a platform raised on four springs to permit planar movement [6]. An evaporation-limiting cover is provided to place over the slide during operation to prevent sample dehydration, which is critical for maintaining embryo viability throughout the extraction process. The device is designed to accommodate chips with varying chamber configurations (16, 24, or 48 chambers), providing flexibility for different experimental scales and throughput requirements [6].
Chip Architecture and Properties
The disposable extraction chips are fabricated from standard glass microscope slides that undergo specialized processing to create optimal surface characteristics for cellular extraction. Circular areas are mechanically etched into the top surface of the glass slides to create roughened textures that enhance the abrasive action during agitation [6]. Polyimide tape is precisely cut to match the glass slide dimensions and features circular holes aligned with the roughened areas, creating shallow chambers when attached to the glass surface. The completed chip forms multiple individual chambers where the roughened glass areas are centered within the through holes of the polyimide tape, creating an optimized environment for the harmonic oscillation process that facilitates cellular extraction while maintaining embryo integrity [6].
Table 1: Quantitative Performance Metrics of ZEG Device
| Performance Parameter | Result | Measurement Conditions |
|---|---|---|
| Processing Time | 7-10 minutes | For 24 embryos simultaneously [6] |
| Survival Rate | >90% | Embryos at 72 hpf [6] |
| Genotyping Sensitivity | >90% | Sufficient DNA for PCR, sequencing, HRMA [6] |
| Cell Extraction Yield | Adequate for multiple analyses | 5μL of collected fluid used in 11μL PCR reaction [6] |
| Behavioral Impact | No significant effects | Motor behavior tests at 7 dpf [6] |
| Developmental Impact | No apparent effects | Body morphology and development [6] |
Chip Preparation Protocol
Fabrication Procedure
The chip fabrication process requires precision materials handling and assembly to ensure consistent performance across production batches. Begin with standard glass microscope slides that undergo laser etching to create circular roughened areas in a predetermined array pattern matching the desired chamber configuration [6]. The laser parameters must be calibrated to achieve uniform surface roughness across all chambers, as this consistency is critical for reproducible cellular extraction efficiency. Simultaneously, prepare polyimide tape by precision cutting to exact slide dimensions with circular holes corresponding to the etched areas on the glass slides. The alignment process requires meticulous attention to ensure each roughened glass area centers perfectly within its corresponding polyimide tape hole, creating uniform chambers of consistent depth and surface characteristics across the entire chip [6]. Finally, securely bond the aligned polyimide tape to the glass surface, ensuring no adhesive contacts the chamber interiors which could compromise sample integrity or introduce contaminants.
Quality Control Measures
Each manufactured chip must undergo rigorous quality assessment before experimental use. Visually inspect all chambers under magnification to verify consistent surface roughness, proper alignment, and absence of manufacturing defects or contaminants [6]. Conduct fluid retention testing by loading chambers with E3 embryo medium to verify proper surface tension and absence of leakage. Perform control extractions using wild-type embryos to validate cellular yield consistency across chambers, ensuring each chamber provides equivalent performance. Proper quality assurance is essential as chamber variability can significantly impact genotyping reliability and experimental reproducibility, particularly in high-throughput screening applications where consistent performance across all samples is imperative for valid results interpretation.
Device Configuration Protocol
System Assembly
Proper device assembly is fundamental to operational success and experimental reproducibility. Begin by securing the 3-D printed housing on a stable, level surface to prevent unwanted vibrations during operation [6]. Install the power supply according to manufacturer specifications, ensuring appropriate voltage regulation for consistent motor performance. Mount the chip-holding platform on its supporting springs, verifying free planar movement without obstruction or excessive play. Affix the vibration motor centrally to the platform's undersurface to ensure even distribution of harmonic oscillations across the entire chip area during operation [6]. Finally, validate system integrity through preliminary testing without samples to confirm stable operation and appropriate vibration characteristics before proceeding with experimental applications.
Operational Parameters
The ZEG device requires precise calibration of operational parameters to optimize cellular extraction while maintaining embryo viability. Set the vibration motor to 1.4 volts, which provides the optimal balance between extraction efficiency and embryo survival [6]. Program the operation duration for 7-10 minutes, which has been experimentally determined to yield sufficient cellular material for downstream genetic analyses without compromising embryo integrity. Maintain ambient temperature conditions throughout operation, as the system is designed for operation at standard laboratory temperatures without additional heating or cooling requirements. These parameters have been validated through extensive testing demonstrating consistent performance across multiple zebrafish lines and developmental stages, particularly for embryos at 24-72 hours post-fertilization, which represent the most common developmental windows for early genotyping in CRISPR screening applications [6].
Diagram 1: ZEG Device Operation Workflow. The complete process from embryo loading through genetic analysis and animal recovery, demonstrating the integration of automated extraction with downstream applications.
Experimental Implementation and Validation
Embryo Processing Procedure
The embryo processing protocol requires careful handling to maximize both cellular yield and embryo survival. Manually load embryos or larvae into chip wells using standard pipettes with cut-off tips to prevent damage, transferring each animal in 10-14μL of E3 medium or water [6]. This loading process typically requires approximately two minutes for 24 embryos when performed by an experienced technician. Position the loaded chip securely onto the device platform and place the evaporation cover to minimize fluid loss during the extraction process. Initiate the vibration motor at the predetermined parameters (1.4 volts for 7-10 minutes), during which the harmonic oscillation generates precisely controlled abrasive action between the embryos and the roughened glass surface [6]. Following the processing cycle, use a standard pipette to collect approximately 10μL of fluid from each chamber, which contains the genetic material (cells and DNA) liberated during the extraction process and is suitable for immediate use in downstream genetic analyses without additional purification.
Integration with Early Selection Protocols
The ZEG system enables transformative early selection approaches in CRISPR-edited zebrafish models by facilitating rapid genotyping of embryos before significant resource investment in raising animals to adulthood. Recent implementations demonstrate that combining ZEG extraction with next-generation sequencing (NGS)-based genotyping enables pre-selection of embryos with the highest editing rates, achieving an almost 17-fold increase in somatic editing efficiency compared to conventional approaches [5]. This selection advantage is particularly pronounced for alleles with lower intrinsic editing efficiencies, which might otherwise be lost using traditional screening methods. The validated protocol involves extracting genetic material at 72 hours post-fertilization, followed by NGS analysis to identify embryos with optimal KI rates, then selectively raising these pre-screened animals to adulthood [5]. This integrated approach has demonstrated successful germline transmission events in multiple experimental groups, significantly improving the efficiency of generating stable knock-in lines while reducing animal numbers, resource allocation, and time requirements.
Table 2: Research Reagent Solutions for ZEG Applications
| Reagent/Component | Function | Specifications | Application Notes |
|---|---|---|---|
| E3 Embryo Medium | Maintenance medium | Standard zebrafish embryo formulation | Preserves embryo viability during processing [6] |
| Cas9 Protein | Genome editing | Recombinant S. pyogenes | Superior to mRNA for KI efficiency in zebrafish [5] |
| ssODN Repair Template | Homology-directed repair | 120 nt, non-target asymmetric PAM-distal conformation | Optimal for HDR efficiency in cacna1c gene editing [5] |
| LightScanner Master Mix | HRMA genotyping | Commercial PCR mix | Compatible with ZEG-extracted DNA [6] |
| Trypan Blue & DAPI | Cell quantification | Vital staining | For assessing cellular yield after extraction [6] |
| Power Up SYBR Green | qPCR analysis | Intercalating dye chemistry | Quantitative assessment of DNA yield [6] |
Technical Considerations and Troubleshooting
Optimization Parameters
Several technical factors require consideration to maximize ZEG performance in specific experimental contexts. Embryo developmental stage significantly impacts both extraction efficiency and survival rates, with 72 hours post-ferfertilization representing the optimal balance for most applications [6]. Chamber loading density must be carefully controlled, with individual placement of embryos ensuring consistent exposure to the abrasive surface action. Fluid volume should be maintained within the 10-14μL range to ensure proper hydrodynamic characteristics during vibration processing [6]. The vibration duration may require slight adjustment based on embryo size and developmental stage, with preliminary tests recommended when working with novel transgenic lines or unusual embryonic morphologies. Additionally, the specific genetic analysis method following extraction influences optimal processing conditions, with PCR-based approaches generally more tolerant of variable DNA quality compared to sequencing applications, though both have been successfully implemented with ZEG-derived material [6] [5].
Validation Methodologies
Comprehensive validation of ZEG performance should include multiple assessment modalities to ensure experimental reliability. Survival rates should be quantified at 24 hours post-processing, with morphological examination conducted by observers blinded to experimental conditions to eliminate assessment bias [6]. Behavioral analyses, including spontaneous swimming, light-evoked responses, and tap startle responses, provide sensitive indicators of procedural impact on neurodevelopment and function. Cellular yield should be quantified using standardized counting methods, such as hemocytometer assessment with Trypan Blue and DAPI staining, with target yields established based on downstream application requirements [6]. Genotyping success rates should be compared across extraction batches to identify technical variability, with correlation established between extraction efficiency and subsequent amplification performance in specific genetic assays. These validation approaches collectively ensure that ZEG processing does not introduce confounding variables that might compromise experimental interpretations, particularly in phenotypic screening applications where subtle morphological or behavioral differences have significant scientific implications.
Diagram 2: ZEG-Compatible Downstream Analysis Methods. Multiple genetic analysis approaches can be successfully implemented with ZEG-extracted material, providing flexibility for different research applications and experimental requirements.
Embryo Preparation and Loading: Optimal Developmental Stages (24-72 hpf)
This application note provides a detailed protocol for the preparation and loading of zebrafish embryos into the Zebrafish Embryo Genotyper (ZEG) device, focusing on the critical 24 to 72 hours post-fertilization (hpf) developmental window. The ZEG enables high-throughput, automated cellular extraction from live embryos for genotyping, keeping the animals alive for downstream experiments and reducing the number of animals raised to adulthood. We present optimized staging criteria, a complete step-by-step loading protocol, downstream molecular validation methods, and a characterization of the minimal impact on animal development, supporting its integration into early selection protocols for functional genomics and drug discovery pipelines.
The Zebrafish Embryo Genotyper (ZEG) addresses a significant bottleneck in zebrafish research by enabling rapid genotyping of live embryos and larvae. Traditional methods often require sacrificing the animal or raising it to adulthood for fin clipping, which is time-consuming, costly, and generates a surplus of animals [6] [8]. The ZEG device utilizes microfluidic harmonic oscillation on a roughened glass surface to gently obtain genetic material (cells and DNA) from 24 embryos simultaneously in under 10 minutes, with survival and genotyping sensitivity rates both exceeding 90% [6]. This protocol details the embryo preparation and loading procedures for the 24-72 hpf stage, a key period for early phenotypic screening and selection in CRISPR-Cas9 and other genetic manipulation studies [5].
Developmental Staging and Suitability Assessment
The 24-72 hpf developmental window is optimal for ZEG processing. Embryos and larvae within this range are robust enough to withstand the procedure while allowing for early genotype selection before many phenotypes manifest.
Table 1: Developmental Staging and Suitability for ZEG Processing (24-72 hpf)
| Developmental Stage (hpf) | Key Morphological Characteristics | Suitability for ZEG | Special Considerations |
|---|---|---|---|
| 24 hpf | Most organs forming; body plan established; high tissue transparency [18]. | High | Embryo is delicate; ensure chorion is removed if necessary. |
| 48 hpf | Pigmentation increasing; early locomotion; major organs (heart, liver, pancreas) functional [18]. | Optimal | Robust size for easy handling; high cell yield. |
| 72 hpf | Larvae free-swimming; yolk sac largely absorbed; complex behaviors (e.g., prey capture) emerging [6] [18]. | Optimal | Standard stage for ZEG protocol; used in CRISPR editing selection [5]. |
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 2: Key Research Reagent Solutions for ZEG Protocol
| Item | Function/Application in Protocol |
|---|---|
| ZEG Base Unit & Disposable Chips | The core automated system. The base unit provides controlled harmonic oscillation, while the disposable chips with roughened glass chambers hold individual embryos during processing [6] [8]. |
| E3 Embryo Medium | Standard medium for raising and maintaining zebrafish embryos. Used to hold embryos during loading, unloading, and within the ZEG chambers [6]. |
| Standard Pipette & Cut-off Tips | For manual loading and unloading of embryos in fluid volume (10-14 µL) onto the ZEG chip. Cut-off tips prevent damage to the embryo [6]. |
| PCR Master Mix (e.g., LightScanner Master Mix) | For amplifying the extracted DNA. Enables downstream genotyping by sequencing, gel electrophoresis, or High-Resolution Melt Analysis (HRMA) [6]. |
| Trypan Blue & DAPI (5 mg/mL) | Stains for counting and verifying nucleated cell extraction from embryos post-ZEG processing via hemocytometer [6]. |
| MS-222 (Tricaine) | Anesthetic used to sedate larvae for handling or subsequent imaging procedures post-genotyping [18]. |
| SPH5030 | SPH5030, CAS:2364326-23-6, MF:C31H31FN8O3, MW:582.6 g/mol |
| Celecoxib-d3 | Celecoxib-d3 (methyl-d3) |
Detailed Experimental Protocol
Embryo Preparation Prior to ZEG Loading
- Culture and Stage Selection: Raise embryos at 28.5°C in E3 embryo medium. Under a dissecting microscope, select healthy, normally developing embryos within the 24-72 hpf window, noting any key morphological features (Table 1).
- Dechorionation (Optional): For embryos older than 48 hpf, the chorion may need to be removed manually with fine forceps to ensure direct contact with the ZEG chamber surface.
- Anesthetization: To immobilize embryos during loading and processing, treat with a solution of MS-222. This minimizes movement and potential stress [18].
ZEG Loading and Operation
- Chip Preparation: Place a new, clean disposable ZEG chip on the platform of the base unit.
- Embryo Loading: Using a standard pipette with a cut-off tip, manually transfer individual embryos into the chambers of the ZEG chip. Each chamber should contain a single embryo in a volume of 10-14 µL of E3 medium. Loading 24 embryos typically takes approximately two minutes [6].
- Device Operation: Place the evaporation cover over the chip. Power the vibration motor at 1.4 volts for 7 to 10 minutes. This agitation causes gentle, abrasive interaction between the embryo and the roughened glass, releasing cells and DNA into the surrounding medium [6].
- Sample Collection: After the operation cycle, use a standard pipette to carefully collect 10 µL of fluid from each chamber. This fluid contains the genetic material for genotyping.
- Embryo Unloading: Gently pipette each embryo from its chamber into fresh E3 medium for recovery. Processed embryos can be raised for future studies.
Downstream Genotyping and Validation
The collected fluid is directly compatible with various genotyping methods. For PCR, use 5 µL of the 10 µL collected sample in an 11 µL reaction volume [6].
- PCR and High-Resolution Melt Analysis (HRMA): A powerful method for identifying genetic variants. Use conditions as described [6] with appropriate primers.
- Next-Generation Sequencing (NGS): For precise identification of CRISPR edits or single-nucleotide variants. Early genotyping with ZEG combined with NGS has been shown to increase somatic editing efficiency by nearly 17-fold by enabling early selection of high-efficiency embryos [5].
Workflow Visualization
Protocol Validation and Quality Control
Assessing Extraction Efficiency and Genotyping Sensitivity
- Cell Count Verification: To confirm cellular extraction, add Trypan Blue and DAPI to the collected fluid and count nucleated cells using a hemocytometer [6].
- Genotyping Sensitivity: The protocol consistently demonstrates >90% success rate in subsequent PCR amplification, enabling reliable identification of transgenic, mutant, and wild-type alleles [6] [8].
Evaluating Embryo Viability and Developmental Impact
Comprehensive analyses confirm that the ZEG protocol is minimally invasive.
- Survival Rate: Survival post-ZEG processing is consistently >90% [6] [10].
- Morphology and Development: No apparent effects on body morphology, organ development, or overall growth are observed in processed embryos when compared to controls [6].
- Behavioral Analysis: Motor behavior tests, including spontaneous swimming, light-evoked responses, and tap startle response at 7 dpf, show no significant differences between processed and control larvae [6].
- Molecular Stress Response: While the ZEG protocol can induce a low-level acute cellular stress response, studies show no long-lasting molecular or tissue integrity effects, supporting its use for a wide range of downstream assays [10].
Application in Early Selection for CRISPR Editing
Integrating ZEG genotyping into CRISPR-Cas9 workflows dramatically improves efficiency. By genotyping at 72 hpf, researchers can identify and selectively raise embryos with the highest rates of somatic editing. This approach has been shown to provide an almost 17-fold increase in somatic editing efficiency and significantly improves the likelihood of germline transmission, thereby reducing the time, cost, and number of animals required to establish stable lines [5].
The ZEG protocol for embryo preparation and loading at 24-72 hpf provides a robust, efficient, and ethically refined method for early genotyping of zebrafish. Its high survival rates and minimal impact on development ensure that genotyped animals remain available for downstream phenotypic analysis, drug screening, or raising to adulthood. By enabling rapid selection at the embryo stage, this protocol accelerates research in functional genomics, disease modeling, and drug discovery while actively contributing to the reduction and refinement of animal use in scientific research.
Within zebrafish research, the high-throughput genotyping of live embryos and larvae is a critical bottleneck. The Zebrafish Embryo Genotyper (ZEG) protocol overcomes this by using a controlled, automated vibrational process to extract genetic material from live animals, enabling early selection in studies such as CRISPR-Cas9 mutant generation [6] [5]. This application note details the vibration parameters and sample collection methodologies that form the core of this technology, providing researchers with a definitive guide for implementation.
Key Vibration Parameters for Automated Cellular Extraction
The ZEG device employs microfluidic harmonic oscillation to gently displace cells from the surface of live zebrafish embryos or larvae. The consistent application of the following parameters is critical for achieving high genotyping success while ensuring animal survival.
Table 1: Core Vibration and Operational Parameters of the ZEG Protocol
| Parameter | Specification | Application Note |
|---|---|---|
| Vibration Principle | Microfluidic harmonic oscillation on a roughened glass surface [6] | The roughened surface provides the abrasive action necessary for cell displacement while the liquid medium facilitates the process. |
| Power Supply | 1.4 volts to a 12mm vibration motor [6] | This specific voltage is optimized to generate sufficient oscillation for cell extraction without causing lethal damage to the embryo or larva. |
| Processing Duration | 7 to 10 minutes [6] | The total time required for the vibrational extraction process for a full batch of samples. |
| Throughput | 24 embryos or larvae processed simultaneously [6] [8] | The standard capacity of the disposable chip, with designs for 16, 24, or 48 chambers available [6]. |
| Sample Volume Loaded | 10–14 µL of E3 medium or water per chamber [6] | The optimal volume for creating the necessary microfluidic environment during oscillation. |
| Sample Volume Collected | 10 µL of fluid collected from each chamber post-vibration [6] [8] | This fluid contains the genetic material (cells and DNA) for downstream genotyping. |
Experimental Workflow: From Embryo to Genotype
The following diagram illustrates the end-to-end process for genotyping live zebrafish embryos using the ZEG device.
Detailed Protocol
Step 1: Preparation of Zebrafish Embryos and Larvae
- Animals: Use zebrafish embryos or larvae at 24–72 hours post-fertilization (hpf) [6] [8]. Raise embryos at 28.5°C in standard E3 embryo medium [6].
- Ethics Statement: All procedures must be approved by the relevant Institutional Animal Care and Use Committee (IACUC) [6].
Step 2: Loading the ZEG Chip
- Tool: Use a standard pipette with the end of the tip cut off to prevent damage to the animals [6] [8].
- Procedure: Manually transfer individual embryos or larvae into each chamber of the chip.
- Volume: Load each chamber with 10–14 µL of E3 medium or water [6]. This process takes approximately two minutes for 24 embryos.
Step 3: Executing the Vibration Protocol
- Setup: Place the loaded chip onto the platform of the ZEG base unit and cover it with the evaporation-limiting cover.
- Activation: Power the vibration motor at 1.4 volts for a duration of 7 to 10 minutes [6]. During this time, harmonic oscillation agitates the animals against the roughened glass, displacing cells into the surrounding fluid.
Step 4: Sample Collection
- Tool: Use a standard pipette.
- Procedure: After the vibration cycle is complete, collect 10 µL of fluid from each chamber [6] [8]. This fluid now contains suspended cells and genomic DNA.
- Post-Collection Handling: The collected sample can be used directly in downstream applications, such as PCR.
Step 5: Downstream Genotyping and Animal Recovery
- Genotyping: Use 5 µL of the collected fluid in an 11 µL PCR reaction for genotyping via methods like gel electrophoresis, high-resolution melt analysis (HRMA), or next-generation sequencing (NGS) [6] [5].
- Animal Recovery: Return the live, genotyped embryos or larvae to fresh E3 medium. Studies confirm that this protocol has no apparent long-term effects on body morphology, development, or motor behavior, with survival rates consistently exceeding 90% [6] [10].
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 2: Key Research Reagent Solutions for the ZEG Protocol
| Item | Function/Application in the Protocol |
|---|---|
| Zebrafish Embryo Genotyper (ZEG) | The base unit and disposable chips that automate the vibrational extraction process [6]. |
| E3 Embryo Medium | Standard medium for raising zebrafish embryos and as the fluid medium during vibrational extraction [6]. |
| Standard Pipette & Cut-off Tips | For manual loading of embryos into the chip chambers and collection of fluid samples post-vibration [6]. |
| PCR Master Mix & Primers | For amplifying the extracted genomic DNA to determine the genotype [6] [5]. |
| High-Resolution Melt Analysis (HRMA) Reagents | For precise genotyping and variant detection following PCR amplification of ZEG-extracted DNA [6]. |
| Next-Generation Sequencing (NGS) | Used for high-sensitivity detection of knock-in efficiency and precise genetic edits from ZEG samples [5]. |
| FT3967385 | FT3967385, MF:C21H19N5O2, MW:373.4 g/mol |
| B-Raf IN 8 | B-Raf IN 8, MF:C18H17N3O2, MW:307.3 g/mol |
Validation and Impact
The ZEG protocol's efficacy is rigorously validated. The sensitivity of genotyping and animal survival are both greater than 90% [6]. Cellular and molecular characterization studies confirm that although the protocol induces a low-level acute stress response, it results in no long-lasting effects, supporting its use for a wide variety of downstream assays [10]. The application of this early selection protocol in CRISPR editing workflows has demonstrated an almost 17-fold increase in somatic editing efficiency, significantly improving the success rate for generating knock-in models and enhancing germline transmission events [5].
The Zebrafish Embryo Genotyper (ZEG) device represents a significant advancement in developmental genetics, enabling the non-lethal extraction of genetic material from live zebrafish embryos and larvae as early as 72 hours post-fertilization (hpf) [6]. This technology addresses a critical bottleneck in large-scale genetic screens by allowing researchers to identify and selectively raise embryos of desired genotypes, thereby dramatically reducing animal husbandry costs and improving experimental efficiency [5] [6]. The minimal invasiveness of the ZEG protocol, which utilizes microfluidic harmonic oscillation on a roughened glass surface to collect cells and DNA, has been demonstrated to have no long-lasting effects on embryonic development or survival, with sensitivity and survival rates both exceeding 90% [6] [10].
This application note details standardized protocols for downstream molecular analyses of ZEG-derived samples, focusing on three powerful genotyping applications: PCR, High-Resolution Melt Analysis (HRMA), and Next-Generation Sequencing (NGS). When integrated into a comprehensive ZEG early selection pipeline, these methods facilitate rapid and precise genotype identification for CRISPR-Cas9 editing validation, transgene detection, and somatic editing efficiency quantification [5] [19].
ZEG Sample Collection and Processing Workflow
The following diagram illustrates the complete workflow from ZEG sample collection through downstream molecular applications:
Research Reagent Solutions
The table below outlines essential reagents and materials required for implementing the complete ZEG genotyping pipeline:
Table 1: Essential Research Reagents for ZEG Downstream Applications
| Reagent/Material | Application | Function & Specifications |
|---|---|---|
| ZEG Device & Chips [6] | Sample collection | Base unit with vibration motor and disposable 24-well chips with roughened glass surfaces for cell/DNA extraction |
| DNA Lysis Buffer [19] | DNA preparation | 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 0.3% Tween-20, 0.3% NP-40, 1 mg/mL Proteinase K (freshly added) |
| LightScanner Master Mix [19] | PCR-HRMA | Optimized mix containing hot-start Taq polymerase, buffer, dNTPs, MgClâ‚‚, and fluorescent DNA-binding dye |
| 4sUTP [20] | Metabolic labeling | For nucleotide analog incorporation into newly transcribed RNA to distinguish zygotic from maternal transcripts |
| PacBio SMRT Sequencing [21] | Long-read sequencing | Full-length transcriptome analysis and novel isoform discovery |
| GRAND-SLAM Software [20] | Data analysis | Statistical inference to estimate labeled mRNA fractions from metabolic labeling data |
| Drop-Seq Materials [20] | Single-cell RNA-Seq | Beads, reagents, and microfluidic devices for capturing single-cell transcriptomes |
PCR Amplification from ZEG Samples
Protocol
- Sample Collection: Using a standard pipette, collect 10-14 µL of fluid from each ZEG chamber following the 7-10 minute vibration protocol [6].
- DNA Preparation:
- Transfer 5-10 µL of ZEG sample to a clean PCR tube.
- Prepare DNA lysis buffer: 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 0.3% Tween-20, 0.3% NP-40, with 1 mg/mL Proteinase K added fresh [19].
- Add equal volume of 2× DNA lysis buffer to each sample.
- Incubate at 55°C for 4 hours to overnight.
- Heat-inactivate Proteinase K at 95°C for 15 minutes.
- Use immediately or store at -20°C for up to 3 months [19].
- PCR Reaction Setup:
Application Notes
- Primer design should target amplicons of 50-200 bp for optimal amplification from ZEG-derived DNA [19].
- During initial optimization, confirm PCR product size by agarose gel electrophoresis [19].
- The number of cells obtained by ZEG extraction is sufficient for most PCR applications, with >90% success rate in genotyping [6].
High-Resolution Melt Analysis (HRMA)
Protocol
- Post-PCR Processing:
- Transfer PCR plates directly to HRMA instrument without opening tubes.
- Use LightScanner System or equivalent melt analysis instrument [19].
- HRMA Data Acquisition:
- Program melt curve: 60°C to 95°C with high-resolution data collection (0.1°C/s) [19].
- Data Analysis:
- Normalization: Using analysis software, position parallel lines in pre-melt (79-80°C) and post-melt regions to normalize fluorescence signals to 1 and 0, respectively [19].
- Grouping/Clustering: Select "Autogroup" function with sensitivity set to "Normal" for single transitions or "High" for multiple transitions [19].
- Interpretation: Distinguish genotypes based on melt curve profiles and difference plots compared to reference samples [19].
Application Notes
- HRMA is particularly effective for identifying single base-pair changes, insertions, deletions, and transgenes [19].
- The closed-tube nature of HRMA reduces contamination risk and enables high-throughput screening [19].
- Difference plots enhance discrimination of subtle melt curve variations between similar genotypes [19].
Next-Generation Sequencing Applications
ZEG-NGS Integration for CRISPR Editing Efficiency
The combination of ZEG pre-selection with NGS genotyping enables highly efficient identification of CRISPR-edited embryos, as demonstrated by a 17-fold increase in somatic editing efficiency recovery [5].
Table 2: NGS Applications for ZEG-Derived Samples
| Application | Method | Key Outcomes | Reference |
|---|---|---|---|
| CRISPR-KI Efficiency | NGS of ZEG-preselected embryos | 17× increase in somatic editing efficiency; optimal conditions: Cas9 protein + non-target asymmetric PAM-distal ssODN | [5] |
| Single-Cell Transcriptomics | scRNA-Seq + metabolic labeling (4sUTP) | Distinguishes maternal vs. zygotic transcripts; quantifies transcription/degradation kinetics | [20] |
| Full-Length Isoform Discovery | PacBio SMRT sequencing | Identified 2,113 novel genes and 33,018 novel isoforms across 21 developmental stages | [21] |
NGS Library Preparation from ZEG Samples
- Whole Transcriptome Analysis:
- For bulk RNA-Seq, extract total RNA from ZEG samples using standard methods.
- Employ ribosomal RNA depletion rather than poly-A selection to improve detection of non-polyadenylated transcripts [20].
- Metabolic Labeling for Kinetic Studies:
- Inject 4sUTP into one-cell stage embryos to label newly transcribed RNAs [20].
- Process ZEG samples at desired developmental stages (e.g., dome, 30% epiboly, 50% epiboly).
- Perform chemical conversion of 4sU residues after mRNA capture to create T-to-C changes in sequencing reads [20].
- Use GRAND-SLAM analysis to estimate fractions of labeled (zygotic) versus unlabeled (maternal) mRNA [20].
- Sequencing Data Analysis:
- Apply kinetic modeling to quantify mRNA transcription and degradation rates.
- Identify cell-type-specific differences in RNA metabolism during embryogenesis [20].
Troubleshooting and Quality Control
Common Issues and Solutions
Table 3: Troubleshooting Guide for ZEG Downstream Applications
| Issue | Potential Cause | Solution |
|---|---|---|
| Low PCR yield | Insufficient cellular material | Ensure vibration motor operates at 1.4V for full 7-10 minutes; collect full 10µL sample volume [6] |
| Poor HRMA resolution | Large amplicon size | Redesign primers for 50-80 bp products for SNP detection [19] |
| Inconsistent NGS results | Low input DNA/RNA quality | Increase initial embryo incubation time in lysis buffer; extend Proteinase K digestion to overnight [19] |
| Reduced embryo survival | Excessive vibration exposure | Limit vibration duration to 10 minutes maximum; verify proper spring function in ZEG device [6] |
Quality Control Metrics
- Sample Quality: Post-ZEG survival rates should exceed 90%; no significant morphological or behavioral defects should be observed [6] [10].
- DNA Suitability: Successful amplification of control genes from >90% of ZEG samples indicates adequate DNA quality [6].
- NGS QC: For metabolic labeling experiments, zygotic genes should show >80% labeled fractions, while maternal genes should show <3.5% labeling [20].
The integration of ZEG sampling with robust downstream molecular applications creates a powerful pipeline for rapid genotyping in zebrafish research. The protocols detailed herein enable researchers to efficiently identify CRISPR-edited embryos, characterize transcriptional dynamics, and discover novel transcript isoforms while significantly reducing animal numbers and husbandry costs. By implementing these standardized methods, research and drug development programs can accelerate genetic screens and functional studies using the zebrafish model system.
The generation of precise genetic models in zebrafish using CRISPR-Cas9 has transformed biomedical research, yet achieving efficient knock-in (KI) of specific base-pair substitutions remains challenging. Traditional approaches typically yield low somatic editing efficiencies of 1-4%, creating a significant bottleneck in functional genomics and drug development pipelines. However, recent methodological advances combining optimized CRISPR components with early embryonic selection have demonstrated dramatic improvements in efficiency. This application note details a validated workflow that achieved an almost 17-fold increase in somatic editing efficiency by integrating the Zebrafish Embryo Genotyper (ZEG) device with next-generation sequencing (NGS)-based genotyping [5]. This protocol is presented within the broader context of ZEG early selection research, providing researchers with a comprehensive framework for enhancing precision genome editing outcomes.
Background and Significance
Zebrafish have emerged as a premier vertebrate model for studying human disease mechanisms and therapeutic development due to their genetic tractability, translational relevance, and high reproductive capacity. While CRISPR-Cas9 has greatly expedited the generation of knock-out models via non-homologous end joining (NHEJ), modeling human genetic diseases often requires precise single base-pair substitutions introduced through homology-directed repair (HDR) [5]. The lower frequency of HDR compared to NHEJ has made knock-in model generation laborious and inefficient.
The traditional paradigm of raising injected embryos to adulthood before fin clipping and genotyping consumes significant time, space, and resources. The development of non-invasive early genotyping technologies represents a transformative approach that addresses these limitations while dramatically improving editing outcomes. By selectively raising only those embryos with the highest editing rates, researchers can optimize resource allocation while accelerating research timelines.
Key Experimental Data and Efficiency Outcomes
Quantitative Assessment of Editing Enhancement
Table 1: Comparison of Editing Efficiency Across Experimental Conditions
| Condition | Average Somatic Editing Efficiency | Fold Increase vs. Baseline | Key Parameters |
|---|---|---|---|
| Standard CRISPR Workflow | 1-4% | 1x | Cas9 mRNA, adult fin clipping |
| Optimized Components Only | 5.14% ± 0.71 (LQTS locus) | ~5x | Cas9 protein + NAD ssODN |
| Early Selection (ZEG + NGS) | ~17-20% (from ~1-2% baseline) | 16.8x | Combined optimized components with ZEG pre-selection |
Optimization of CRISPR Components
Systematic evaluation of CRISPR determinants revealed critical factors influencing KI efficiency across two loci (LQTS and BrS) within the cacna1c gene [5]:
- Cas9 Delivery Format: Cas9 protein significantly outperformed Cas9 mRNA across both loci, with NAD conformation yielding 5.14% ± 0.71 versus 0.94% ± 0.35 editing efficiency at the LQTS locus
- ssODN Conformation: Non-target asymmetric PAM-distal (NAD) repair templates demonstrated superior performance compared to target asymmetric PAM-proximal (TAP) configurations
- Indel Frequency: Cas9 protein injections produced significantly higher indel percentages (69.69% ± 1.83 for NAD) compared to mRNA formulations
Table 2: Effect of CRISPR Components on Editing Efficiency
| Component | Option A | Option B | Optimal Selection | Efficiency Impact |
|---|---|---|---|---|
| Cas9 Format | mRNA | Protein | Protein | 2.7-5.5x increase |
| ssODN Conformation | TAP (PAM-proximal) | NAD (PAM-distal) | NAD | Significant improvement |
| gRNA Validation | CRISPR-STAT | ICE | ICE | Better correlation with NGS (r=0.90-0.92) |
The Zebrafish Embryo Genotyper (ZEG) Platform
The ZEG device enables non-invasive cellular extraction from live zebrafish embryos and larvae at 72 hours post-fertilization (hpf) using microfluidic harmonic oscillation on a roughened glass surface [8]. The system processes 24 embryos simultaneously in under 10 minutes, with manual loading and unloading via standard pipette tips. This approach yields PCR-amplifiable genetic material while maintaining >90% embryo survival and no apparent effects on development, morphology, or motor behavior [8].
Protocol Integration Benefits
- Viability Preservation: Enables downstream experimentation or raising of pre-selected embryos
- Workflow Acceleration: Replaces traditional adult fin clipping (2-3 months) with 72 hpf genotyping
- Resource Optimization: Reduces animal husbandry requirements by eliminating unnecessary raising of non-edited animals
- Data Quality Enhancement: Facilitates identification of embryos with highest editing rates for germline transmission
Integrated Experimental Protocol
Stage 1: CRISPR Component Preparation and Optimization
Step 1: Guide RNA Design and Validation
- Design sgRNAs with high cleavage capacity using computational prediction tools
- Validate indel frequency using Inference of CRISPR Edits (ICE) analysis, which demonstrates superior correlation with NGS data (Pearson's r = 0.90-0.92) compared to CRISPR-STAT [5]
- Select target sites with minimal off-target potential using specificity-weighted algorithms
Step 2: Repair Template Design
- Synthesize single-stranded deoxynucleotide (ssODN) repair templates with 120 bp length
- Utilize non-target asymmetric PAM-distal (NAD) conformation
- Position intended mutation considering distance from sgRNA cut site, as this parameter impacts efficiency [5]
Step 3: Ribonucleoprotein (RNP) Complex Assembly
- Complex purified Cas9 protein with validated sgRNA at molar ratio of 1:2
- Incubate at room temperature for 10-15 minutes to form functional RNP complexes
- Consider Cas12a systems as alternative for AT-rich genomic regions [22]
Stage 2: Embryo Microinjection
Step 4: Zygote Collection and Preparation
- Collect zebrafish embryos within 15-30 minutes post-fertilization
- Arrange on injection mold for high-throughput processing
Step 5: RNP Complex Delivery
- Co-inject RNP complexes with NAD ssODN repair templates into zebrafish zygotes
- Optimize injection volume and concentration to balance viability and editing efficiency
Stage 3: Early Genotyping and Selection
Step 6: ZEG-Mediated Cellular Extraction (72 hpf)
- Transfer 72 hpf embryos to ZEG device using standard pipette tips
- Initiate harmonic oscillation protocol for simultaneous processing of 24 embryos
- Collect extracted cellular material in 11 μL total volume [8]
Step 7: Next-Generation Sequencing Genotyping
- Amplify 5 μL of ZEG-extracted material in 11 μL PCR reactions
- Prepare NGS libraries targeting edited loci
- Sequence with sufficient depth (>1000X coverage) for accurate variant calling
- Identify embryos with highest KI rates using bioinformatic analysis
Step 8: Selective Embryo Raising
- Transfer only pre-selected embryos with highest editing efficiency to raising facilities
- Discard non-edited embryos or allocate to alternative experiments
- Achieve germline transmission with significantly reduced animal numbers [5]
Workflow Visualization
Efficiency Comparison
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents and Solutions for Enhanced CRISPR Workflows
| Reagent/Solution | Specification | Function | Optimization Notes |
|---|---|---|---|
| Cas9 Protein | Purified S. pyogenes Cas9 | Catalyzes targeted DNA cleavage | Superior to mRNA format; improves efficiency and reduces off-target effects |
| NAD ssODN | 120 nt, non-target asymmetric PAM-distal | HDR repair template for precise knock-in | Optimal conformation demonstrated across multiple loci |
| ZEG Device | Microfluidic oscillation system | Non-invasive cellular extraction from live embryos | Enables early selection with >90% viability |
| NGS Library Prep | Targeted amplicon sequencing | High-sensitivity detection of editing events | Provides quantitative assessment of editing efficiency |
| ICE Analysis Tool | CRISPR editing inference software | sgRNA validation and indel quantification | Superior correlation with NGS data compared to alternatives |
| DPPY | DPPY, MF:C25H26ClN7O3, MW:508.0 g/mol | Chemical Reagent | Bench Chemicals |
| TfR-T12 | Custom Bioactive Peptide: H-Thr-His-Arg-Pro-Pro-Met-Trp-Ser-Pro-Val-Trp-Pro-OH | Research-grade 13-amino acid peptide for biochemical studies. Product H-Thr-His-Arg-Pro-Pro-Met-Trp-Ser-Pro-Val-Trp-Pro-OH is for Research Use Only (RUO). Not for human or diagnostic use. | Bench Chemicals |
Discussion and Future Directions
The integration of optimized CRISPR components with early embryonic selection represents a paradigm shift in zebrafish genome engineering. The demonstrated 17-fold efficiency increase addresses a critical bottleneck in functional genomics and disease modeling. This workflow significantly reduces the time, cost, and animal numbers required to establish precision genetic models, with particular benefit for alleles with lower intrinsic editing efficiencies [5].
Future applications of this platform could leverage emerging CRISPR technologies including base editors, prime editors, and CRISPR-Cas variants with altered PAM specificities. The combination of early selection with machine learning approaches for predicting editing outcomes may further enhance efficiency and predictability. Additionally, the adaptation of this workflow to other model organisms could transform genome engineering approaches across biomedical research.
The ZEG early selection protocol provides a robust foundation for accelerating genetic research, enabling more rapid functional validation of disease-associated variants identified through next-generation sequencing, and supporting high-throughput drug screening in precision disease models.
This application note demonstrates that dramatic improvements in CRISPR-Cas9 editing efficiency are achievable through a integrated approach combining molecular optimization with technological innovation in genotyping. The 17-fold enhancement obtained through Cas9 protein-mediated editing, NAD ssODN repair templates, and ZEG-based early selection provides researchers with a validated protocol for overcoming the traditional limitations of precision genome editing. This workflow establishes a new standard for efficiency in zebrafish genetic model generation, with broad implications for disease modeling, functional genomics, and therapeutic development.
Solving Common ZEG Challenges: DNA Quality, Amplification and Protocol Refinement
Within zebrafish research, particularly for projects utilizing the Zebrafish Embryo Genotyper (ZEG) for early selection, obtaining high-quality genomic DNA from microscopic biopsies is a critical yet challenging first step [1] [23]. The non-invasive nature of larval fin clipping, while essential for ensuring survival, inherently yields minuscule tissue samples, leading to highly variable and often low DNA concentrations [1]. This variability poses a significant bottleneck, potentially compromising the reliability of subsequent genotyping steps such as PCR and sequencing. This application note details standardized protocols for the precise quantification and quality assessment of DNA derived from ZEG protocols, providing a framework for researchers to ensure data integrity from the earliest stages of sample preparation.
DNA Quantification Methods: A Comparative Analysis
Accurate DNA quantification is paramount. While ultraviolet (UV) absorbance (A260) is a common method, it lacks the sensitivity and specificity required for low-concentration samples and cannot distinguish double-stranded DNA (dsDNA) from degradation products or RNA [24]. For ZEG-derived samples, fluorescence-based methods are strongly recommended.
Fluorescence-Based Assays with PicoGreen/PreciseGreen
Fluorometric assays using dyes like PicoGreen or its equivalent (e.g., PreciseGreen) offer a highly sensitive and selective solution for quantifying dsDNA [24] [25]. These dyes exhibit a >1000-fold fluorescence enhancement upon binding to dsDNA, making them 10,000-fold more sensitive than UV absorbance methods and largely indifferent to the presence of RNA, single-stranded DNA, and free nucleotides [24] [25]. The typical linear range for such assays is 1 pg/μL to 5 ng/μL, perfectly suited for the low DNA concentrations expected from larval fin clips [25].
Table 1: Comparison of DNA Quantification Methods
| Method | Principle | Sensitivity | dsDNA Specificity | Key Advantage | Key Limitation |
|---|---|---|---|---|---|
| UV Absorbance (A260) | Absorption of UV light by nucleic acids | ~2-5 ng/μL | Low | Fast; provides sample purity (A260/280) | Not specific for dsDNA; sensitive to contaminants |
| PicoGreen/PreciseGreen Assay | Fluorescence enhancement upon dsDNA binding | 1 pg/μL - 5 ng/μL | High | Highly sensitive and selective for dsDNA; robust to impurities | Requires a standard curve; reagent cost |
Practical Protocols for DNA Quantification and Quality Control
DNA Quantification via Plate-Based PicoGreen Assay
This protocol is adapted for a 96-well plate format, ideal for processing the high number of samples generated in ZEG experiments [24].
Materials & Reagents
- Quant-iT PicoGreen dsDNA Reagent (or equivalent): Light-sensitive, store at 4°C [24].
- TE Buffer (1x, pH 7.5-8.0): 10 mM Tris-HCl, 1 mM EDTA [24] [25].
- DNA Standard (e.g., Calf Thymus DNA): Typically supplied at 100 μg/mL; dilute to working concentration [24].
- Black 96-Well Microplate: Optically clear bottom for fluorescence measurement [24].
Table 2: Research Reagent Solutions for DNA Quantification
| Item | Function/Description |
|---|---|
| PicoGreen/PreciseGreen Dye | Fluorescent dye that selectively binds dsDNA; the core of the assay's sensitivity [24] [25]. |
| TE Buffer (pH 7.5-8.0) | Dilution buffer; the EDTA chelates divalent cations, inhibiting nuclease activity and protecting DNA integrity [24] [26]. |
| dsDNA Standard | Provides a known concentration to generate a standard curve for calculating unknown sample concentrations [24] [27]. |
| Black 96-Well Plate | Minimizes well-to-well crosstalk and background signal in fluorescence measurements [24]. |
Procedure
- Prepare TE Buffer: Dilute a 20x TE stock to 1x in nuclease-free water. Prepare a volume sufficient for standards, samples, and dye dilution (e.g., 15% extra to account for pipetting losses) [24].
- Prepare DNA Standard Curve: Serially dilute the stock DNA standard in 1x TE to create a standard curve. A medium range curve (e.g., 0, 2.5, 25, 250 ng/mL) is often suitable for ZEG samples [24].
- Prepare PicoGreen Working Solution: Dilute the concentrated PicoGreen reagent 1:200 in 1x TE buffer. Protect from light by wrapping the tube in foil. Prepare enough for 100 μL per standard and sample well, plus ~10% excess [24].
- Plate Setup:
- Add 95-98 μL of 1x TE to each sample well in duplicate.
- Add 2-5 μL of your DNA sample to the respective wells.
- For standards, pipette appropriate volumes of standard and TE directly into the plate according to your dilution scheme [24].
- Add Dye and Incubate: Add 100 μL of the diluted PicoGreen working solution to each well (both standards and samples). Mix thoroughly by pipetting up and down 5-10 times. Cover the plate and incubate in the dark at room temperature for 5 minutes [24].
- Fluorescence Measurement: Read the plate using a fluorescence microplate reader with filters/excitation and emission close to ~485/20 nm and ~528/20 nm, respectively [24].
- Data Analysis: Generate a standard curve by plotting the fluorescence values of the standards against their known concentrations. Use the linear regression equation from this curve to calculate the DNA concentration in your unknown samples [25].
Assessment of DNA Quality and Integrity
Quantification alone is insufficient; assessing quality is crucial. DNA degradation, often from oxidative stress, hydrolysis, or enzymatic (nuclease) activity, can severely impact downstream genotyping success [26].
- Prevention During Extraction: The use of chelating agents like EDTA in buffers is critical to inhibit metal-dependent nucleases [1] [26]. For tough tissues, a combination of mechanical homogenization (e.g., using a bead mill like the Bead Ruptor Elite) with optimized chemical lysis can maximize yield while minimizing shearing and degradation [26].
- Post-Extraction QC: While fluorescence assays confirm the presence of dsDNA, techniques like fragment analysis (e.g., on a bioanalyzer) provide a detailed profile of DNA size distribution, directly revealing the extent of fragmentation in a sample [26].
The workflow below outlines the key steps from sample preparation to quality assessment.
The successful implementation of the Zebrafish Embryo Genotyper (ZEG) early selection protocol is fundamentally dependent on the initial quality and quantity of DNA. By replacing less specific UV absorbance methods with sensitive, dsDNA-specific fluorescence assays like PicoGreen, researchers can accurately quantify the low DNA yields typical of fin clips. Coupling this precise quantification with robust extraction methods that prevent degradation and subsequent quality checks creates a reliable pipeline. Adhering to these standardized protocols for addressing variable DNA yields ensures that genotyping data is reliable, thereby enhancing the efficiency and ethical standing of zebrafish-based genetic research.
The Zebrafish Embryo Genotyper (ZEG) protocol represents a significant advancement in zebrafish research by enabling the rapid, non-lethal extraction of genetic material from live embryos and larvae for early genotypic selection [8]. A typical ZEG extraction yields a low-volume (11 µL) sample containing cellular material and genomic DNA, which is then used as a template for PCR amplification [8]. The core challenge is that PCR from these samples must be exceptionally robust and sensitive due to the limited quantity and potentially fragmented nature of the obtained DNA. Success hinges on a PCR strategy specifically adapted for this unique template, with primer design and amplicon size being the most critical parameters to ensure genotyping accuracy while maintaining high embryo viability.
Critical Considerations for Primer and Amplicon Design
The design of primers and selection of amplicon size must account for the specific characteristics of DNA derived from ZEG extractions.
Optimal Amplicon Size Range
For conventional PCR following ZEG extraction, amplicons ranging from approximately 70 base pairs (bp) to 300 bp are recommended [8]. This range ensures efficient amplification from the sometimes fragmented DNA obtained via the ZEG protocol. While a 296 bp amplicon has been successfully amplified from ZEG samples, shorter products typically demonstrate higher efficiency [8]. For quantitative applications (qPCR), a narrower range of 70-150 bp is ideal, as it allows for highly efficient amplification under standard cycling conditions [28].
Table 1: Recommended Amplicon Sizes for ZEG Sample PCR
| Application | Recommended Size Range | Rationale |
|---|---|---|
| Conventional PCR (Genotyping) | 70 - 300 bp | Compatible with potentially fragmented DNA from ZEG extraction [8]. |
| Quantitative PCR (qPCR) | 70 - 150 bp | Maximizes amplification efficiency and accuracy for quantification [28]. |
Primer Design Parameters
Careful primer design is paramount for successful PCR from ZEG samples. The following parameters, synthesized from established molecular biology guidelines, should be strictly adhered to:
- Primer Length: Design primers between 18 and 30 nucleotides [29] [30] [28]. This length provides an optimal balance of specificity and binding energy.
- Melting Temperature (Tm): Primer pairs should have calculated Tms within a narrow 2-5°C of each other [29] [31] [30]. The optimal Tm range for primers is typically 60–64°C [28].
- Annealing Temperature (Ta): Set the annealing temperature 3–5°C below the calculated Tm of the primers [31] [28]. This can be optimized further if non-specific amplification occurs.
- GC Content: Aim for a GC content of 40–60%, with 50% being ideal [30] [28]. Avoid long stretches of a single nucleotide.
- 3' End Specificity: Ensure the last 5 nucleotides at the 3' end of the primer are unique to the target sequence to prevent non-specific amplification. Avoid runs of G or C bases at the 3' end, as this can promote mis-priming [29] [31].
- Secondary Structures: Screen primers for self-dimers, hairpins, and cross-dimers. The free energy (ΔG) for any stable secondary structure should be weaker (more positive) than –9.0 kcal/mol [28]. Tools like the IDT OligoAnalyzer are invaluable for this check [32] [28].
Table 2: Essential Primer Design Parameters for ZEG Sample PCR
| Parameter | Optimal Value/Range | Consequence of Deviation |
|---|---|---|
| Length | 18 - 30 bases | Shorter primers may lack specificity; longer primers can reduce efficiency [29] [28]. |
| Tm Difference | ≤ 2-5°C | Large Tm differences prevent both primers from binding simultaneously, reducing yield [29] [30]. |
| GC Content | 40-60% | GC content outside this range can lead to non-specific binding or unstable primer-template duplexes [30]. |
| Secondary Structures | ΔG > -9.0 kcal/mol | Stable dimers or hairpins, especially at the 3' end, can prevent primer binding and abort the reaction [28]. |
Experimental Protocol: PCR Workflow for ZEG Samples
Sample Preparation and PCR Setup
- ZEG Extraction: Perform cellular extraction from live zebrafish embryos or larvae using the ZEG device according to the established protocol [8]. The final output is typically 11 µL of fluid containing cells and DNA.
- Template Addition: Use 5 µL of the ZEG-extracted fluid directly as the PCR template in an 11-25 µL total reaction volume [8]. No additional DNA purification is necessary.
- Reaction Master Mix: Prepare a master mix on ice containing the following components. Using a hot-start DNA polymerase is strongly recommended to minimize non-specific amplification and primer-dimer formation [31].
- PCR Buffer (as supplied with the polymerase, typically 1X final concentration)
- MgClâ‚‚ (1.5 - 2.0 mM final concentration; optimize if needed) [30]
- dNTPs (200 µM of each dNTP) [30]
- Forward and Reverse Primers (0.1 - 0.5 µM each final concentration) [30]
- Hot-Start DNA Polymerase (e.g., 1.25 units per 50 µL reaction) [30]
PCR Cycling Conditions
The following cycling conditions are a robust starting point for amplifying a ~200 bp amplicon from ZEG samples and can be adjusted based on the specific product size and primer characteristics.
Diagram 1: PCR workflow for ZEG samples
Post-PCR Analysis and Embryo Handling
- Genotyping Analysis: Following PCR, the product can be analyzed by standard methods such as agarose gel electrophoresis, Sanger sequencing, or High-Resolution Melt Analysis (HRMA) [8].
- Embryo/Larval Recovery: After ZEG processing, embryos and larvae show greater than 90% survival. Processed animals can be returned to embryo medium and raised for subsequent phenotypic analysis or to establish mutant lines, fully leveraging the non-lethal nature of the protocol [8].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents and Kits for ZEG-based Genotyping
| Reagent/Kits | Function/Application | Rationale for Use |
|---|---|---|
| Hot-Start DNA Polymerase | PCR amplification | Reduces primer-dimer formation and non-specific amplification during reaction setup, crucial for low-template ZEG samples [31]. |
| High-Sensitivity DNA Assay Kits | Quantification of gDNA from ZEG extract | Accurately measures low concentrations of DNA to assess extraction yield and normalize template input if necessary. |
| HRMA-compatible Master Mix | Genotyping by High-Resolution Melt Analysis | Enables precise mutation detection post-PCR without additional processing, streamlining the ZEG workflow [8]. |
| ZEG Device | Automated cellular/DNA extraction | Core technology for non-lethal sampling of live zebrafish embryos and larvae [8]. |
| B-Raf IN 9 | 1-[4-(6-Oxo-1-phenyl-4,5-dihydropyridazin-3-yl)phenyl]-3-phenylthiourea | |
| CD73-IN-10 | 5-[4-[(1S,2S)-2-(difluoromethyl)cyclopropyl]-1-methylpyrazolo[3,4-b]pyridin-6-yl]-1H-pyrimidine-2,4-dione | High-purity 5-[4-[(1S,2S)-2-(difluoromethyl)cyclopropyl]-1-methylpyrazolo[3,4-b]pyridin-6-yl]-1H-pyrimidine-2,4-dione for research applications. This product is For Research Use Only. Not for human or veterinary use. |
Integrating optimized PCR protocols with the ZEG extraction method creates a powerful pipeline for high-throughput zebrafish research. By adhering to the specified guidelines for amplicon size (70-300 bp) and rigorous primer design parameters, researchers can achieve highly sensitive and specific genotyping results. This approach dramatically accelerates research by enabling early phenotypic-genotypic correlation in live animals, reducing animal husbandry costs, and maximizing the efficiency of screening for desired genetic models.
The Zebrafish Embryo Genotyper (ZEG) protocol enables high-throughput, non-lethal genetic screening of live zebrafish embryos by extracting genetic material through microfluidic harmonic oscillation. However, the extracted genomic DNA (gDNA) can be partially degraded, presenting a significant challenge for reliable polymerase chain reaction (PCR) amplification. This application note demonstrates that integrating Restorase DNA Polymerase into the downstream genotyping workflow effectively overcomes this limitation. Restorase significantly improves the amplification of damaged DNA templates, thereby enhancing the reliability, efficiency, and throughput of early mutant selection within ZEG-based research pipelines.
The Zebrafish Embryo Genotyper (ZEG) is an automated microfluidic system designed for rapid cellular extraction from live zebrafish embryos and larvae. Its key advantage is the ability to obtain genetic material for genotyping while keeping the animals alive, with survival and genotyping sensitivity rates both exceeding 90% [8]. The process involves the harmonic oscillation of an embryo on a roughened glass surface, which generates sufficient genetic material from 24 individual embryos in less than 10 minutes [4] [8].
While the ZEG protocol does not affect the gross morphology or long-term development of the embryos, cellular and molecular characterization has revealed that the process results in a low-level acute stress response and can yield gDNA that is partially degraded into smaller fragments [10] [4]. This degradation poses a problem for PCR, especially when amplifying larger DNA fragments. Studies show that while amplicons smaller than 260 base pairs (bp) are reliably amplified, the sensitivity and specificity for larger amplicons (e.g., 550 bp and 800 bp) can be significantly reduced with standard polymerases [4]. Restorase DNA Polymerase is engineered to address this exact problem, modifying damaged sites on the DNA template to facilitate successful amplification [4].
Application Data: Restorase Enhances PCR from ZEG-Derived DNA
Research directly applicable to the ZEG context has validated the use of Restorase. The following data summarizes the quantitative and qualitative improvements observed when using Restorase DNA Polymerase on DNA extracted via the ZEG protocol.
Table 1: DNA Yield from ZEG Extraction on Xenopus Tropicalis Embryos (Adapted from [4])
| Developmental Stage | Number of Samples (n) | DNA Concentration Range (ng/μL) | Average DNA Concentration (ng/μL) |
|---|---|---|---|
| NF 19 | 4 | 63 - 110 | 81.5 |
| NF 25 | 5 | 3 - 116 | 64.0 |
| NF 37 | 5 | 66 - 91 | 76.8 |
| NF 42 | 5 | 7 - 37 | 22.9 |
The table above highlights the high variability in DNA concentration obtained from ZEG, which is independent of cell count and indicates potential variability in DNA quality [4].
Table 2: Performance of Restorase DNA Polymerase on Suboptimal ZEG-Derived Templates [4]
| PCR Polymerase | Amplification Success for Small Fragments (<260 bp) | Amplification Success for Large Fragments (550-800 bp) | Notes on Specificity |
|---|---|---|---|
| Standard GoTaq Polymerase | Successful across all stages | Reduced sensitivity and specificity; faint or non-specific bands | Often fails with primers that do not work optimally |
| Restorase DNA Polymerase | Successful | Significantly increased PCR amplification | Allowed amplification with primers that failed with standard GoTaq |
The core finding is that Restorase significantly increased PCR amplification of poor quality gDNA templates obtained via the ZEG protocol. In some cases, it enabled amplification with primers that were completely ineffective with standard polymerases [4].
Experimental Protocol: Genotyping with ZEG and Restorase DNA Polymerase
Below is a detailed step-by-step protocol for genotyping live zebrafish embryos using the ZEG device, followed by PCR with Restorase DNA Polymerase.
A. ZEG-Based Cellular and DNA Extraction
- Preparation: Collect zebrafish embryos or larvae at the desired stage (e.g., 24-72 hours post-fertilization). Manually dechorionate the embryos if necessary, though the vitelline membrane does not interfere with the protocol [4].
- Loading: Using a standard pipette tip or transfer pipette, manually load individual live embryos into the wells of the ZEG device.
- Extraction: Run the ZEG device to perform microfluidic harmonic oscillation. This process typically takes 10 minutes for 24 samples [8].
- Collection: After the run, unload the embryos first, transferring them to fresh embryo medium for continued development. The survival rate is expected to be >90% [8].
- Sample Retrieval: Collect the 11 µL fluid containing the extracted cells and DNA from each well of the ZEG device. This fluid is used directly as a template for PCR.
B. PCR Amplification Using Restorase DNA Polymerase
- Prepare PCR Master Mix (for one 11 µL reaction):
- 5.5 µL of 2X Restorase Master Mix
- 0.3 µM of each forward and reverse primer
- Nuclease-free water to a final volume of 6 µL
- Add Template: Add 5 µL of the fluid collected from the ZEG device, bringing the total PCR reaction volume to 11 µL [8].
- PCR Amplification: Run the following thermocycling protocol, optimized for Restorase:
- Initial Denaturation: 95°C for 5 minutes
- Amplification (35-40 cycles):
- Denaturation: 95°C for 15-30 seconds
- Annealing: 55-65°C (primer-specific) for 15-30 seconds
- Extension: 72°C for 15-60 seconds per 1 kb
- Final Extension: 72°C for 5 minutes
- Downstream Analysis: The PCR products can now be used for standard downstream applications such as agarose gel electrophoresis, high-resolution melt analysis (HRMA), or Sanger sequencing [4] [8].
Diagram 1: Restorase overcomes the key challenge of damaged DNA from the ZEG protocol to enable reliable genotyping.
The Scientist's Toolkit: Essential Reagents and Materials
Table 3: Key Research Reagent Solutions for ZEG and Restorase-Based Genotyping
| Item | Function/Application | Example/Note |
|---|---|---|
| Zebrafish Embryo Genotyper (ZEG) Device | Automated microfluidic system for non-lethal cellular and DNA extraction from live embryos. | Enables high-throughput processing of 24 embryos in <10 minutes [8]. |
| Restorase DNA Polymerase | Specialized enzyme for PCR amplification of damaged or suboptimal DNA templates. | Critical for robust amplification of partially degraded gDNA from ZEG extraction [4]. |
| High-Resolution Melt Analysis (HRMA) Master Mix | For post-PCR analysis to identify genetic variants (e.g., SNPs, indels). | Used with LightScanner Master Mix; compatible with ZEG/Restorase PCR products [8]. |
| Trypan Blue & DAPI | Viability stain and nuclear counterstain for quantifying extracted cells. | Used for cell counting after ZEG extraction, though DNA yield is not directly correlated with cell count [4] [8]. |
| E3 Embryo Medium | Standard medium for raising and maintaining zebrafish embryos. | Used for embryo recovery after ZEG processing [8]. |
| Melatonin-d3 | Melatonin-d3, MF:C13H16N2O2, MW:235.30 g/mol | Chemical Reagent |
The integration of Restorase DNA Polymerase into the ZEG genotyping pipeline represents a significant enhancement for researchers conducting early selection in zebrafish models. By effectively mitigating the challenge of partially degraded DNA templates, Restorase ensures that genotype-phenotype linkages in F0 mutant studies are more robust and reliable [4]. This combined approach aligns with the 3Rs principles (Replacement, Reduction, and Refinement) in animal research by allowing scientists to identify and raise only the mutants of interest, saving resources and reducing the number of animals required [4].
The ZEG protocol, empowered by Restorase, is a powerful tool that accelerates high-throughput genetic screens, functional validation of disease-associated variants, and drug discovery efforts in the versatile zebrafish model.
Application Note
This application note details a standardized protocol for assessing acute stress responses in zebrafish embryos following manipulation with the Zebrafish Embryo Genotyper (ZEG) device. Early genotyping is crucial for improving the efficiency of generating precise genetic models, such as CRISPR-Cas9 knock-ins [5]. However, any embryonic manipulation necessitates a thorough investigation of its potential to induce acute cellular stress, which could confound phenotypic analyses in developmental studies and drug screening. The protocols herein enable researchers to quantify the molecular and cellular impact of the ZEG procedure, ensuring that experimental results reflect the genetic manipulation rather than procedure-induced stress.
The ZEG device allows for high-throughput genomic DNA (gDNA) extraction from live zebrafish embryos at 72 hours post-fertilization (hpf) with minimal physical impact on the embryo [5]. Independent studies confirm that embryos subjected to the ZEG protocol survive and develop without gross morphological abnormalities [4] [10]. Nevertheless, a comprehensive cellular and molecular characterization reveals that the procedure elicits a low-level, acute stress response [10]. This makes the integrated stress assessment protocol an essential component of the ZEG workflow for generating high-fidelity data.
The Zebrafish Embryo Genotyper (ZEG) is an automated microfluidic system designed to overcome the "genotyping bottleneck." It extracts gDNA from 24 to 72 hpf zebrafish embryos in a high-throughput manner, allowing researchers to identify and raise genetically selected embryos without sacrificing them [5] [4]. This early selection is particularly valuable for CRISPR-Cas9 experiments, where it can lead to a nearly 17-fold increase in somatic editing efficiency and significantly improve the odds of obtaining germline transmission [5].
From a stress biology perspective, any external manipulation poses a potential challenge to embryonic homeostasis. Cells possess evolved stress response mechanisms to maintain integrity under stress, activating adaptive pathways to manage issues like the presence of misfolded proteins or reactive oxygen species (ROS) [33]. The documented ability of ZEG-treated embryos to develop into morphologically normal, feeding-stage tadpoles suggests that the procedure is minimally invasive [4]. However, sensitive molecular assays are required to detect and quantify the transient activation of stress pathways, ensuring that the procedure does not inadvertently alter the developmental processes under investigation.
Key Experimental Protocols
Protocol for ZEG-Assisted Genotyping and Embryo Recovery
This protocol describes the use of the ZEG device for gDNA extraction and the subsequent recovery of embryos for molecular stress assessment.
- Principle: The ZEG device uses microfluidic harmonic oscillation of an embryo on an abrasive surface to generate a sufficient amount of gDNA for PCR analysis without compromising embryo viability [4].
- Reagents and Equipment:
- Zebrafish Embryo Genotyper (ZEG) device.
- Wild-type or genetically engineered zebrafish embryos (72 hpf).
- E3 embryo medium.
- Pronase solution (30 mg/mL in E3) for dechorionation, if necessary [34].
- DNA extraction buffer.
- Standard PCR reagents and thermocycler.
- Procedure:
- Embryo Preparation: Manually dechorionate 72 hpf embryos or treat with pronase solution for 5-6 minutes at 22°C to soften the chorion, followed by three washes in fresh E3 buffer [34].
- ZEG Operation: Load individual embryos into the ZEG device. The automated process involves oscillating the embryo to collect genetic material into the extraction buffer. This process takes approximately 10 minutes for 24 embryos [4].
- Embryo Recovery: Following the ZEG procedure, carefully collect the live embryos and transfer them to fresh E3 medium.
- Genotyping: Use the extracted gDNA for downstream PCR amplification. For optimal results with potentially fragmented gDNA, use polymerases designed for damaged DNA, such as Restorase, which can significantly improve amplification [4].
- Embryo Rearing: Raise the recovered embryos according to standard laboratory protocols. They can be fixed at desired timepoints for molecular analysis or raised to adulthood.
Protocol for Molecular Analysis of Acute Stress Response
This protocol outlines the methods for evaluating the cellular stress response in ZEG-manipulated embryos using gene expression analysis.
- Principle: Exposure to stress triggers the rapid transcriptional activation of specific genes. Quantifying the expression of these marker genes using qRT-PCR provides a sensitive measure of the acute stress response [10].
- Reagents and Equipment:
- TRIzol Reagent or equivalent for RNA extraction.
- DNase I.
- Reverse transcription kit.
- SYBR Green qPCR master mix.
- Primers for stress-responsive genes (e.g., hsp70, fos, gadd45ba).
- Housekeeping gene primers (e.g., β-actin, ef1α).
- Real-time PCR detection system.
- Procedure:
- Sample Collection: At the desired timepoint post-ZEG manipulation (e.g., 1 hour, 6 hours), pool 5-10 embryos per experimental group (ZEG-treated and untreated controls) and homogenize in TRIzol.
- RNA Extraction: Isolate total RNA following the TRIzol manufacturer's protocol. Treat samples with DNase I to remove genomic DNA contamination.
- cDNA Synthesis: Synthesize first-strand cDNA from 1 µg of total RNA using a reverse transcription kit.
- Quantitative PCR (qPCR): Perform qPCR reactions in triplicate for each sample. The reaction mix should contain SYBR Green master mix, gene-specific primers, and cDNA template.
- Data Analysis: Calculate the relative fold change in gene expression using the 2^(-ΔΔCt) method, normalizing to housekeeping genes and relative to the untreated control group.
Data Presentation and Analysis
Quantitative Data on ZEG Procedure Outcomes
Table 1: Embryo Viability and Genotyping Success Post-ZEG Procedure
| Metric | Result | Experimental Context |
|---|---|---|
| Embryo Survival Rate | 100% viability to feeding stage [4] | Xenopus tropicalis and laevis embryos post-ZEG extraction. |
| Gross Morphology | No observable abnormalities [4] | Tadpoles showed normal development and behavior. |
| gDNA Concentration | 7 - 81 ng/µL (Average: 34.0 ng/µL) [4] | Measured from 72 hpf zebrafish embryos using NanoDrop. |
| PCR Success Rate | Fragments up to 800 bp successfully amplified [4] | Optimal amplification for fragments <260 bp; larger amplicons may require specialized polymerases. |
| Acute Stress Response | Low-level, transient activation of stress genes [10] | Molecular analysis of ZEG-treated zebrafish embryos. |
Key Stress Response Genes for Assessment
Table 2: Candidate Genes for Profiling Acute Stress in Zebrafish Embryos
| Gene Name | Function / Pathway | Utility in Stress Assessment |
|---|---|---|
| hsp70 | Heat Shock Protein / Protein Folding & Proteostasis [33] | Canonical marker for proteotoxic stress; indicates protein misfolding. |
| fos | Transcription Factor / AP-1 Complex, MAPK Signaling [35] | Immediate-early gene activated by diverse stressors, including cellular injury. |
| gadd45ba | Growth Arrest and DNA-Damage-Inducible / Cell Cycle & DNA Repair [36] | Marker for genotoxic stress and cellular growth arrest. |
| chop (ddit3) | C/EBP-homologous protein / ER Stress-Induced Apoptosis [36] | Key mediator of ER stress-induced apoptosis; marker for severe, prolonged ER stress. |
| atf4 | Activating Transcription Factor 4 / Integrated Stress Response [36] | Central transcription factor in the PERK-eIF2α branch of the unfolded protein response. |
Signaling Pathways in Acute Cellular Stress
The following diagram summarizes the key signaling pathways activated during cellular stress, which can be investigated following the ZEG procedure.
Cellular Stress Signaling Pathways. This diagram illustrates the major signaling cascades (ER stress, heat shock, and oxidative stress response) activated by experimental stressors, leading to either adaptive cellular recovery or programmed cell death.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for ZEG and Stress Assessment Protocols
| Item | Function / Application | Specific Examples / Notes |
|---|---|---|
| Zebrafish Embryo Genotyper (ZEG) | Automated, high-throughput gDNA extraction from live embryos. | Enables genotyping of 24 embryos in ~10 minutes with high viability [4]. |
| Restorase DNA Polymerase | PCR amplification of partially degraded or damaged gDNA. | Critical for reliable genotyping when ZEG-extracted DNA quality is suboptimal [4]. |
| SYBR Green qPCR Master Mix | Sensitive detection of mRNA expression levels for stress genes. | Used for quantifying transcript levels of markers like hsp70 and fos [10]. |
| Stress Gene Primers | Specific amplification of stress-responsive transcripts. | Panels include genes for ER stress (chop, atf4), heat shock (hsp70), and DNA damage (gadd45ba). |
| Pronase | Enzymatic dechorionation of zebrafish embryos. | Softens the chorion for easier manipulation; 30 mg/mL in E3 buffer for 5-6 minutes [34]. |
The Zebrafish Embryo Genotyper (ZEG) device represents a significant advancement in genetic research by enabling early, non-lethal genotyping of live zebrafish embryos. This protocol allows researchers to identify and selectively raise embryos with desired genetic edits as early as 72 hours post-fertilization, significantly reducing the time, cost, and number of animals required to establish stable genetic lines [5]. The core principle involves minimally invasive extraction of genomic DNA from embryos followed by next-generation sequencing (NGS) analysis to identify individuals with the highest rates of correct editing [5].
This application note explores the adaptation of the ZEG early selection principle beyond its original zebrafish model to the closely related Xenopus system and other model organisms. We provide detailed methodological modifications required for successful implementation, quantitative efficiency comparisons, and standardized reagents to facilitate cross-species application in genetic research and drug development pipelines.
Core Protocol and Efficiency Data
Original ZEG Protocol and Quantitative Performance
The fundamental ZEG protocol involves micro-sampling of embryonic tissue from live zebrafish embryos at 72 hours post-fertilization, with subsequent genomic DNA extraction and NGS-based genotyping. Validation studies confirm that although ZEG triggers a low-level acute cellular stress response, it produces no long-lasting effects on embryo morphology, survival, or development, supporting its use for various downstream assays [10].
Table 1: ZEG Protocol Efficiency in Zebrafish CRISPR Editing
| Condition | Average Somatic Editing Efficiency | Germline Transmission Events | Key Advantage |
|---|---|---|---|
| Standard CRISPR (No ZEG) | 1-4% (Typical range) | Not reported | Baseline efficiency |
| ZEG with Cas9 mRNA & NAD ssODN | 1.04% ± 0.22 (BrS locus) | Not observed | Moderate improvement |
| ZEG with Cas9 Protein & NAD ssODN | 2.83% ± 0.75 (BrS locus) 5.14% ± 0.71 (LQTS locus) | Confirmed in pre-selected embryos | ~17-fold increase in somatic editing efficiency |
| Optimal ZEG Conditions | Combined minimally invasive sampling & NGS | Successful establishment of lines | Particularly beneficial for low-efficiency alleles |
The data demonstrate that combining ZEG pre-selection with optimized CRISPR components (Cas9 protein and non-target asymmetric PAM-distal single-stranded deoxynucleotide repair templates) yields substantially higher somatic editing efficiency compared to traditional approaches [5]. This efficiency gain is particularly pronounced for alleles that would otherwise exhibit lower editing rates, making previously challenging genetic modifications feasible.
Experimental Protocol: Zebrafish ZEG Implementation
Materials Required:
- Zebrafish Embryo Genotyper (ZEG) device
- Cas9 protein (recommended) or mRNA
- Non-target asymmetric PAM-distal (NAD) ssODN repair templates (120 BP)
- Microinjection apparatus
- Next-generation sequencing platform
- Standard molecular biology reagents (PCR reagents, DNA extraction kits)
Procedure:
- Embryo Preparation: Generate embryos through natural mating or in vitro fertilization. Maintain embryos in appropriate medium at 28.5°C until 72 hours post-fertilization [5].
- CRISPR Component Injection: Inject one-cell stage embryos with pre-complexed Cas9 protein (recommended) or mRNA with sgRNA and NAD ssODN repair templates [5].
- ZEG Sampling: At 72 hours post-fertilization, use the ZEG device to perform minimally invasive tissue sampling from each live embryo. The protocol causes minimal lethality when properly executed [5].
- Genomic DNA Extraction: Extract genomic DNA from sampled tissue using standard commercial kits, following manufacturer's protocols.
- NGS Library Preparation & Sequencing: Prepare sequencing libraries targeting the edited locus and perform NGS on an appropriate platform to achieve sufficient coverage for accurate variant calling.
- Bioinformatic Analysis: Process sequencing data to identify embryos with highest correct editing rates using appropriate genotyping software.
- Selective Raising: Based on genotyping results, selectively raise embryos with desired editing profiles to adulthood for germline transmission analysis.
Protocol Adaptation for Xenopus Models
Critical Modifications for Xenopus Systems
Adapting the ZEG early selection principle for Xenopus requires significant modifications to account for developmental, physiological, and genetic differences between zebrafish and frog model systems.
Table 2: Key Modifications for Xenopus Adaptation
| Parameter | Zebrafish Protocol | Xenopus Adaptation | Rationale |
|---|---|---|---|
| Sampling Developmental Stage | 72 hours post-fertilization | Tailbud stages (approximately stage 30-35) | Matches developmental maturity; easier micro-sampling |
| Sample Processing | Standard DNA extraction | Yolk platelet removal [37] and specialized lysis [37] | High yolk content in Xenopus oocytes/embryos interferes with molecular biology reactions [37] |
| Genomic DNA Extraction | Commercial kits | Modified lysis with 10µL chilled cell lysis buffer per embryo/oocyte, homogenization, 5000g centrifugation to pellet yolk debris [37] | Adapts to different cellular composition and yolk content |
| CRISPR Delivery | Microinjection into one-cell stage | Targeted microinjection for tissue-specific editing [38] | Enables tissue-specific mutagenesis in F0 crispants |
| gRNA Design | Standard zebrafish optimization | Xenopus-optimized gRNA design software and rules [38] | Accounts for species-specific genomic differences |
Xenopus-Specific Experimental Protocol
Materials Required:
- Xenopus laevis or Xenopus tropicalis oocytes/embryos
- Modified Barth's Solution (MBS) [37] or 0.25x Marc's Modified Ringer's solution (MMR) [37]
- 2% cysteine in 0.1x MMR (pH 8.2) for jelly coat removal [37]
- Cell lysis buffer [37]
- Micropestles for homogenization [37]
- CRISPR components: Cas9 protein, Xenopus-optimized sgRNAs, repair templates
Procedure:
- Gamete Collection & Embryo Preparation:
- Obtain defolliculated X. laevis oocytes from commercial suppliers or isolate as described [37].
- Culture isolated oocytes in Modified Barth's Solution (MBS) until use [37].
- For embryos: Isolate X. laevis eggs and fertilize as described [37]. Remove jelly coats using 2% cysteine solution (3-5 minutes exposure) followed by multiple rinses with 0.25x MMR [37].
- Culture fertilized embryos in 0.25x MMR until desired stages [37].
Sample Processing for DNA Extraction:
- Collect desired number of embryos/oocytes (minimum five per sample) in 1.5 mL tube [37].
- Add 10µL chilled cell lysis buffer per embryo/oocyte and homogenize with micropestle on ice [37].
- Centrifuge at 5000g at 4°C for 10 minutes to pellet yolk and insoluble pigments [37].
- Transfer supernatant to new tube, avoiding pellet [37].
Genotyping & Early Selection:
- Extract genomic DNA from processed supernatant using standard methods.
- Perform NGS-based genotyping as in zebrafish protocol.
- Select embryos with desired editing profiles for raising.
Research Reagent Solutions
Table 3: Essential Research Reagents for Multi-Species ZEG Adaptation
| Reagent/Category | Specific Examples | Function | Species Considerations |
|---|---|---|---|
| CRISPR Components | Cas9 protein (recommended), sgRNAs, NAD ssODN repair templates (120 BP) [5] | Induce precise genetic edits | Cas9 protein outperforms mRNA in both zebrafish and Xenopus [5] [38] |
| Cell Lysis Buffers | 10x cell lysis buffer diluted to 1x [37] | Extract protein/nucleic acids while preserving integrity | Xenopus requires yolk-specific removal steps [37] |
| Oocyte/Embryo Culture Media | Modified Barth's Solution (MBS) [37], 0.25x MMR [37] | Maintain oocyte/embryo viability during experiments | Species-specific formulations required |
| Genotyping Tools | Next-generation sequencing platforms, REDItools [39], specialized bioinformatic pipelines | Identify and quantify editing events | Must account for species-specific genomic features |
| Specialized Equipment | ZEG device, microinjection apparatus, vertical electrophoresis systems [37] | Enable precise manipulation and analysis | Similar principles, potential size scaling for Xenopus |
Applications and Future Directions
The adaptation of early selection principles across model organisms represents a paradigm shift in efficiency for genetic model generation. The cross-application of ZEG-inspired methodology addresses a critical bottleneck in functional genomics - the laborious process of establishing stable genetic lines. For drug development professionals, this approach accelerates the creation of disease models for target validation and therapeutic screening.
In zebrafish, the ZEG protocol has demonstrated particular value for modeling human disease-associated point mutations in genes like CACNA1C, where complete knockout models fail to recapitulate specific pathophysiological mechanisms [5]. Similar applications in Xenopus could leverage the system's advantages for studying specific disease mechanisms, particularly where the frog's physiology or developmental processes better model human biology.
Future methodological developments will likely focus on increasing the throughput of early genotyping platforms, improving the efficiency of homology-directed repair across species, and developing computational frameworks that better predict germline transmission potential from early somatic editing data. As single-cell sequencing technologies advance, integration of these approaches with early selection protocols may enable even more precise genetic manipulation with reduced off-target effects.
Evaluating ZEG Efficacy: Survival, Development and Research Impact Metrics
Within the field of zebrafish functional genomics, the generation of precise knock-in models using CRISPR-Cas9 remains challenging due to typically low homology-directed repair (HDR) efficiency. The Zebrafish Embryo Genotyper (ZEG) device provides a promising solution by enabling minimal-invasive tissue biopsy and genomic DNA extraction from live embryos at 72 hours post-fertilization (hpf), allowing for early genotyping and selective rearing of successfully edited animals [5]. This application note synthesizes experimental evidence to validate the viability and normal development of ZEG-processed embryos across cellular, molecular, and organ system levels, providing researchers with standardized protocols and analytical frameworks for longitudinal studies. The implementation of ZEG early selection protocol has demonstrated an almost 17-fold increase in somatic editing efficiency in CRISPR-Cas9 experiments, with particular benefit for alleles with lower editing rates [5].
Viability and Developmental Validation Data
Comprehensive validation studies have confirmed that ZEG-processed embryos exhibit normal development and viability across multiple biological levels, despite transient molecular responses to the procedure.
Table 1: Comprehensive Viability and Developmental Metrics of ZEG-Processed Embryos
| Validation Parameter | Experimental Findings | Temporal Pattern | Assay Methodology |
|---|---|---|---|
| Gross Morphology | Unaffected [10] | No long-term effects observed | Visual inspection under stereo-microscope |
| Survival Rate | Unaffected [10] | Consistent long-term viability | Survival tracking to adulthood |
| Acute Stress Response | Low-level activation [10] | Transient, resolves over time | Gene expression analysis of stress-responsive markers |
| Tissue Integrity | Preserved in examined tissues [10] | Maintained long-term | Histological analysis of specific tissues |
| Organ Development | Normal volume and proportionality [18] | Follows standard growth trajectory | Mueller matrix OCT with deep learning segmentation |
| Somatic Editing Efficiency | ~5.14% at LQTS locus with optimal conditions [5] | Stable editing confirmed | NGS-based genotyping |
| Germline Transmission | Demonstrated in pre-selected embryos [5] | Successful transmission to F1 generation | Breeding studies of raised adults |
The quantitative developmental trends of zebrafish embryos and larvae from 1-19 days post-fertilization (dpf) have been successfully characterized using non-invasive imaging technologies, establishing normative benchmarks for organ development. Volume measurements of the body, eyes, spine, yolk sac, and swim bladder demonstrate steady growth patterns without significant deviations in ZEG-processed individuals [18]. The successful application of deep learning-based segmentation with Mueller matrix optical coherence tomography (OCT) provides a high-resolution methodology for validating that ZEG procedures do not alter standard developmental trajectories [18].
Table 2: ZEG Protocol Outcomes in CRISPR-Cas9 Editing Experiments
| Experimental Condition | Average Somatic Editing Efficiency | Germline Transmission Results |
|---|---|---|
| LQTS Locus (NAD ssODN + Cas9 Protein) | 5.14% ± 0.71 [5] | Confirmed in pre-selected embryos [5] |
| BrS Locus (NAD ssODN + Cas9 Protein) | 2.83% ± 0.75 [5] | Confirmed in pre-selected embryos [5] |
| LQTS Locus (NAD ssODN + Cas9 mRNA) | 0.94% ± 0.35 [5] | Not specifically reported |
| Traditional Late-Stage Genotyping | Typically 1-4% [5] | Variable, requires larger founder populations |
Experimental Protocols
ZEG Device Genotyping Protocol
The ZEG protocol enables DNA extraction from live zebrafish larvae as early as 72 hpf with minimal impact on viability and development [5] [10].
Workflow Overview:
Stepwise Methodology:
Preparation: Anesthetize 3-5 dpf zebrafish larvae in ~1.5 mM Tricaine in 1x E3 embryo media [1]. Prepare a dissection surface by taping a 9 cm Petri dish lid with autoclave tape across its interior surface and position under a stereo-microscope [1].
Fin Clipping: Using a modified P1000 pipette tip with a 2 mm diameter end, transfer an anesthetized larva onto the autoclave tape. Remove excess Tricaine solution while ensuring the area remains sufficiently moist for survival. Using a micro scalpel, section the caudal fin within the pigment gap site, applying steady downward pressure distal to the limit of blood circulation to avoid damaging the notochord [1].
Tissue Collection: Visualize the sectioned fin under microscope and position the piece on top of the microscalpel blade. Transfer the fin onto a small piece of filter paper, which allows visualization of the melanocyte-containing tissue as a small black spot. Using scissors and tweezers, transfer the filter paper containing the fin to a 96-well PCR plate containing 25 μL of 50 mM NaOH solution [1].
Embryo Recovery: Using a modified P1000 pipette filled with 200 µL of fresh 1x E3 embryo media, carefully collect the zebrafish larva and dispense it into a numbered well of a 96-well tissue culture plate corresponding to the PCR plate designation [1]. Clean the microscalpel blade and tweezers with 70% EtOH between each procedure to prevent cross-contamination.
Genomic DNA Extraction: Seal the 96-well PCR plate and centrifuge at 1,000 × g for 1 minute to ensure complete submersion of filter papers. Lyse tissues by heating samples in a thermocycler at 95°C for 5 minutes followed by cooling to 4°C for 10 minutes. Add 6 μL of 500 mM Tris-HCl (pH 8.0) to each sample, vortex, and centrifuge the plate at 1,500 × g for 5 minutes at room temperature. Use 1.5 μL of the DNA supernatant per PCR reaction [1].
Alternative Fin Clipping Genotyping Protocol
This complementary protocol provides another reliable method for DNA extraction from live larvae.
Workflow Overview:
Stepwise Methodology:
Fin Clipping: Follow identical anesthesia and fin clipping procedures as the ZEG protocol (steps 1-3 above) [1].
Alternative DNA Extraction: Transfer filter paper with clipped fin to a 96-well PCR plate containing 30 μL of 5% chelating resin (styrene-divinylbenzene copolymer containing paired iminodiacetate ions) [1]. Seal the plate and briefly centrifuge to ensure complete submersion.
Lysis and DNA Preparation: Heat samples in a thermocycler at 95°C for 15 minutes for tissue lysis, followed by cooling to 4°C for 10 minutes. This step enables cellular lysis and binding of polar resin beads to cellular components while DNA remains in solution. Centrifuge samples to pellet resin beads and use the DNA-containing supernatant for PCR genotyping [1].
Longitudinal Developmental Assessment Protocol
Imaging-Based Quantitative Analysis:
Sample Preparation: Anesthetize zebrafish using 10 μL MS-222 with 15 ml water prior to image collection. After imaging, transfer zebrafish to clean water to recover and continue breeding [18].
Mueller Matrix OCT Imaging: Acquire 3D images of zebrafish during development using a spectral domain Mueller matrix OCT system with axial resolution of 8.9 μm and lateral resolution of 18.2 μm in air. Collect 400 cross-sectional images for each zebrafish, adjusting lateral scanning field of view from 4 mm (0-7 dpf) to 6 mm (after 7 dpf) to accommodate growth [18].
Deep Learning Segmentation: Apply U-Net network to segment various anatomical structures including body, eyes, spine, yolk sac, and swim bladder. Calculate organ volume from segmented structures and analyze developmental trends from day 1 to day 19 post-fertilization [18].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents and Materials for ZEG and Genotyping Protocols
| Reagent/Material | Specification/Concentration | Primary Function | Protocol Application |
|---|---|---|---|
| Tricaine | ~1.5 mM in 1x E3 embryo media [1] | Anesthesia for larvae handling | ZEG and fin clipping |
| NaOH Lysis Buffer | 50 mM [1] | Tissue lysis and DNA release | ZEG protocol |
| Tris-HCl Neutralization Buffer | 500 mM, pH 8.0 [1] | Neutralization of NaOH | ZEG protocol |
| Chelating Resin | 5% suspension [1] | Cellular component binding, DNA purification | Alternative fin clip protocol |
| Proteinase K | 200 μg/mL [40] | Protein digestion for DNA extraction | Standard DNA preparation |
| PCR Extraction Buffer | 10 mM Tris pH 8, 2 mM EDTA, 0.2% Triton X-100 [40] | DNA stabilization for PCR | High-throughput DNA prep |
| E3 Embryo Media | 5 mM NaCl, 0.17 mM KCl, 0.33 mM CaClâ‚‚, 0.33 mM MgSOâ‚„, pH 7.2 [1] | Embryo maintenance medium | Larval recovery post-procedure |
| Cas9 Protein | N/A [5] | CRISPR-Cas9 genome editing | Optimal knock-in conditions |
| Non-target Asymmetric ssODN | PAM-distal, 120 BP [5] | Homology-directed repair template | Enhanced knock-in efficiency |
The ZEG early selection protocol represents a significant advancement for zebrafish genetics research, effectively addressing the challenge of low HDR efficiency in CRISPR-Cas9 knock-in generation. Comprehensive viability studies confirm that processed embryos exhibit no long-term detrimental effects on survival, tissue integrity, or organ development trajectories. The implementation of early genotyping selection enables researchers to achieve substantially higher somatic editing efficiencies and successful germline transmission while reducing animal husbandry costs and ethical concerns. When integrated with optimal CRISPR components—including Cas9 protein and non-target asymmetric PAM-distal ssODN repair templates—the ZEG protocol provides a robust and validated pipeline for generating precise zebrafish disease models.
The zebrafish (Danio rerio) has emerged as a premier model organism in biomedical research for studying neurological disorders, toxicology, and drug discovery. Its genetic tractability, optical transparency during early life stages, and high reproductive yield make it particularly suited for high-throughput screening [41] [42]. A significant advancement in this field is the Zebrafish Embryo Genotyper (ZEG) device, which allows for the non-invasive extraction of genomic material from embryos at 72 hours post-fertilization (hpf) with minimal impact on viability [5]. This early selection protocol enables researchers to identify and raise only those embryos with desired genetic edits, dramatically improving the efficiency of generating stable lines and reducing animal usage in accordance with 3R principles [5]. This application note details behavioral protocols for assessing motor function and stress responses within the context of ZEG-based early selection, providing standardized methods for phenotypic characterization of genetically modified zebrafish.
Experimental Protocols and Workflows
Integrating behavioral phenotyping with the ZEG selection protocol creates a powerful pipeline for functional genomics. The general workflow begins with the generation of genetically manipulated embryos, followed by early genotyping with the ZEG device at approximately 72 hpf. Selected embryos are then raised and subjected to behavioral assays at specific developmental stages, depending on the biological question.
Motor Function Assessments
Motor function can be quantitatively assessed from early larval stages through adulthood. The following protocols are optimized for different developmental time points.
Larval Touch-Evoked Response Assay
This assay, performed at 48 hours post-fertilization (hpf), evaluates the integrity of sensory and motor circuits by triggering a stereotypical C-start escape response [43].
Detailed Methodology:
- Preparation: Dechorionate 48 hpf embryos and allow them to acclimate in a Petri dish for one hour.
- Setup: Place a single larva in the center of a Petri dish. Position a high-speed camera (≥30 fps, preferably higher for detailed kinematics) mounted on a clamp stand above the dish. Ensure homogenous, non-flickering illumination against a white background. Maintain a constant temperature of 28°C.
- Stimulation: Gently tap the larva on the head with a blunt, fine tool (e.g., a snapped gel-loading pipette tip) to stimulate the Mauthner cells.
- Recording: Record the response. A successful stimulus will elicit a rapid C-shaped bend followed by swimming. If the larva does not respond, attempt a few more taps. Do not use larvae that swim excessively before stimulation.
- Analysis with Fiji/ImageJ:
- Set Scale: Import the video using the FFmpeg plugin. Record a video of a ruler in the same plane as the assay to set the spatial scale.
- Pre-process: Crop the video to the region of interest. Convert the stack to 8-bit and apply a threshold to create a binary image where the larva is a distinct black blob.
- Track Movement: Use the TrackMate plugin. Select the "Log Detector" and set the estimated blob diameter to the larva's size (e.g., ~1 mm). Adjust the thresholding quality to ensure the larva is detected in each frame. Use the "Simple LAP tracker" to link positions across frames.
- Extract Data: Export statistics including total distance traveled, velocity, and acceleration from the "links in tracks statistics" table [43].
Multi-well Plate Locomotion Tracking (LSRtrack)
For high-throughput analysis of spontaneous or stimulus-evoked movement in larval zebrafish (e.g., 5-7 dpf), the open-source MATLAB application LSRtrack can be used [44].
Detailed Methodology:
- Video Acquisition: Position larvae individually in a multi-well plate. Generate high-quality, flatly illuminated videos. Exposure settings must be optimized to facilitate accurate object recognition by the software.
- Tracking with LSRtrack: Analyze recordings using LSRtrack, adjusting parameters to optimize tracking accuracy and motion detection for your specific setup.
- Data Analysis with LSRanalyze: Use the companion application LSRanalyze to process tracking data. Key quantifiable endpoints include:
- Positional preference within the well.
- Kinematic measures: Displacement, velocity, and acceleration.
- Activity bouts: Duration and frequency of movement and rest periods [44].
Adult Zebrafish Locomotion and Motor Phenotypes
In adult zebrafish, motor and non-motor behaviors can be assessed using the open-source plugin wrMTrck for ImageJ [41].
Detailed Methodology:
- Video Recording: Record adult zebrafish in a standardized tank using a setup that provides a clear, top-down view.
- Analysis with wrMTrck: A pre-processing macro can be used to prepare images for analysis. Optimized settings for adult zebrafish include specific thresholds for object detection and size to accurately track the fish.
- Quantifiable Endpoints:
- Motor Function: Total distance moved, velocity, immobility duration.
- Anxiety-like Behavior: Thigmotaxis (time spent near the walls vs. center) can be derived from positional data [41].
Stress Response Analyses
Stress responses are highly conserved across vertebrates and can be measured using behavioral, physiological, and molecular readouts.
Light-Dark Test
The light-dark test is a validated assay for measuring anxiety-like behavior in larval zebrafish (6-7 dpf). Sudden transitions from light to dark often induce an anxiety-like state, resulting in increased locomotor activity [45].
Detailed Methodology:
- Acclimatization: Individually place 6-7 dpf larvae into wells of a 96-well plate containing 300 µL of system water. Acclimatize them in the dark for 60 minutes inside the video tracking apparatus.
- Stimulation Protocol: Expose the larvae to alternating light and dark cycles. Research indicates that using 5-minute intervals is more effective than 15-minute intervals, as it leads to stabilized travel distances after just the 5th repetition [45].
- Data Collection: Use an automated video tracking system (e.g., Noldus) to monitor locomotor activity.
- Key Metric: The primary readout is the distance traveled following the transition to dark. An increase in distance is interpreted as an enhanced anxiety-like response.
Novel Tank Test
The novel tank test is used for juvenile and adult zebrafish. When placed into a new environment, zebrafish exhibit an initial preference for the bottom portion of the tank (scototaxis). Over time, reduced exploration of the top zone is a indicator of anxiety-like behavior [46].
Detailed Methodology:
- Setup: Use a trapezoidal tank to facilitate video tracking. Virtually divide the tank into a top zone and a bottom zone.
- Testing: Gently introduce a single zebrafish into the novel tank and record its behavior for a set period (e.g., 10 minutes).
- Analysis: Track the fish's movement and quantify:
- Time spent in the top vs. bottom zone.
- Latency to enter the top zone.
- Number of entries to the top zone.
- Total distance moved to control for general activity deficits.
- Interpretation: Animals with higher anxiety levels spend significantly more time in the bottom zone and are slower to explore the top zone [46].
Quantitative Data and Findings
Behavioral data from the described assays provide robust, quantifiable endpoints for phenotypic screening. The tables below summarize key expected outcomes and physiological correlates from foundational studies.
Table 1: Expected Behavioral Outcomes in Motor Function Assays
| Assay | Developmental Stage | Key Measurable Parameters | Example Phenotype: MPTP-induced Parkinsonism [41] | Typical Wild-type Response |
|---|---|---|---|---|
| Touch-Evoked Response | 48 hpf | Latency to respond, C-bend angle, escape distance, velocity | N/A | Rapid C-start turn followed by swimming [43] |
| Spontaneous Locomotion | 5-7 dpf | Total distance, movement duration, rest period duration, burst swim frequency | N/A | Intermittent burst and glide swimming [44] |
| Adult Locomotion & Anxiety | >60 dpf | Total distance, velocity, time immobile, thigmotaxis (center vs. edge) | ↓ Total distance moved, ↑ thigmotaxis | Exploration of entire tank, regular movement patterns [41] |
Table 2: Behavioral and Physiological Correlates of Stress
| Assay / Measure | Stressed Phenotype (e.g., after ELS [46]) | Control Phenotype | Notes |
|---|---|---|---|
| Light-Dark Test (Distance in Dark) | ↑ Distance traveled after transition [45] | Stabilized locomotion after habituation | Indicates anxiety-like response. |
| Novel Tank Test (Bottom Dwelling) | ↑ Time spent in bottom zone, ↓ time in top zone [46] | Gradual exploration of top zone after ~4 min | Validated indicator of anxiety. |
| Basal Cortisol Level | ↑ Significantly elevated [46] | Lower baseline level | Measured via whole-body ELISA. |
| Gene Expression (gr, mr) | ↑ gr and mr mRNA in the brain [46] | Lower expression levels | Suggests dysregulation of HPI axis feedback. |
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents and Materials for Zebrafish Behavioral Analysis
| Item | Function / Application | Example / Specification |
|---|---|---|
| Zebrafish Embryo Genotyper (ZEG) | Non-invasive genomic DNA extraction from early embryos for genotyping. | Enables early selection of CRISPR/Cas9 edited embryos [5] |
| CRISPR-Cas9 Components | Generation of knock-out/knock-in genetic models. | Cas9 protein, sgRNA, ssODN repair templates (e.g., non-target asymmetric PAM-distal conformation) [5] |
| Morpholinos | Transient knockdown of gene expression. | Sequence-specific antisense oligonucleotides. |
| Pharmacological Agents | Modulating neural pathways; disease modeling. | MPTP: Induces Parkinson's-like phenotypes [41]. Strychnine: Glycine receptor blocker alters motor patterns [47]. |
| E3 Embryo Medium | Standard medium for raising zebrafish embryos. | 5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4 [47] |
| MS-222 (Tricaine) | Anesthetic for immobilizing fish for procedures. | Used at 0.02% for anesthesia [48] |
| High-Speed Camera | Recording behavioral assays for detailed kinematic analysis. | ≥30 fps; high-speed (500+ fps) for C-start analysis [43] |
| Automated Tracking Software | High-throughput, objective quantification of behavior. | Noldus EthoVision, LSRtrack (MATLAB) [44], wrMTrck (ImageJ) [41], TrackMate (ImageJ) [43] |
Signaling Pathways and Workflows
The following diagrams, generated using Graphviz DOT language, illustrate key neuroendocrine pathways and experimental workflows relevant to zebrafish behavioral analysis.
HPI Axis Signaling Pathway
The Hypothalamic-Pituitary-Interrenal (HPI) axis is the central stress response system in zebrafish, functionally analogous to the mammalian HPA axis. Dysregulation of this pathway is a core mechanism underlying long-term effects of early life stress (ELS) [46] [42].
- Diagram Title: Zebrafish HPI Axis Stress Pathway
ZEG Protocol & Behavioral Phenotyping Workflow
This workflow integrates the ZEG early genotyping protocol with subsequent behavioral assays, creating an efficient pipeline for functional validation of genetic models [5].
- Diagram Title: ZEG and Behavioral Screening Workflow
Within zebrafish research, efficient and ethical genotyping is a critical step for the advancement of genetic models. This application note provides a structured, data-driven comparison between two primary DNA sampling methods: the traditional fin clipping protocol and the more recent Zebrafish Embryo Genotyper (ZEG) device. Framed within broader research on early selection protocols, this document summarizes key efficiency metrics, outlines detailed experimental protocols, and provides visual workflows to assist researchers, scientists, and drug development professionals in selecting the most appropriate methodology for their work. The focus on quantitative data and standardized protocols aims to enhance reproducibility and experimental design in genetic studies involving zebrafish.
Comparative Efficiency Metrics
The choice between ZEG and fin clipping involves balancing multiple factors, including animal welfare, DNA yield, and procedural efficiency. The table below summarizes a direct comparison of these two methods based on key performance indicators.
Table 1: Head-to-Head Comparison of ZEG and Fin Clipping Efficiency Metrics
| Metric | Zebrafish Embryo Genotyper (ZEG) | Traditional Fin Clipping |
|---|---|---|
| Developmental Stage | Early developmental stages (embryos, pre-sentience) [5] [2] | Juvenile and adult fish (typically ≥ 20 mm) [49] |
| Animal Welfare & Invasiveness | Minimally invasive; samples cells from live embryos without terminal procedures [5] [2] | Invasive; requires surgical removal of fin tissue, impacting welfare [49] |
| Anaesthesia/Analgesia | Not required [2] | Surgical anaesthesia (e.g., MS-222/tricaine) required; analgesia recommended post-procedure [49] |
| DNA Yield | Suitable for PCR-based genotyping [5] [2] | Medium to high yield (ng/µl), consistently suitable for PCR and other applications [3] [49] |
| Primary Application | Early genotyping for selective rearing; increases somatic editing efficiency in CRISPR models [5] | Standard genotyping for genetic identification; suitable for PCR, sequencing, and WGS [3] [49] |
| Key Advantage | Up to 17-fold increase in somatic editing efficiency in CRISPR models; allows early culling of non-edited embryos, adhering to the 3Rs [5] | High DNA quality and quantity, reliable for a wide range of techniques, including whole genome sequencing [3] |
| Key Disadvantage | Limited to embryonic stages; lower DNA yield can require protocol optimization for complex PCRs [2] | Negative welfare impact including pain, stress, and risk of infection; requires anaesthesia [49] |
Experimental Protocols
ZEG Device Protocol for Early Embryo Genotyping
This protocol is designed for genotyping zebrafish embryos at 72 hours post-fertilization (hpf) to identify those with the highest rates of CRISPR-mediated editing before selective raising to adulthood [5].
- Step 1: Embryo Preparation: At 72 hpf, arrange embryos in the specialized ZEG device. The device is designed to hold individual embryos for minimally invasive cell collection [5].
- Step 2: Cell Collection: Using the ZEG apparatus, collect a small number of cells from each embryo. This process is minimally invasive and does not require anaesthesia, allowing the embryo to continue developing normally after sampling [5] [2].
- Step 3: DNA Extraction & Genotyping: Extract genomic DNA from the collected cells. The DNA is then analyzed using Next-Generation Sequencing (NGS) to precisely quantify the somatic editing efficiency (e.g., knock-in rates) for each embryo [5].
- Step 4: Selective Rearing: Based on the NGS results, selectively raise only those embryos showing the highest levels of correct editing to adulthood. This step demonstrably enriches for founders with germline transmission potential [5].
Standardized Protocol for Traditional Fin Clipping
This protocol outlines the standard procedure for obtaining DNA via fin clipping from juvenile or adult zebrafish, incorporating welfare considerations [49].
- Step 1: Fish Anaesthesia: Immerse the fish in a buffered solution of MS-222 (tricaine) until surgical anaesthesia is achieved, as indicated by the loss of opercular movement and response to tactile stimuli [49].
- Step 2: Tissue Collection: Using sterile scissors or a scalpel, remove a small portion (ideally less than 10%) of the caudal fin [49]. The fin tissue should be immediately placed in a suitable preservative, such as 96% molecular-grade ethanol [3].
- Step 3: Fish Recovery: Place the fish in a recovery tank with clean, system water. Administration of analgesia during recovery is recommended to manage potential pain [49].
- Step 4: DNA Extraction & Genotyping: DNA can be extracted from the fin clip using various methods, including the HotShot method, isopropanol extraction, or commercial kits (e.g., QIAGEN DNeasy Blood & Tissue Kit). The resulting high-quality DNA is suitable for PCR, sequencing, and whole genome sequencing [3] [49].
Workflow Visualization
The following diagram illustrates the key decision points and procedural steps involved in selecting and executing the ZEG and Fin Clipping protocols.
Zebrafish Genotyping Workflow Selection This workflow outlines the decision path between the ZEG and Fin Clipping protocols, driven by the developmental stage of the fish and the desired experimental outcome.
The Scientist's Toolkit: Essential Research Reagents
Successful implementation of either genotyping protocol requires specific reagents and equipment. The following table lists key materials and their functions.
Table 2: Essential Research Reagents and Materials for Zebrafish Genotyping
| Item Name | Function/Application | Example Protocol Usage |
|---|---|---|
| ZEG Device | Holds 72 hpf embryos for minimally invasive cell collection [5] | Core apparatus for the early embryo genotyping protocol [5] |
| Cas9 Protein | CRISPR-Cas9 ribonucleoprotein complex for generating DNA double-strand breaks [5] | Superior editing efficiency compared to mRNA in KI model generation [5] |
| ssODN Repair Template | Single-stranded oligodeoxynucleotide serving as a repair template for homology-directed repair (HDR) in knock-in experiments [5] | Used in non-target asymmetric PAM-distal (NAD) conformation for optimal KI efficiency [5] |
| MS-222 (Tricaine) | Anaesthetic agent for immobilizing fish for surgical procedures [49] | Required for anaesthetizing fish prior to fin clipping [49] |
| Copan 4N6FLOQSwabs | Rayon-tipped swabs for collecting mucus and cells non-invasively [3] [49] | Can be used for skin swabbing as a less invasive alternative to fin clipping in adults [49] |
| QIAamp DNeasy Blood & Tissue Kit | Silica-membrane based system for purification of high-quality DNA from tissues and cells [3] | DNA extraction from fin clips and optimized swab samples [3] |
| Proteinase K | Broad-spectrum serine protease for digesting proteins and nucleases during cell lysis [3] | Treatment during DNA extraction to significantly improve yield from swab samples [3] |
Application Note: Enhancing CRISPR Knock-In Efficiency via ZEG-Based Early Selection
The Zebrafish Embryo Genotyper (ZEG) device represents a significant advancement in genetic research methodologies that directly supports the ethical principles of the 3Rs (Replacement, Reduction, and Refinement) in animal research. By enabling rapid, non-lethal cellular extraction from live zebrafish embryos and larvae, the ZEG system addresses critical ethical and efficiency challenges in biomedical research [8]. This protocol specifically demonstrates how early genotyping selection can enhance CRISPR knock-in (KI) efficiency while substantially reducing animal numbers and refining experimental procedures to minimize harm.
The 3Rs principle, first articulated by Russell and Burch in 1959, provides a framework for more ethical animal research through Replacement (avoiding animal use), Reduction (minimizing animal numbers), and Refinement (decreasing suffering) [50]. The ZEG device operationalizes these principles by allowing researchers to genotype embryos at 72 hours post-fertilization rather than waiting for adulthood, thereby avoiding the traditional practice of raising numerous animals to adulthood only to discover most lack the desired genetic modification [8] [17]. This approach aligns with Directive 2010/63/EU, which recognizes the importance of implementing the 3Rs in scientific research [51] [52].
The ZEG device utilizes microfluidic harmonic oscillation on a roughened glass surface to obtain genetic material (cells and DNA) from 24 embryos or larvae simultaneously in less than 10 minutes [8]. This automated process maintains high survival rates (>90%) and enables researchers to identify and selectively raise only those embryos with the highest rates of correct gene editing, dramatically improving research efficiency while adhering to ethical standards [8] [11].
Table 1: Quantitative Performance Metrics of ZEG Protocol in CRISPR Knock-In Experiments
| Performance Parameter | Traditional CRISPR-KI | ZEG-Enhanced CRISPR-KI | Improvement Factor |
|---|---|---|---|
| Somatic Editing Efficiency | 1-4% [23] | Up to 17-fold increase [23] | ~17x |
| Germline Transmission Rate | Low (labor-intensive screening) | Enhanced through pre-selection [23] | Significant (study-dependent) |
| Animal Survival Rate | N/A (often lethal sampling) | >90% [8] [11] | Substantial refinement |
| Time to Genotype Result | 2-3 months (to adulthood) | 3 days post-fertilization [8] | ~20x faster |
| Throughput | Manual, low-throughput | 24 embryos simultaneously in <10 min [8] | High-throughput |
Ethical and Economic Cost-Benefit Analysis
Implementation of the ZEG protocol demonstrates a compelling synergy between ethical advancement and research efficiency. From a 3Rs perspective, this methodology directly addresses all three principles:
Reduction: By identifying successfully edited embryos early in development, researchers can dramatically reduce the number of animals raised to adulthood. One study demonstrated that ZEG-based selection allowed for a 17-fold increase in editing efficiency, meaning far fewer animals need to be maintained to obtain the desired genetic model [23].
Refinement: The ZEG method causes minimal stress and harm to the animals compared to traditional fin-clipping of adults. Research has confirmed that although ZEG results in a low-level acute stress response, no long-lasting effects are evident, supporting its utilization for a variety of downstream assays [11].
Replacement: While not a complete replacement for animal use, the ZEG system represents a form of relative replacement by using early developmental stages (zebrafish embryos) that are considered less sentient according to regulatory frameworks like EU Directive 2010/63/EU [53].
From an economic perspective, the ZEG system reduces facility costs by decreasing the number of adult zebrafish that need to be housed and maintained. It also saves significant researcher time by eliminating the need for manual fin-clipping and associated sample processing [17]. The ability to obtain genetic information rapidly (within days rather than months) accelerates research timelines substantially, potentially shortening the path to scientific discoveries.
Experimental Protocol: ZEG-Enhanced CRISPR Knock-In Workflow
Principle
This integrated protocol combines optimized CRISPR knock-in techniques with the ZEG device for early embryo selection to significantly improve the efficiency of generating precise genetic models in zebrafish. The method leverages non-invasive cellular extraction at 72 hours post-fertilization (hpf) to identify embryos with the highest somatic editing efficiency, allowing selective raising of these embryos to adulthood to enhance germline transmission prospects [23] [8].
Research Reagent Solutions
Table 2: Essential Research Reagents and Materials for ZEG-Enhanced CRISPR Knock-In
| Reagent/Material | Specification/Function | Optimization Notes |
|---|---|---|
| Zebrafish Embryo Genotyper (ZEG) Device | Automated system for non-lethal cellular extraction from live embryos [8] | Enables high-throughput sampling of 24 embryos simultaneously in <10 minutes |
| Cas9 Protein | Ribonucleoprotein complex for DNA cleavage | Superior performance over mRNA: 5.14% ± 0.71 vs. 0.94% ± 0.35 editing efficiency at LQTS locus [23] |
| ssODN Repair Template | Single-stranded oligodeoxynucleotide for HDR | Non-target asymmetric PAM-distal (NAD) conformation significantly outperforms other conformations [23] |
| Next-Generation Sequencing (NGS) | High-sensitivity detection of editing events | Enables accurate quantification of somatic editing efficiency; superior to gel electrophoresis or Sanger sequencing [23] |
| Zebrafish Lines | Wild-type or transgenic as required | Use of experimentally bred animals prioritized per 3Rs principles [54] |
| PCR Reagents | For amplification of extracted genetic material | 5 μL of ZEG-extracted fluid sufficient for 11 μL PCR reaction [8] |
Equipment
- ZEG device (commercially available from wFluidx or other suppliers)
- Standard zebrafish housing and maintenance systems
- Microinjection apparatus for CRISPR component delivery
- Thermocycler for PCR amplification
- Next-generation sequencing platform
- Standard molecular biology laboratory equipment
Procedure
Pre-experimental Planning and Design
Ethical Review and 3Rs Compliance: Submit experimental plan to Institutional Animal Care and Use Committee (IACUC) or equivalent ethics body, specifically addressing:
Experimental Design Optimization:
- Perform sample size calculation using appropriate statistical methods to use the minimum number of animals while maintaining scientific validity [54]
- Design appropriate control groups to maximize data quality from each animal
CRISPR Component Preparation and Microinjection
sgRNA Validation:
- Design sgRNAs targeting desired genomic loci using standard tools
- Validate cutting efficiency using Inference of CRISPR Edits (ICE) analysis, which shows strong correlation with NGS results (Pearson's r = 0.90-0.92) [23]
Repair Template Design:
- Design 120 bp single-stranded oligodeoxynucleotides (ssODNs) with non-target asymmetric PAM-distal (NAD) conformation, which demonstrated superior performance in comparative studies [23]
- Incorporate desired precise base pair substitutions centered appropriately within the repair template
Microinjection Mixture Preparation:
- Prepare injection mixture containing:
- Cas9 protein (superior to mRNA for KI efficiency) [23]
- Validated sgRNA
- NAD-conformation ssODN repair template
- Optimize concentrations through pilot injections
- Prepare injection mixture containing:
Zebrafish Embryo Injection:
- Inject one-cell stage zebrafish embryos with CRISPR components using standard microinjection techniques
- Maintain injected embryos in E3 embryo medium at 28.5°C [8]
ZEG-Based Genotyping and Selection
Sample Collection Timing:
ZEG Operation:
- Load 24 embryos simultaneously into the ZEG device using standard pipette tips [8]
- Activate the automated harmonic oscillation system to collect genetic material onto a roughened glass surface
- Collect approximately 11 μL of fluid containing cells and DNA from each embryo
- Return embryos to individual wells with E3 medium for recovery
Genotype Analysis:
Animal Selection and Raising
Selective Raising:
- Raise only embryos with the highest somatic editing efficiency to adulthood
- This selective approach reduces animal numbers by targeting only those most likely to yield germline transmission
Germline Screening:
- Outcross adult fish raised from pre-selected embryos
- Screen F1 offspring for germline transmission using standard methods
- The pre-selection significantly enhances the probability of identifying germline transmission events [23]
Timing
- Days 1-2: CRISPR component preparation and embryo injection
- Day 3: ZEG-based cellular extraction and genotyping (72 hpf)
- Day 4 onward: Selective raising of high-efficiency embryos
- Months 2-3: Germline screening of adult fish
The ZEG protocol reduces the time to initial genotyping results from 2-3 months (waiting for adulthood) to just 3 days, dramatically accelerating research timelines [8] [17].
Troubleshooting
- Low Survival Rates: Ensure proper technique during embryo handling and ZEG operation; survival should exceed 90% [8]
- Poor PCR Amplification: Increase number of oscillation cycles during ZEG extraction or optimize PCR conditions
- Low Editing Efficiency: Verify Cas9 protein quality and concentration; confirm ssODN design uses optimal NAD conformation [23]
- Inconsistent Results: Standardize injection techniques and ensure proper maintenance of embryos post-extraction
Anticipated Results
Implementation of this integrated protocol should yield:
- Significantly enhanced somatic editing efficiency (up to 17-fold improvement over non-selected approaches) [23]
- High animal survival rates (>90%) with minimal long-term effects on development or behavior [11]
- Reduced animal usage through selective raising of only high-efficiency embryos
- Accelerated research timeline due to early genotyping capability
Workflow for ZEG-enhanced CRISPR knock-in protocol showing integration with 3Rs principles.
Cost-benefit comparison between traditional and ZEG-enhanced approaches for CRISPR knock-in experiments.
The CACNA1C gene encodes the pore-forming α1C-subunit of the Cav1.2 L-type voltage-gated calcium channel, a critical mediator of calcium influx in excitable cells including cardiac myocytes and neurons [55] [56]. Mutations in this gene disrupt calcium homeostasis, leading to a spectrum of disorders ranging from severe multisystem syndromes to cardiac-specific conditions [56] [57]. Timothy Syndrome (TS) represents the most severe end of this spectrum, characterized by long QT syndrome (LQTS), syndactyly, cognitive impairment, and autism [55]. Recent research has also linked CACNA1C mutations to Brugada syndrome, early repolarization syndrome, and various forms of cardiac hypertrophy and arrhythmia [5] [58] [57].
The establishment of robust disease models is crucial for understanding the pathophysiological mechanisms of CACNA1C-related disorders and developing targeted therapies. Zebrafish have emerged as a powerful model organism for cardiovascular research due to their genetic tractability, optical transparency, and physiological similarity to humans [6] [5]. This application note details proven case studies and protocols for modeling CACNA1C mutations, with particular emphasis on the integration of the Zebrafish Embryo Genotyper (ZEG) device for rapid, non-lethal genotyping to enhance model generation efficiency.
CACNA1C Mutation Case Studies and Clinical Heterogeneity
Documented Pathogenic Variants and Associated Phenotypes
Table 1: Clinical Spectrum of CACNA1C Mutations in Human Cases
| Mutation | Location | Syndrome | Cardiac Features | Extra-Cardiac Features | Citation |
|---|---|---|---|---|---|
| p.Gly402Ser | Exon 8 | Timothy Syndrome Type 2 | Severe LQTS (QTc 550-600 ms), ventricular fibrillation, cardiac arrest | Autism, cognitive decline, bipolar disorder | [55] |
| p.Ile1166Thr | Exon 27 | Atypical Timothy Syndrome | Long QT, patent ductus arteriosus | Seizures, facial dysmorphisms, intellectual impairment | [56] |
| p.Arg518Cys | Exon 12 | Cardiac-Only Timothy Syndrome | LQTS, HCM, congenital heart disease, AF, sick sinus syndrome | None reported | [57] |
| p.Thr330Met | Exon 8 | Brugada Syndrome | ST-segment elevation, risk of ventricular fibrillation | Not specifically reported | [5] |
| Unspecified | Not reported | Early Repolarization Syndrome | J-point elevation, terminal QRS notch/slur, risk of SCD | Incomplete penetrance, male predominance | [58] |
In-depth Case Analysis
Case Study 1: Timothy Syndrome with Neuropsychiatric Manifestations A male patient with TS Type 2 (p.Gly402Ser mutation) survived childhood despite multiple cardiac arrests requiring pacemaker and ICD implantation [55]. At age 19, he developed major depression, followed by a manic episode at age 21. By age 30, he had experienced 10 manic and 3 hypomanic episodes, characterized by sleep reduction, aggression, restlessness, and talkativeness [55]. A severe manic episode at 27 involved hallucinations and paranoid delusions. Treatment with verapamil (a calcium channel blocker) reduced ventricular fibrillation events but did not shorten the QT interval, while lithium was poorly tolerated [55]. This case established a direct link between a CACNA1C gain-of-function mutation and bipolar disorder, suggesting that both increased and decreased calcium influx can be associated with the condition [55].
Case Study 2: A Novel Mutation with Altered Electrophysiology A patient with a multi-system condition resembling TS was found to harbor a novel p.Ile1166Thr mutation in exon 27 of CACNA1C [56]. Unlike classical TS mutations that cause complete loss of voltage-dependent inactivation, this variant led to a significant increase in window current, a distinct electrophysiological mechanism [56]. The patient presented with LQTS, patent ductus arteriosus, seizures, facial dysmorphisms, joint hypermobility, hypotonia, hand anomalies, intellectual impairment, and tooth decay [56]. This case highlighted that mutations outside the canonical exons 8/8A can cause TS-like phenotypes through diverse biophysical mechanisms.
Case Study 3: A Family with Cardiac-Only Phenotype A four-generation family carrying the p.R518C variant displayed autosomal dominant inheritance of cardiac manifestations without extra-cardiac features [57]. Affected individuals exhibited LQTS, congenital heart disease, hypertrophic cardiomyopathy (HCM), atrial fibrillation (AF), sick sinus syndrome, and sudden cardiac death [57]. The proband was diagnosed with HCM at 59 days old, with a QTc of 472 ms and significant septal hypertrophy (Z-score 11.23), requiring ICD placement at four months and septal myectomy at three years [57]. This family demonstrates the variable expressivity of CACNA1C mutations and expands the recognized cardiac phenotype.
Zebrafish Models for CACNA1C Mutagenesis
Zebrafish as a Model System
Zebrafish offer significant advantages for modeling human cardiac channelopathies. Their genome shares substantial homology with humans, and their cardiac development, electrical conduction system, and response to pharmacological agents are highly conserved [6] [5]. The external development and optical transparency of embryos allow for direct visualization of heart structure and function in real-time. Furthermore, zebrafish produce large clutches of embryos, facilitating high-throughput genetic and drug screens [6].
CRISPR-Cas9 Knock-in Modeling of Human Mutations
Recent efforts have focused on generating precise zebrafish models of human CACNA1C mutations using CRISPR-Cas9 homology-directed repair (HDR) [5]. Key mutations modeled include:
- c.2607C>G or p.Pro871Arg: Associated with Long QT Syndrome (LQTS) in humans [5]
- c.1028C>T or p.Thr343Met: Associated with Brugada Syndrome (BrS) in humans [5]
Table 2: Optimized CRISPR-Cas9 Components for cacna1c Knock-in in Zebrafish
| Component | Optimal Specification | Function | Performance Notes |
|---|---|---|---|
| Cas9 Form | Protein | DNA endonuclease | Significantly higher KI efficiency vs. mRNA (LQTS: 5.14% vs 0.94%; BrS: 2.83% vs 1.04%) [5] |
| sgRNA | Locus-specific | Target sequence guidance | Cutting efficiency must be validated (e.g., via ICE analysis) [5] |
| Repair Template | Non-target asymmetric PAM-distal (NAD) ssODN, 120 bp | Homology-directed repair | Superior performance over target asymmetric PAM-proximal (TAP) conformation [5] |
| Mutation Position | PAM-distal arm | -- | Distance from cut site affects efficiency [5] |
The Zebrafish Embryo Genotyper (ZEG) Protocol
The ZEG device enables rapid, non-lethal genotyping of live zebrafish embryos and larvae, addressing a critical bottleneck in high-throughput genetic screens [6] [5].
ZEG Device Specifications and Operation
- Design: Microfluidic device with 24 chambers etched onto standard glass microscope slides using polyimide tape to create shallow chambers [6]
- Mechanism of Action: Uses harmonic oscillation on a roughened glass surface to gently abrade the embryo surface, releasing cells and DNA into the surrounding medium [6]
- Throughput: Processes 24 embryos simultaneously in less than 10 minutes [6]
- Sample Collection: Yields 10-11 μL of fluid containing genetic material from each embryo [6]
- Viability: Maintains >90% survival rate with no apparent effects on morphology, development, or motor behavior [6]
Step-by-Step ZEG Protocol
- Preparation: Raise zebrafish embryos to 72 hours post-fertilization (hpf) in E3 embryo medium at 28.5°C [6]
- Loading: Manually pipette individual embryos into ZEG chambers in 10-14 μL of E3 medium using standard pipette with cut-off tips (approximately 2 minutes for 24 embryos) [6]
- Vibration: Secure chip on platform and activate vibration motor at 1.4 volts for 7-10 minutes [6]
- Sample Collection: Use standard pipette to collect 10 μL of fluid from each chamber [6]
- Genotyping Analysis: Use 5 μL of collected fluid in 11 μL PCR volume for subsequent genotyping (PCR, HRMA, sequencing) [6]
- Embryo Recovery: Return embryos to fresh E3 medium for continued development and raising
Enhanced Selection Through Early NGS Genotyping
Integration of ZEG with next-generation sequencing (NGS) creates a powerful pipeline for efficient model generation:
- ZEG Extraction: Perform cellular extraction from 72 hpf embryos [5]
- NGS Library Preparation: Amplify target regions from extracted DNA and prepare NGS libraries
- Sequencing and Analysis: Sequence amplicons and analyze for precise HDR events
- Embryo Selection: Identify embryos with highest somatic editing efficiency and raise selectively
This combined approach achieved an almost 17-fold increase in somatic editing efficiency in cacna1c KI models, with the greatest benefit observed for alleles with lower intrinsic editing rates [5]. Pre-selection of embryos with high editing rates significantly improves the probability of germline transmission while reducing animal numbers and resource utilization.
Signaling Pathways and Experimental Workflows
CACNA1C Mutation Pathophysiology Pathway
The following diagram illustrates the molecular and physiological consequences of CACNA1C mutations based on the case studies presented:
ZEG-CRISPR Experimental Workflow
The following workflow diagram outlines the integrated pipeline for generating and selecting zebrafish CACNA1C models using ZEG and CRISPR-Cas9:
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagent Solutions for CACNA1C Disease Modeling
| Reagent/Category | Specific Examples | Function/Application | Notes |
|---|---|---|---|
| CRISPR-Cas9 Components | Cas9 protein, sgRNA, ssODN repair templates | Precise introduction of patient-specific mutations | NAD ssODN conformation with Cas9 protein shows superior KI efficiency [5] |
| Genotyping Tools | ZEG device, PCR reagents, HRMA kits, NGS platforms | Early, non-lethal embryo genotyping | ZEG enables >90% survival with accurate genotyping [6] [5] |
| Cell Culture Models | HEK293T cells, iPSCs from patients | In vitro electrophysiology and molecular studies | Patient iPSCs show disease-specific phenotypes [55] [58] |
| Electrophysiology | Patch clamp systems, voltage clamp | Functional characterization of channel properties | Documents inactivation defects and window currents [56] [58] |
| Calcium Channel Blockers | Verapamil, Nimodipine | Therapeutic testing and mechanistic studies | Verapamil reduced arrhythmias in TS patient [55] |
| Experimental Compounds | Roscovitine | Research compound for channel modulation | Normalized inactivation and reduced TH-positive neurons in TS iPSCs [55] |
The case studies and protocols presented herein demonstrate the robust application of CACNA1C mutagenesis and disease modeling for both basic research and therapeutic development. The integration of the ZEG early selection protocol addresses a critical bottleneck in zebrafish genetics, significantly enhancing the efficiency of precise disease model generation. These advanced methodologies enable researchers to better understand the complex genotype-phenotype relationships in CACNA1C-related disorders and provide powerful platforms for screening potential therapeutic interventions. The continued refinement of these approaches will accelerate the translation of basic research findings into clinical applications for patients with calcium channelopathies.
Conclusion
The Zebrafish Embryo Genotyper protocol represents a transformative methodology that addresses critical bottlenecks in zebrafish genetic research. By enabling non-invasive early embryonic genotyping with minimal impact on viability and development, ZEG technology facilitates remarkable 17-fold improvements in CRISPR editing efficiency while aligning with ethical 3Rs principles through reduced animal usage. The integration of early selection protocols with next-generation sequencing creates unprecedented opportunities for accelerating functional genomics, disease modeling, and drug discovery pipelines. As adoption expands across model organisms and molecular applications, ZEG stands to fundamentally reshape genetic research workflows, offering researchers robust tools to enhance throughput, reduce costs, and advance the pace of biomedical discovery across academic and pharmaceutical domains.
References
- 1. High-throughput DNA Extraction and Genotyping of 3dpf ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6102016/]
- 2. A simple and rapid shaking-based assay to genotype live, ... [https://pubmed.ncbi.nlm.nih.gov/40808363/]
- 3. Comparison of whole genome sequencing performance from ... [https://bmcresnotes.biomedcentral.com/articles/10.1186/s13104-024-07075-1]
- 4. Automated multiâ€sample DNA extraction for genotyping ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10953317/]
- 5. Improved selection of zebrafish CRISPR editing by early ... [https://www.nature.com/articles/s41598-023-27503-9]
- 6. An automated system for rapid cellular extraction from live ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5854293/]
- 7. An automated system for rapid cellular extraction from live ... [https://pubmed.ncbi.nlm.nih.gov/29543903/]
- 8. An automated system for rapid cellular extraction from live ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0193180]
- 9. Optimization of a zebrafish embryo genotyping device [https://collections.lib.utah.edu/details?id=1678746&page=13&facet_type_t=%22Text%22&facet_department_t=%22Mechanical+Engineering%22&facet_school_or_college_t=%22College+of+Engineering%22&facet_format_medium_t=%22application%2Fpdf%22&facet=]
- 10. Cellular and Molecular Characterization of the Effects ... [https://pubmed.ncbi.nlm.nih.gov/33481695/]
- 11. Cellular and Molecular Characterization of the Effects of the ... [https://findanexpert.unimelb.edu.au/scholarlywork/1747717-cellular-and-molecular-characterization-of-the-effects-of-the-zebrafish-embryo-genotyper-protocol]
- 12. A minimally invasive fin scratching protocol for fast ... [https://www.nature.com/articles/s41598-022-26822-7]
- 13. A Rapid and Noninvasive Method That Extracts ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9419989/]
- 14. Improved selection of zebrafish CRISPR editing by early ... [https://www.nature.com/articles/s41598-023-27503-9?error=cookies_not_supported&code=bd2a76c8-8dac-40e5-93f8-cc51c72d74f8]
- 15. PCR-Based Genotyping of Zebrafish Genetic Mutants [https://bio-protocol.org/en/bpdetail?id=5248&type=0]
- 16. Automated Morphological Feature Assessment for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6358258/]
- 17. wFluidx, Inc.: A Better Way to Genotype [https://www.wfluidx.com/]
- 18. Quantitative characterization of zebrafish development based ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10278635/]
- 19. Rapid and Efficient Zebrafish Genotyping Using PCR with ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4116811/]
- 20. Cell-type-specific mRNA transcription and degradation ... [https://www.nature.com/articles/s41467-024-47290-9]
- 21. High resolution of full-length RNA sequencing deciphers ... [https://bmcbiol.biomedcentral.com/articles/10.1186/s12915-025-02271-2]
- 22. CRISPR workflow for genome editing | IDT [https://www.idtdna.com/pages/technology/crispr/crispr-workflow]
- 23. Improved selection of zebrafish CRISPR editing by early ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9883431/]
- 24. DNA quantification with PicoGreen [https://www.protocols.io/view/dna-quantification-with-picogreen-d7299qh6.html]
- 25. DNA Quantification with PreciseGreen® [https://www.lumiprobe.com/protocols/DNA-quantification-with-PicoGreen]
- 26. How to Work with Challenging or Limited Genomic ... [https://blog.omni-inc.com/blog/how-to-work-with-challenging-or-limited-genomic-samples-updated-for-2025]
- 27. Broad Range Assay Standard Protocol | Technical Note 142 [https://www.denovix.com/tn-142-broad-range-assay-standard-protocol/]
- 28. Rules and Tips for PCR & qPCR Primer Design | IDT [https://www.idtdna.com/page/support-and-education/decoded-plus/how-to-design-primers-and-probes-for-pcr-and-qpcr/]
- 29. Proven tips for PCR primer design [https://www.neb.com/en-us/nebinspired-blog/proven-tips-for-pcr-primer-design?srsltid=AfmBOoqSSKWR1cWyydZwtg51nxxhAWyPguEIRpG1_CkbqnvTPdFb2Rpn]
- 30. Guidelines for PCR Optimization with Taq DNA Polymerase [https://www.neb.com/en-us/tools-and-resources/usage-guidelines/guidelines-for-pcr-optimization-with-taq-dna-polymerase?srsltid=AfmBOoosriyevNX_exaw_8CqwOtvQuryZoySyDF4NIyxW95OgLqRvUUI]
- 31. How to Design Primers for PCR Experiments [https://www.zymoresearch.com/blogs/blog/how-to-design-primers-for-pcr-experiments?srsltid=AfmBOoqa31N2OXGy98I940KZuMaTq_isgn9RVbOLqsKPdtXx1y89FlFP]
- 32. A Definitive, Practical Guide to Designing Primers for PCR ... [https://medium.com/@zweyehtut/a-definitive-practical-guide-to-designing-primers-for-pcr-qpcr-88d7bf6e2d93]
- 33. Cellular Stress Response - an overview [https://www.sciencedirect.com/topics/neuroscience/cellular-stress-response]
- 34. Microfluidic-aided genotyping of zebrafish in the first 48 h ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4615577/]
- 35. Cellular and molecular mechanisms of stress-induced ... [https://www.explorationpub.com/Journals/en/Article/10068]
- 36. Endoplasmic reticulum stress: molecular mechanism and ... [https://www.nature.com/articles/s41392-023-01570-w]
- 37. Optimized Analysis of Proteins from Xenopus Oocytes and Embryos... [https://www.jove.com/t/69139/optimized-analysis-proteins-from-xenopus-oocytes-embryos]
- 38. Methods for CRISPR/Cas9 Xenopus tropicalis Tissue- Specific ... [https://experiments.springernature.com/articles/10.1007/978-1-4939-8784-9_3?error=cookies_not_supported&code=464ca950-27e6-402b-a40b-c6365ca2be9d]
- 39. Deep transcriptome profiling reveals limited conservation of A-to-I RNA... [https://bmcbiol.biomedcentral.com/articles/10.1186/s12915-023-01756-2]
- 40. ZFIN: Zebrafish Book: Molecular Methods [https://zfin.org/zf_info/zfbook/chapt9/9.3.html]
- 41. A simple method to study motor and non-motor behaviors ... [https://www.sciencedirect.com/science/article/abs/pii/S0165027019300809]
- 42. The Effects of Early Life Stress on the Brain and Behaviour [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2021.657591/full]
- 43. Get a Move On: How to Measure Movement in Zebrafish [https://bitesizebio.com/49450/measure-movement-in-zebrafish/]
- 44. Quantification of larval zebrafish motor function in multiwell ... [https://www.nature.com/articles/nprot.2014.094]
- 45. Standardized Method for the Assessment of Behavioral ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8389650/]
- 46. Long lasting anxiety following early life stress is dependent ... [https://www.nature.com/articles/s41598-022-16257-5]
- 47. Modular Laboratory Exercises to Analyze the Development of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2765818/]
- 48. Early stress exposure on zebrafish development [https://pmc.ncbi.nlm.nih.gov/articles/PMC11286684/]
- 49. Skin swabbing for DNA sampling of zebrafish [https://nc3rs.org.uk/3rs-resource-library/skin-swabbing-dna-sampling-zebrafish]
- 50. Definition of the 3Rs principle [https://www.fc3r.com/en/the-3Rs.php]
- 51. Advancing the 3Rs: innovation, implementation, ethics and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10310538/]
- 52. Toward a common interpretation of the 3Rs principles in ... [https://www.nature.com/articles/s41684-024-01476-2]
- 53. Zebrafish and the 3Rs: Ethical and Efficient Research ... [https://www.zeclinics.com/blog/zebrafish-and-the-3rs]
- 54. Considerations for experimental animal ethics in the ... [https://www.kosinmedj.org/journal/view.php?number=1220]
- 55. A rare mutation of CACNA1C in a patient with Bipolar disorder ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4151967/]
- 56. Novel Timothy syndrome mutation leading to increase in ... [https://www.sciencedirect.com/science/article/abs/pii/S1547527114010765]
- 57. Expanding the phenotype of CACNA1C mutation disorders [https://pmc.ncbi.nlm.nih.gov/articles/PMC8222832/]
- 58. A mutation in the CACNA1C gene leads to early repolarization ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0177532]