RNAscope Sample Preparation Fixation: A Complete Guide for Reliable Gene Expression Analysis
This comprehensive guide details the critical role of fixation in RNAscope sample preparation for researchers and drug development professionals. It covers foundational principles of RNA preservation, detailed methodological protocols for FFPE and frozen tissues, systematic troubleshooting for common fixation issues, and validation strategies against gold-standard techniques. The article emphasizes how proper fixation practices ensure high sensitivity and specificity in detecting RNA biomarkers, ultimately supporting robust and reproducible research outcomes in biomedical and clinical contexts.
RNAscope Sample Preparation Fixation: A Complete Guide for Reliable Gene Expression Analysis
Abstract
This comprehensive guide details the critical role of fixation in RNAscope sample preparation for researchers and drug development professionals. It covers foundational principles of RNA preservation, detailed methodological protocols for FFPE and frozen tissues, systematic troubleshooting for common fixation issues, and validation strategies against gold-standard techniques. The article emphasizes how proper fixation practices ensure high sensitivity and specificity in detecting RNA biomarkers, ultimately supporting robust and reproducible research outcomes in biomedical and clinical contexts.
The Critical Role of Fixation in RNAscope Technology
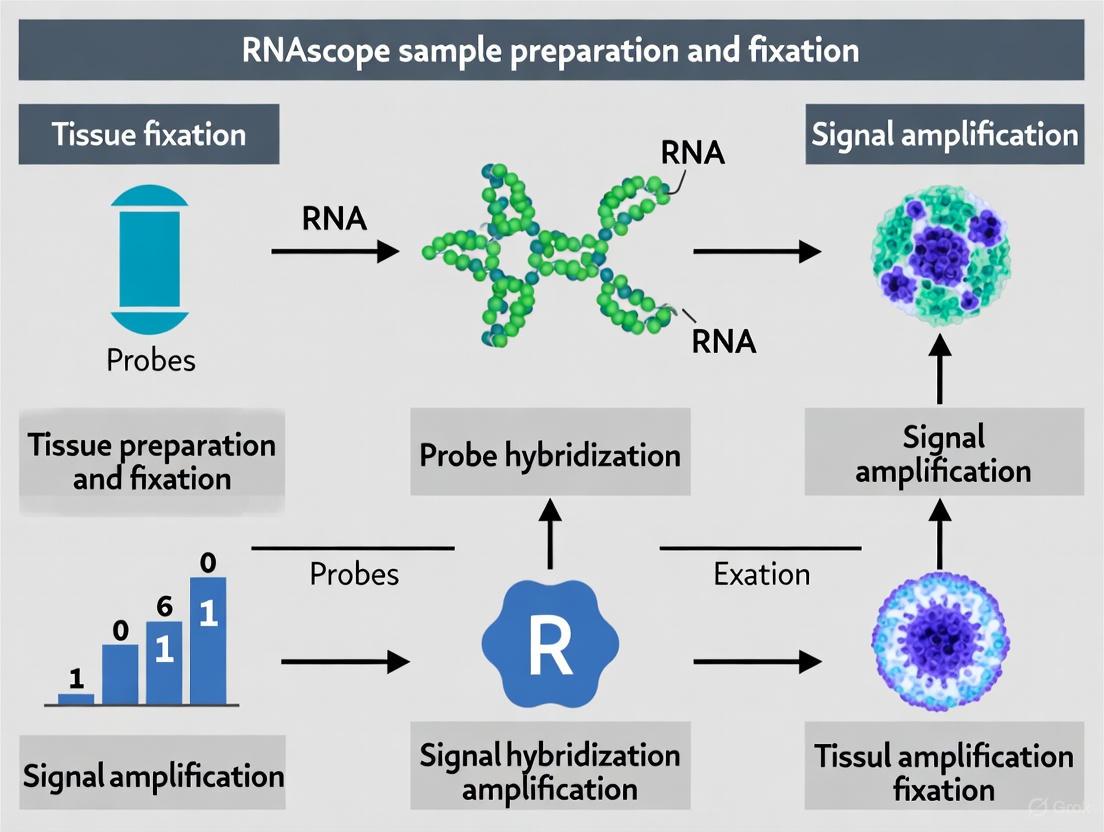
RNAscope represents a groundbreaking advance in the field of spatial genomics, providing researchers with an unprecedented ability to visualize RNA expression within the morphological context of intact cells and tissues. This novel in situ hybridization (ISH) platform addresses the critical limitations of conventional RNA detection methods, which often suffer from insufficient sensitivity and specificity to reliably detect low-abundance RNA biomarkers in clinical and research specimens [1]. Traditional techniques like quantitative RT-PCR, while sensitive, require RNA extraction that destroys valuable tissue architecture and spatial information [1] [2]. Similarly, conventional RNA ISH methods have been largely restricted to detecting highly expressed targets due to challenges with background noise and non-specific hybridization [1] [2].
The fundamental innovation of RNAscope lies in its unique probe design strategy that enables simultaneous signal amplification and background suppression, achieving single-molecule visualization while preserving tissue morphology [3] [1]. This technological leap has positioned RNAscope as a powerful tool for translating RNA biomarker discoveries into clinically applicable diagnostic assays, particularly in the fields of cancer research, neuroscience, and drug development [1] [4] [2]. The compatibility of RNAscope with routine formalin-fixed, paraffin-embedded (FFPE) tissue specimens, the standard for clinical pathology archives, further enhances its utility for both retrospective and prospective studies [1].
The Core Technology: Z-Probes and Signal Amplification
The Double Z Probe Design
At the heart of the RNAscope technology is the proprietary double Z probe design, a concept that fundamentally improves the signal-to-noise ratio of RNA in situ hybridization. This design strategy employs pairs of target-specific probes (double Z probes) that must hybridize in tandem to the target RNA sequence for signal amplification to occur [3] [5].
Each individual Z probe contains three distinct structural elements:
- The lower region consists of an 18-25 base sequence complementary to the target RNA, selected for specific hybridization properties [3]
- A spacer sequence that links the target-binding region to the tail sequence [3]
- The upper tail region featuring a 14-base tail sequence [3]
The critical innovation is that two independent Z probes must bind adjacent to each other on the target RNA molecule, with their combined tail sequences forming a single 28-base binding site for the pre-amplifier molecule [3] [1]. This requirement dramatically reduces background noise because it is statistically improbable that two independent probes would nonspecifically bind to a non-target sequence in the correct orientation and proximity to form the functional pre-amplifier binding site [3] [2]. This design ensures that only target-specific signals undergo amplification, while non-specific hybridization events remain undetectable [3].
For each target RNA, approximately 20 double Z probe pairs are designed to hybridize along a 1-kilobase region of the target molecule [3] [1]. This multi-probe approach provides robustness against variable target accessibility or partial RNA degradation, which is particularly valuable when working with archived clinical specimens [3] [5].
Signal Amplification Cascade
The RNAscope signal amplification system operates through a cascade of sequential hybridization events that build upon the foundation established by the double Z probes:
Pre-amplifier Binding: Once a double Z probe pair hybridizes to the target RNA, the combined 28-base tail sequence serves as a binding site for the pre-amplifier molecule [3] [1]
Amplifier Assembly: Each pre-amplifier contains 20 binding sites for amplifier molecules, which subsequently bind to their complementary sites [3]
Label Probe Attachment: Each amplifier provides 20 binding sites for label probes that are conjugated with either fluorescent molecules or chromogenic enzymes [3]
This multi-stage amplification system can theoretically generate up to 8,000 labels for each target RNA molecule, providing the sensitivity necessary for single-molecule detection [1] [2]. The label probes can be conjugated with various detection moieties, including fluorescent dyes for multiplex analysis or enzymes such as horseradish peroxidase (HRP) or alkaline phosphatase for chromogenic detection compatible with standard bright-field microscopy [1].
Table 1: Key Advantages of RNAscope Probe Design and Amplification Strategy
| Feature | Technical Basis | Practical Benefit |
|---|---|---|
| High Sensitivity | 20×20×20 amplification strategy enables detection of single RNA molecules [3] | Single-molecule visualization; requires only 3 probe pairs for detection [3] |
| Exceptional Specificity | Double Z probe design prevents amplification of non-specific signals [3] | Distinguishes single-base differences; minimal background noise [3] [2] |
| Degraded Sample Compatibility | Short target regions (40-50 bases) work with partially degraded RNA [3] | Effective with archival FFPE tissues and suboptimal specimens [3] [1] |
| Quantitative Capability | Discrete punctate signals correspond to individual RNA molecules [3] | Enables cell-by-cell manual counting or automated quantification [3] [6] |
RNAscope Workflow and Protocol
The standard RNAscope procedure follows a systematic workflow that can be implemented manually or automated using platforms such as the Leica BOND RX system [7] [4]. The protocol varies slightly depending on sample type (FFPE vs. frozen tissue), but core steps remain consistent.
Sample Preparation and Pretreatment
Proper sample preparation is critical for successful RNAscope analysis. For FFPE tissues, sections of 5μm thickness are standard [1]. The pretreatment process involves:
- Deparaffinization and Dehydration: Slides are deparaffinized in xylene followed by ethanol series [1]
- Antigen Retrieval: Tissue sections are incubated in citrate buffer (10 mmol/L, pH 6) at boiling temperature (100-103°C) for 15 minutes [1]
- Protease Digestion: Treatment with protease (typically 10 μg/mL) at 40°C for 30 minutes to unmask target RNA and permeabilize cells [1]
For frozen tissues, optimal preparation includes perfusion fixation with 4% paraformaldehyde (PFA), cryoprotection with sucrose gradients, and embedding in OCT compound before sectioning at 10μm thickness [8]. Frozen sections may undergo post-fixation in 4% PFA, ethanol dehydration, and protease treatment appropriate for the specific tissue type [8].
Hybridization and Amplification
The core detection phase involves sequential hybridization steps:
Target Probe Hybridization: RNAscope probes (approximately 20 double Z probe pairs targeting 1kb of the RNA sequence) are hybridized to the target RNA in a specialized hybridization buffer at 40°C for 2-3 hours [1]
Signal Amplification Cascade:
Between each hybridization step, slides are washed with a stringent wash buffer to remove unbound reagents [1]
Detection and Visualization
The final detection depends on the label type used:
- Chromogenic Detection: For bright-field microscopy, HRP-based label probes are typically used with DAB or Fast Red substrates, followed by counterstaining with hematoxylin [1]
- Fluorescent Detection: For fluorescence microscopy, label probes are directly conjugated with fluorophores such as Alexa Fluor dyes [1]
The resulting signals appear as discrete punctate dots, with each dot representing an individual RNA molecule that can be visualized under standard microscopy [3] [1].
Table 2: Essential Research Reagent Solutions for RNAscope
| Reagent/Category | Specific Examples | Function and Application |
|---|---|---|
| Probe Systems | Target-specific Z-probes, Positive control (PPIB, Polr2A, UBC), Negative control (dapB) [2] | Target detection; assay validation and quality control [2] |
| Detection Reagents | HRP- or AP-based label probes, DAB, Fast Red, Alexa Fluor dyes [1] | Signal generation for chromogenic or fluorescent detection [1] |
| Pretreatment Kits | RNAscope Pretreatment Kit, Protease solutions [3] [1] | Tissue permeabilization and target unmasking [3] |
| Automation Systems | Leica BOND RX with RNAscope integration [7] | High-throughput, consistent automated processing [7] |
| Analysis Software | HALO, QuPath, CellProfiler, Aperio [3] [2] [6] | Image analysis and quantitative signal quantification [3] [6] |
Quality Control and Validation
Robust quality control measures are essential for reliable RNAscope results. The technology incorporates built-in control systems to validate assay performance:
- Positive Controls: Housekeeping genes such as PPIB (peptidylprolyl isomerase B) for moderately expressed genes, Polr2A (RNA polymerase II subunit A) for low expression targets, and UBC (Ubiquitin C) for highly expressed genes verify tissue RNA integrity and assay procedure [2]
- Negative Controls: The bacterial gene dapB (dihydrodipicolinate reductase) confirms the absence of background noise in animal tissues [1] [2]
Proper control implementation is particularly crucial when working with clinical specimens, where RNA degradation or suboptimal fixation can impact results [1] [2]. The quality control system ensures that both false positives and false negatives are minimized, with studies demonstrating concordance rates of 81.8-100% with qPCR, qRT-PCR, and DNA ISH methods [2].
Advanced Applications and Methodological Variations
Multiplex Detection and Co-localization Studies
RNAscope supports sophisticated experimental designs for complex research questions:
Multiplex RNA Detection: Using multiple probe sets with different fluorophores, RNAscope can simultaneously detect up to four different RNA targets in a single sample [1] [2]. The Leica BOND RX system with software version 7.0 extends this capability to visualize up to six different markers on a single slide [7]
RNA-Protein Co-detection: Advanced protocols enable simultaneous detection of RNA and protein targets on the same tissue section through combined RNAscope and immunohistochemistry (IHC) [2] [9]. Novel tissue blocking reagents and detection strategies facilitate this integration without cross-reactivity [9]
BaseScope for Short Sequences: A variant technology called BaseScope is optimized for detecting shorter RNA sequences (50-300 bases), making it particularly suitable for identifying splice variants, single nucleotide polymorphisms, and highly homologous sequences [8]
Automated Platforms for High-Throughput Applications
For laboratories requiring higher throughput and standardization, automated platforms like the Leica BOND RX provide fully integrated RNAscope solutions [7]. These systems offer:
- Processing of up to 30 slides per run with total hands-off operation [7]
- Consistent application of reagents through proprietary Covertile technology that maintains tissue morphology [7]
- Flexible programming for custom protocols and integration with emerging technologies [7]
Quantitative Image Analysis Approaches
Accurate quantification of RNAscope results leverages both manual and computational methods:
- Manual Counting: Direct counting of punctate dots within defined cell regions under microscopy [3]
- Automated Analysis: Software platforms including HALO, QuPath, and CellProfiler enable high-throughput quantitative analysis of RNAscope data [3] [2] [6]
These analytical tools can identify cells, count RNA dots per cell, and generate quantitative expression data, with specific pipelines developed for different assay formats (chromogenic vs. fluorescent) [6]. The discrete nature of RNAscope signals (each dot represents a single RNA molecule) makes it particularly amenable to robust quantification compared to traditional diffuse ISH signals [3] [2].
Troubleshooting and Technical Considerations
Successful implementation of RNAscope requires attention to several technical aspects:
- Protease Optimization: Protease concentration and incubation time must be titrated for different tissue types to balance signal intensity with tissue morphology [1] [8]
- Fixation Conditions: Standardized fixation in 10% neutral buffered formalin for 6-72 hours following ASCO/CAP guidelines ensures optimal RNA preservation [1]
- Hybridization Conditions: Temperature, pH, and buffer composition must be carefully controlled during hybridization and amplification steps [1]
- Signal-to-Noise Optimization: Appropriate use of controls and titration of detection reagents helps maximize specific signal while minimizing background [3] [2]
For challenging samples with partial RNA degradation, the robust probe design (20 independent probe pairs targeting different regions of the RNA) provides redundancy that maintains detection capability even when some target regions are inaccessible [3] [5].
RNAscope technology represents a transformative approach to spatial RNA analysis, combining exceptional sensitivity and specificity through its innovative double Z probe design and signal amplification system. Its ability to provide single-molecule detection in the context of preserved tissue architecture makes it uniquely powerful for both basic research and clinical applications. As spatial genomics continues to advance, RNAscope stands as a cornerstone technology that enables researchers to bridge the gap between molecular discoveries and their biological context, ultimately driving advances in understanding disease mechanisms and developing targeted therapies.
The quality of sample fixation is the most critical determinant of success in RNAscope in situ hybridization assays. Proper fixation preserves RNA integrity, maintains tissue morphology, and enables accurate detection of RNA molecules at single-cell resolution. Within the broader context of RNAscope sample preparation research, this protocol establishes the foundational principles for achieving reliable and reproducible results across diverse experimental conditions. Suboptimal fixation represents a primary point of failure in RNA ISH workflows, leading to either degraded RNA signals or compromised tissue architecture that undermines subsequent spatial analysis. This application note details the gold-standard fixation methodology using Fresh 10% Neutral Buffered Formalin (NBF) for 16-32 hours at room temperature, providing researchers with evidence-based protocols to ensure data validity in gene expression studies, biomarker validation, and drug development research.
Core Protocol: FFPE Sample Preparation Using 10% NBF
Materials and Equipment
Table 1: Essential Materials for RNAscope Sample Preparation
| Item | Specification | Purpose |
|---|---|---|
| Fixative | Fresh 10% Neutral Buffered Formalin (NBF) | Preserve RNA and tissue morphology |
| Tissue Processing | Standard ethanol series, xylene, paraffin wax | Dehydrate, clear, and embed tissue |
| Microtome | Standard histological equipment | Cut 5 ± 1 μm sections |
| Water Bath | 40-45°C | Float paraffin ribbons |
| Microscope Slides | SuperFrost Plus (Fisher Scientific #12-550-15) | Prevent tissue detachment during assay |
| Hydrophobic Barrier Pen | ImmEdge Pen (Vector Labs #310018) | Create reagent containment barriers |
Step-by-Step Fixation and Embedding Protocol
Tissue Dissection and Fixation Initiation
Fixation Duration and Timing
- Maintain fixation for precisely 16-32 hours [10] [12] [11].
- Critical Note: Fixation for less than 16 hours or more than 32 hours will impair RNAscope assay performance [12]. Under-fixation results in protease over-digestion during pretreatment, leading to RNA loss and poor morphology. Over-fixation causes protease under-digestion, resulting in poor probe accessibility, low signal, and compromised signal-to-background ratio despite excellent morphology preservation [10].
Post-Fixation Processing
- Following fixation, wash samples with 1X PBS to remove residual fixative.
- Dehydrate tissue using a standard ethanol series (typically 70%, 95%, 100% ethanol).
- Clear tissue in xylene and embed in paraffin wax using standard histological procedures [12].
Sectioning and Slide Preparation
- Cut embedded tissue into 5 ± 1 μm sections using a microtome [12] [11].
- Float paraffin ribbons in a 40-45°C water bath and mount on SuperFrost Plus slides [12].
- Air dry slides overnight at room temperature.
- Store sections with desiccants at room temperature and use within 3 months for optimal results [12] [11]. For long-term storage (>1 year) of paraffin blocks, 2-8°C with desiccation is recommended [12].
Protocol Extension: Specialized Sample Types
Fresh Frozen Tissue Preparation
For fresh frozen applications, particularly relevant for neuroscience research using rodent brain tissue:
- Deeply anesthetize the animal and perform sacrifice by decapitation.
- Quickly remove the brain from the skull, carefully removing bone and membrane residues.
- Immediately snap-freeze the brain in chilled 2-methylbutane (isopentane) at -30°C for 25 seconds [13].
- Wrap the snap-frozen brain in aluminum foil, avoid any thawing, and immediately store on dry ice or at -80°C.
- Store brains at -80°C for up to 12 months; longer storage is not recommended as mRNA degrades progressively [13].
- Cut 10-20 μm sections using a cryostat and mount on SuperFrost Plus slides [11].
Decalcified Tissue Processing
For calcified tissues (teeth, bone) requiring decalcification before RNAscope:
- Following perfusion and post-fixation in 4% PFA for 3 days at 4°C, subject samples to decalcification.
- Optimal Decalcification Methods: Use either ACD decalcification buffer or Morse's solution to preserve RNA integrity [14].
- Traditional decalcification procedures (e.g., with formic acids or EDTA) may strongly reduce detectable mRNA transcripts and are not recommended [14].
- After decalcification, process through standard dehydration, paraffin embedding, and sectioning at 5 μm thickness.
Figure 1: Comprehensive workflow for RNAscope sample preparation, covering FFPE, fresh frozen, and decalcified tissue processing paths.
Experimental Validation and Quality Control
Control Slide Implementation
- Run a minimum of 3 slides per sample: target marker panel, positive control, and negative control probe [11].
- For FFPE samples, use RNAscope Control Slides (Human: Cat. No. 310045; Mouse: Cat. No. 310023) with appropriate positive and negative control probes [12].
- Positive Control Probes: Use species-specific housekeeping genes (PPIB for single-plex, POLR2A and PPIB for duplex, POLR2A/PPIB/UBC for 3-plex) [11].
- Negative Control Probes: Use bacterial dapB to assess background and non-specific signal [11].
Interpretation of Control Results
- A valid experiment requires the positive control to have a score of 2+ for RNAscope (or 1+ for BaseScope) and the dapB negative control to have a score of 0 [11].
- Only when these control conditions are met can you confidently interpret target RNA expression in the tissue specimen [11].
- For suboptimally fixed tissues, optimization of pretreatment conditions (target retrieval and/or protease digestion) may be required [10].
Troubleshooting Fixation Issues
Table 2: Troubleshooting Fixation-Related Problems in RNAscope
| Problem | Potential Cause | Solution |
|---|---|---|
| Poor tissue morphology | Under-fixation (<16 hrs) | Optimize fixation time; ensure fresh 10% NBF is used |
| Low signal or poor signal-to-background ratio | Over-fixation (>32 hrs) | Reduce fixation time; avoid prolonged fixation |
| RNA degradation | Delayed fixation; improper storage | Begin fixation immediately after dissection; store samples properly |
| Variable staining in TMA | Differential fixation across cores | May require optimization of pretreatment conditions for each core [10] |
| No signal in dental pulp | Improper decalcification method | Use ACD decalcification buffer or Morse's solution [14] |
Essential Research Reagent Solutions
Table 3: Critical Reagents for RNAscope Sample Preparation and Assay
| Reagent/Equipment | Function | Specification |
|---|---|---|
| 10% NBF | Tissue fixation | Fresh, neutral buffered formalin; pH 7.0 |
| RNAscope Target Retrieval Reagents | Antigen retrieval | Restores probe accessibility in over-fixed tissue |
| RNAscope Protease Plus | Tissue permeabilization | Digests proteins to enable probe access; concentration critical |
| HybEZ Oven | Hybridization incubation | Maintains precise humidity and temperature (40°C) control |
| RNAscope Positive/Negative Control Probes | Assay validation | Verify RNA quality and assay performance; species-specific |
| SuperFrost Plus Slides | Tissue adhesion | Special coating prevents tissue detachment during stringent washes |
| ImmEdge Hydrophobic Barrier Pen | Liquid containment | Creates barrier to prevent reagent evaporation and cross-contamination |
Adherence to the gold-standard fixation protocol using Fresh 10% NBF for 16-32 hours at room temperature provides the foundation for successful RNAscope assays. This rigorously validated methodology ensures optimal preservation of both RNA integrity and tissue morphology, enabling precise spatial gene expression analysis across diverse sample types including FFPE, fresh frozen, and decalcified tissues. Implementation of appropriate controls and quality assessment measures, as outlined in this protocol, is essential for generating reliable, reproducible data in research and drug development applications. Through standardized sample preparation practices, researchers can maximize the powerful capabilities of RNAscope technology for single-molecule RNA detection while maintaining experimental rigor and reproducibility.
Sample fixation is a foundational step in the RNAscope in situ hybridization (ISH) workflow, serving as the primary determinant for preserving both tissue architecture and nucleic acid integrity. Within the context of a broader thesis on RNAscope sample preparation, this application note delineates the precise consequences of fixation deviations from established protocols. Proper fixation creates an equilibrium where RNA is sufficiently cross-linked to remain in situ, yet accessible for hybridization with target-specific probes. The industry-standard recommendation for formalin-fixed, paraffin-embedded (FFPE) tissues specifies fixation in fresh 10% neutral buffered formalin (NBF) for 16–32 hours at room temperature [10] [15]. Deviations from this narrow window initiate a cascade of molecular and morphological compromises that directly impact assay sensitivity, specificity, and the validity of experimental conclusions. This document provides a detailed quantitative and qualitative analysis of these effects, supported by experimental data and complemented by protocols for troubleshooting and optimization.
The Direct Impacts of Under-fixation and Over-fixation
Mechanistic Consequences and Morphological Outcomes
The following table summarizes the primary effects of fixation outside the recommended parameters:
| Fixation Type | Impact on Protease Digestion | Effect on RNA & Morphology | Signal & Background Outcome |
|---|---|---|---|
| Under-fixation [10] | Over-digestion | Loss of RNA and poor tissue morphology [10] | Low signal and poor signal-to-background ratio |
| Over-fixation [10] | Under-digestion | Poor probe accessibility while maintaining excellent tissue morphology [10] | Low signal and poor signal-to-background ratio |
The underlying mechanism for these outcomes revolves around the balance formaldehyde achieves between protein-RNA cross-linking and biomolecule preservation. Under-fixation results in an inadequately cross-linked matrix, leaving the tissue vulnerable to over-digestion during the requisite protease pretreatment step. This excessively permeabilizes the tissue, allowing RNA to leach out, thereby diminishing the target pool and disrupting cellular morphology [10]. Conversely, over-fixation creates an excessively dense network of cross-links that, while excellent for preserving morphology, physically blocks the access of RNAscope ZZ probes to their target RNA sequences. The protease step cannot adequately relieve this masking, leading to significantly reduced hybridization efficiency and signal intensity, even though the RNA may be present [10].
Quantitative Evidence of Prolonged Formalin Fixation
While the standard protocol warns against fixation beyond 32 hours, a 2024 study systematically quantified the effect of extremely prolonged formalin fixation on RNAscope signal detection. The research evaluated the signal of a reference gene (16S rRNA) in various tissues fixed in 10% NBF for periods ranging from 1 day to 270 days [16].
The data from this study is summarized in the table below:
| Formalin Fixation Duration | Signal Intensity & Percent Area | Qualitative Detection Outcome |
|---|---|---|
| 1 - 28 days | Progressive but measurable decrease | Detectable signal in all tissues |
| 60 - 90 days | Continued decrease | Signal remains detectable |
| 180 days | Significant decrease | Signal still detectable in tissues |
| 270 days | Signal intensity and percent area decreased to minimal levels | No detectable signal [16] |
This quantitative data demonstrates that while RNAscope is remarkably robust, with detectable signals persisting after 180 days of formalin immersion, a point of irreversible loss is eventually reached. The study attributed this loss to the formation of irreversible covalent bonds and RNA fragmentation over extended fixation periods, which ultimately destroys the target sequences necessary for probe hybridization [16]. This finding is crucial for researchers working with archival samples where fixation history may be unknown.
Visualizing the Effects and Optimization Pathways
The following diagram illustrates the cause-and-effect relationships of improper fixation and the corresponding optimization pathways to rectify these issues.
Experimental Protocol for Fixation Condition Assessment and Optimization
This protocol provides a step-by-step methodology for diagnosing and troubleshooting suboptimal fixation, a critical procedure for validating archival samples in a research thesis.
Materials and Reagents
| Category | Item | Function & Specification |
|---|---|---|
| Essential Controls | RNAscope Control Slides (e.g., Hela or 3T3 cell pellets) [15] | Verify assay performance and reagent integrity. |
| Positive Control Probes (e.g., PPIB, POLR2A, UBC) [15] [11] | Species-specific probes to confirm RNA quality and accessibility. | |
| Negative Control Probe (dapB) [15] [11] | Assess non-specific background and false positives. | |
| Pretreatment Reagents | RNAscope Target Retrieval Reagents [17] | Reverse cross-links formed during fixation to expose target RNA. |
| RNAscope Protease Reagents (Plus, III, IV) [17] | Permeabilize cell membranes and unmask RNA targets; key for optimization. | |
| Lab Supplies | SuperFrost Plus Microscope Slides [15] [11] | Prevent tissue detachment during stringent assay steps. |
| HybEZ II Hybridization System [11] | Provide precise temperature and humidity control for hybridization. |
Step-by-Step Procedure
Sectioning and Slide Preparation: Cut FFPE tissue sections at a thickness of 5 ± 1 µm [15]. Mount sections on SuperFrost Plus slides to ensure adhesion. Air-dry slides and bake at 60°C for 1-2 hours before proceeding [15].
Initial Assay Run with Controls: Process test slides alongside RNAscope control slides (e.g., Hela or 3T3 cell pellets) [15]. Follow the standard RNAscope protocol using the recommended starting conditions for target retrieval and protease digestion from the user manual.
Diagnostic Interpretation: Evaluate the staining results by comparing the target signal with the control probes.
- Assay Validation: The control slides with positive (PPIB) and negative (dapB) probes must perform as expected. Successful staining is defined by a PPIB score ≥2 and a dapB score <1 [15] [18].
- Sample Quality Diagnosis: Score the positive control probe (e.g., PPIB) on your test sample. A low score indicates an issue with RNA integrity or accessibility due to improper fixation [10].
Systematic Pretreatment Optimization: If the control probe signals are suboptimal, titrate the protease digestion time.
- For Suspected Under-fixation: The tissue is overly digested. Reduce the protease treatment time in subsequent runs (e.g., by 50%) to protect RNA and preserve morphology [10].
- For Suspected Over-fixation: The tissue is under-digested. Increase the protease treatment time (e.g., double the time) to break more cross-links and improve probe accessibility [10].
- Note: In cases of extreme over-fixation (e.g., months in formalin), signal retrieval may not be possible due to irreversible RNA damage [16].
Validation of Optimized Conditions: Once a clear signal with high signal-to-background is achieved for the control probes, apply the optimized protocol to the target-of-interest probe.
Discussion and Best Practices
The integrity of RNAscope data is contingent upon pre-analytical variables, with fixation being the most critical. This document establishes that both under- and over-fixation lead to the same ultimate outcome—poor assay signal—but through diametrically opposed mechanisms. A key finding for retrospective studies is that while RNAscope is robust and can detect RNA in FFPE blocks stored for up to 15 years [16], the duration of initial formalin fixation is a more stringent limiting factor. Furthermore, the use of fresh 10% NBF is emphasized, as old or non-neutral formalin can accelerate RNA degradation and introduce artifacts not remedied by standard optimization [10] [11].
For a comprehensive thesis, it is essential to integrate these fixation protocols with downstream analysis. RNAscope data is semi-quantitative, and scoring is based on counting punctate dots per cell, with each dot representing a single RNA molecule [17] [18]. Proper fixation ensures that this dot count accurately reflects true gene expression levels, enabling reliable analysis across diverse scenarios such as heterogeneous expression, co-expression studies, and rare cell detection [18]. By adhering to the prescribed fixation guidelines and employing the outlined troubleshooting protocols, researchers can ensure the generation of high-quality, publication-grade data that faithfully represents the spatial expression of RNA targets within their morphological context.
The RNAscope in situ hybridization (ISH) assay represents a significant advancement in molecular pathology, enabling the sensitive and specific detection of RNA targets within intact cells and tissues. A cornerstone of its reliability is the appropriate selection and preparation of biological samples. The technology is compatible with Formalin-Fixed Paraffin-Embedded (FFPE), fresh-frozen, and fixed-frozen tissues, as well as various cell preparations. The choice of sample type directly influences experimental outcomes by affecting RNA preservation, accessibility for probe hybridization, and tissue morphology. This application note provides a detailed comparative analysis of these sample types, supported by quantitative data and standardized protocols, to guide researchers in making informed decisions tailored to their experimental goals within the broader context of RNAscope sample preparation fixation research [15] [17] [2].
Comparative Analysis of Sample Types
The choice between FFPE and frozen tissues involves a fundamental trade-off between superior morphological detail and superior nucleic acid preservation. The table below summarizes the core characteristics, advantages, and challenges of each compatible sample type for the RNAscope assay.
Table 1: Comprehensive Comparison of RNAscope-Compatible Sample Types
| Sample Type | Core Characteristics | Key Advantages | Primary Challenges & Considerations |
|---|---|---|---|
| FFPE | Fixed in 10% NBF, dehydrated, embedded in paraffin [15] | Superior morphology; stable at room temperature; vast archival availability (biobanks); compatible with long-term storage (>25 years demonstrated) [19] [20] | RNA cross-linking & fragmentation; requires antigen retrieval and protease digestion; fixation process must be standardized [20] |
| Fresh-Frozen | Tissue snap-frozen in liquid nitrogen, stored at ≤ -80°C [21] | Optimal RNA preservation; no cross-linking; simpler/faster protocol; no target retrieval required [22] [21] [20] | Compromised cellular morphology; requires constant ultra-low temperature storage; logistically challenging to collect and store [21] [20] |
| Fixed-Frozen | Tissue fixed prior to or after freezing, then sectioned [23] [17] | Balanced approach: better morphology than fresh-frozen; better RNA preservation than FFPE [23] | Protocol variability; potential for ice crystal formation disrupting architecture [22] |
| Cultured Cells | Adherent or suspension cells, fixed post-culture [17] | Uniform cell population; controlled experimental conditions; ideal for assay optimization [17] | Lack of tissue context and heterogeneity; requires specific fixation and permeabilization [17] |
Quantitative data from next-generation sequencing (NGS) studies highlight the impact of sample type on molecular data quality. One study on colorectal cancer reported a 94.0% concordance in mutation detection between matched FFPE and fresh-frozen tissues using a multi-gene panel, with concordance at the gene level ranging from 73.8% to 100% [24]. Another systematic review found that RNAscope has a high concordance rate (81.8–100%) with gold-standard techniques like qPCR and qRT-PCR [2]. These findings confirm that while FFPE samples can yield highly reliable data, the integrity of RNA in fresh-frozen samples often makes them the gold standard for sensitive detection [21].
Table 2: Key Quantitative Comparisons Between FFPE and Fresh-Frozen Tissues from NGS Studies
| Metric | FFPE Tissues | Fresh-Frozen Tissues | Context & Implications |
|---|---|---|---|
| Mutation Detection Concordance | 94.0% (Variant level) | 94.0% (Variant level) | High concordance supports use of FFPE for validated NGS panels [24] |
| Gene-Level Concordance Range | 73.8% - 100% | 73.8% - 100% | Some genes show perfect agreement, while others require careful validation [24] |
| Mapping Statistics (RNA-Seq) | Comparable to FF | Comparable to FFPE | With optimized protocols, FFPE can achieve mapping quality similar to frozen [21] |
| Gene Detection Overlap | Significant overlap with FF | Significant overlap with FFPE | A large proportion of the transcriptome can be reliably detected in both [21] |
RNAscope Technology Workflow and Principle
The exceptional sensitivity and specificity of the RNAscope assay stem from its patented signal amplification and background suppression technology. Unlike traditional RNA ISH, which uses a single labeled probe, RNAscope employs a novel double-Z probe design [17] [2].
The following diagram illustrates the core mechanism of the RNAscope assay, from probe hybridization to signal amplification.
This multi-step process ensures that a signal is generated only when two Z-probes bind adjacent to each other on the target RNA, drastically reducing background noise. The high level of signal amplification (up to 8,000-fold) enables the detection of even low-abundance RNA molecules with single-molecule resolution [17] [2].
Detailed Experimental Protocols
Standardized protocols are critical for the success and reproducibility of the RNAscope assay. The following sections provide detailed methodologies for different sample types.
FFPE Tissue Section Protocol
Key Considerations: FFPE tissues require careful attention to fixation and de-crosslinking steps. For archival samples where fixation details are unknown, pretreatment optimization is strongly recommended [15] [25].
Workflow Diagram: FFPE and Fresh-Frozen Tissue Protocol
Protocol Steps:
- Sectioning: Cut FFPE tissue blocks into 5 ± 1 μm sections and mount on SuperFrost Plus slides [15].
- Deparaffinization and Dehydration: Bake slides at 60°C for 1-2 hours, then deparaffinize in xylene and dehydrate through a graded series of ethanol [15] [25].
- Pretreatment:
- Target Retrieval: Immerse slides in Target Retrieval solution and incubate at 95-100°C for 15 minutes. This step reverses formalin-induced crosslinks [17] [25].
- Protease Digestion: Apply Protease Plus (or Protease III/IV) and incubate for 15-30 minutes at 40°C. This permeabilizes the tissue and unmasks RNA targets [17] [25].
- Probe Hybridization: Apply the desired probe mix and incubate for 2 hours at 40°C in the HybEZ Oven [17].
- Signal Amplification: Perform a series of amplifications (AMP 1, 2, and 3) as per the kit protocol, with wash steps in between [22] [17].
- Detection & Counterstaining: Develop the signal using the appropriate chromogenic or fluorescent substrates. Counterstain with hematoxylin (chromogenic) or DAPI (fluorescent), and mount with compatible media [22] [25].
Fresh-Frozen and Fixed-Frozen Tissue Protocol
Key Considerations: The main distinction for frozen tissues is the omission of the target retrieval step, as there are no formalin-induced crosslinks to reverse. However, proper fixation is critical to preserve morphology and prevent RNA degradation [22] [17].
Protocol Steps:
- Tissue Preparation:
- Fresh-Frozen: Snap-freeze tissue in liquid nitrogen-cooled isopentane or directly in liquid nitrogen. Store at -80°C until sectioning. Cut sections at 10-20 μm thickness [22] [21].
- Fixed-Frozen: Perfuse or immerse tissue in 4% PFA for fixation (e.g., 2 hours at RT), then cryoprotect in sucrose solution before snap-freezing and sectioning at 7-15 μm [22] [23].
- Section Fixation: Immerse air-dried sections in 4% PFA for 15 minutes to 2 hours at room temperature [22].
- Dehydration: Dehydrate slides through a series of ethanol (50%, 70%, 100%) for 5 minutes each [22].
- Protease Digestion: Apply Protease Plus (or another recommended protease) for 10-30 minutes at 40°C. Note: Protease concentration and time may require optimization based on fixation strength [22] [17].
- Probe Hybridization & Amplification: Follow the same universal RNAscope steps as for FFPE tissues (Steps 4-6 above) [22] [17].
The Scientist's Toolkit: Essential Reagents and Controls
Success with the RNAscope assay depends on using the correct reagents and systematic validation through controls. The following table catalogues essential solutions.
Table 3: Essential Research Reagent Solutions for RNAscope Assays
| Reagent Category | Specific Examples | Critical Function |
|---|---|---|
| Control Probes | Positive: PPIB, POLR2A, UBCNegative: dapB [15] [17] [25] | Validate assay performance and sample RNA quality. PPIB confirms detection of moderate-expression genes [2]. |
| Pretreatment Reagents | Target Retrieval Reagents, Protease Plus, Protease III, Protease IV, Hydrogen Peroxide [23] [17] | Unmask target RNA by reversing crosslinks (FFPE) and permeabilizing tissue/cell membranes. |
| Core Detection Kit | RNAscope Multiplex Fluorescent Reagent Kit v2 [22] | Contains amplifiers (AMP 1-3), HRP blockers, and detection reagents necessary for the signal amplification cascade. |
| Probe Diluent | RNAscope Probe Diluent [22] [25] | Used to dilute concentrated probe stocks to the correct working concentration for hybridization. |
| Specialized Buffers | 50x Wash Buffer, HybEZ Buffer [22] [25] | Maintain optimal stringency and pH during hybridization and wash steps to ensure specific binding. |
| Epischisandrone | Epischisandrone, MF:C21H24O5, MW:356.4 g/mol | Chemical Reagent |
| Humantenidine | Humantenidine, MF:C19H22N2O4, MW:342.4 g/mol | Chemical Reagent |
The robust compatibility of the RNAscope platform with FFPE, fresh-frozen, and fixed-frozen tissues provides researchers with exceptional flexibility for spatial transcriptomics. The decision matrix ultimately balances the superior morphological detail of FFPE samples against the higher integrity of nucleic acids preserved in frozen tissues. By adhering to the detailed protocols and validation frameworks outlined in this document—including the mandatory use of control probes and pretreatment optimization—researchers can reliably generate high-quality, reproducible data. This enables the precise investigation of gene expression within its native tissue context, accelerating discovery in basic research and drug development.
Step-by-Step Fixation Protocols for Different Sample Types
Formalin-Fixed Paraffin-Embedded (FFPE) tissue represents the most widely used archive in pathology, enabling long-term preservation of tissue histomorphology for diagnostic and research purposes [26] [27]. The FFPE preparation process stabilizes tissue components through chemical cross-linking and physical embedding, creating samples that remain viable for years or even decades [27] [19]. Within the context of RNAscope sample preparation fixation research, proper FFPE handling is paramount because the quality of preserved RNA directly impacts the sensitivity and specificity of this advanced in situ hybridization technology [26] [17]. While FFPE biobanking offers practical advantages over fresh frozen tissue (FFT) methods by eliminating the need for ultra-low-temperature storage, it introduces specific challenges for RNA analysis due to nucleic acid cross-linking and fragmentation during fixation [26] [27]. This protocol outlines comprehensive guidelines for FFPE tissue processing optimized for subsequent RNAscope applications, ensuring researchers can reliably preserve RNA integrity for spatial transcriptomic analysis.
FFPE Tissue Preparation Workflow
The diagram below illustrates the complete FFPE tissue preparation pathway, from tissue acquisition to sectioning, highlighting critical steps that influence RNA integrity.
Materials and Reagents
Research Reagent Solutions
| Item | Function | Application Notes |
|---|---|---|
| 10% Neutral Buffered Formalin (NBF) | Primary fixative that cross-links proteins and nucleic acids to preserve tissue architecture | Optimal fixation: 12-24 hours; excessive fixation causes severe RNA fragmentation [26] [27] |
| Ethanol Solutions | Dehydrating agent that removes water from tissue to enable paraffin infiltration | Use graded series (e.g., 70%, 95%, 100%) to prevent excessive tissue hardening [27] |
| Xylene or Isopropanol | Clearing agent that removes alcohol and fat, creating compatibility with paraffin | Xylene is common but toxic; isopropanol is a less toxic alternative [27] |
| Paraffin Wax | Embedding medium that provides structural support for microtomy | Maintain at ~60°C for embedding; higher temperatures may damage RNA [27] |
| RNAscope Target Retrieval | Buffer system to reverse formalin-induced cross-links and expose RNA targets | Used during pre-treatment; critical for RNAscope assay performance [17] |
| RNAscope Protease Reagents | Enzymes that permeabilize cell membranes and unmask RNA targets | Protease Plus, III, or IV selected based on tissue type and fixation [17] |
Step-by-Step Protocol
Tissue Acquisition and Fixation
Procedure:
- Obtain fresh tissue specimens via biopsy or dissection, minimizing ischemic time (duration between tissue removal and fixation) as prolonged ischemia causes cellular degradation and RNA compromise [27].
- Immediately immerse tissue in adequate volume of 10% Neutral Buffered Formalin (NBF) (typically 10:1 fixative-to-tissue ratio) to ensure complete penetration [27].
- Maintain fixation at room temperature for 6-24 hours, with optimal fixation for breast cancer tissues documented at 12-24 hours [26] [27].
- Avoid extreme fixation durations: insufficient fixation causes inadequate preservation, while excessive fixation promotes extensive nucleic acid cross-linking [27].
Technical Note: For RNAscope applications, controlled fixation timing is critical as formalin fixation induces spontaneous cytosine deamination and RNA fragmentation, potentially complicating subsequent RNA in situ hybridization [26].
Dehydration and Clearing
Procedure:
- Following fixation, rinse tissue briefly in buffer to remove excess formalin.
- Process tissue through a graded ethanol series (e.g., 70%, 95%, 100%) for dehydration, with incubation times adjusted according to tissue size and type [27].
- Transfer tissue to clearing agent (xylene or isopropanol) to displace ethanol and remove lipid components, creating hydrophobicity necessary for paraffin infiltration [27].
- Ensure complete clearing; insufficient clearing results in inadequate paraffin penetration and compromised structural support for sectioning.
Precaution: When using isopropanol as a less toxic alternative to xylene, note that embedding must be performed with higher-temperature paraffin wax [27].
Paraffin Embedding
Procedure:
- Transfer cleared tissues to molten paraffin wax maintained at approximately 60°C in an embedding center [27].
- Conduct multiple wax changes to ensure complete tissue infiltration.
- Orient tissue appropriately in embedding molds and fill with fresh paraffin.
- Cool blocks rapidly on chilled plates to facilitate wax solidification.
- Store finished FFPE blocks at room temperature; properly prepared blocks remain stable for years to decades [27] [19].
Technical Note: Improper embedding can introduce artifacts in tissue sections, affecting accuracy in downstream morphological and molecular analyses [27].
Sectioning
Procedure:
- Cool FFPE blocks on ice before sectioning to improve wax rigidity.
- Cut sections at 4-7µm thickness using a microtome, optimizing thickness based on tissue type and intended applications [26].
- Float sections on a warm water bath (40-45°C) to remove wrinkles.
- Mount sections on positively charged slides (e.g., Superfrost Plus) to enhance adhesion during subsequent processing [26].
- Dry mounted sections thoroughly overnight at 37°C or for 1-2 hours at 60°C before storage or use [26].
Application Note: For RNAscope assays, section thickness should be optimized experimentally; FFPET typically uses 4µm sections, while FFT may require 7µm sections [26].
Quality Assessment for RNAscope
Housekeeping Gene Validation
For research utilizing RNAscope technology, assessment of RNA integrity is essential following FFPE processing. This is typically accomplished using housekeeping gene (HKG) probes that target constitutively expressed transcripts [26] [17]. The table below summarizes quantitative data from FFPE samples using RNAscope HKG probes.
Table 1: Housekeeping Gene Expression in FFPE Tissue for RNAscope Quality Control
| Housekeeping Gene | Expression Level | Average Spots/Cell in Tumor Regions | RNA Integrity Assessment | Archival Stability |
|---|---|---|---|---|
| UBC | High expressor | >15 spots/cell [28] | Most sensitive to degradation; significant archival duration-dependent reduction [26] | Not recommended for long-archived samples |
| PPIB | High expressor | >8 spots/cell [28] | Shows pronounced degradation over time (R²=0.33-0.35) [26] | Moderate archival stability |
| POLR2A | Moderate to low expressor | ≥2 spots/cell [28] | More stable with archival time; minimal degradation impact [26] | Recommended for older samples |
| HPRT1 | Low expressor | Data not available | Relatively stable expression pattern [26] | Good archival stability |
RNA Integrity Verification Protocol
Experimental Methodology:
- Perform RNAscope Multiplex Fluorescent Assay according to manufacturer protocols using four HKG probes (UBC, PPIB, POLR2A, and HPRT1) alongside negative control bacterial dapB [26] [17].
- Apply appropriate pre-treatment steps: bake FFPET slides (2 hours at 60°C) or fix FFT slides (4% PFA, 20 minutes RT) [26].
- Conduct antigen retrieval for FFPET samples at 98°C-102°C using RNAscope Target Retrieval reagents [26] [17].
- Hybridize with target probes, amplify signals, and develop using fluorescent or chromogenic detection methods [17].
- Image stained slides using automated quantitative pathology imaging systems (e.g., Vectra Polaris) within 2 weeks of assay completion [26].
Interpretation Guidelines:
- Successful RNA preservation demonstrated by PPIB/POLR2A score ≥2 or UBC score ≥3 [17].
- Negative control (dapB) should score <1, indicating minimal background noise [17].
- Samples showing adequate HKG expression are considered "fit-for-purpose" for test biomarker analysis using RNAscope [28].
Troubleshooting and Optimization
Table 2: Common FFPE Preparation Challenges and Solutions for RNAscope Applications
| Issue | Potential Cause | Solution | Impact on RNAscope |
|---|---|---|---|
| Excessive RNA degradation | Prolonged ischemic time, improper fixation, extended archival duration | Minimize ischemic time, optimize fixation duration, use POLR2A for older samples | Reduced signal intensity, potential false negatives [26] |
| Incomplete penetration | Large tissue thickness, inadequate processing | Limit tissue thickness to <3-4mm, ensure sufficient processing time | Variable staining across tissue sections |
| Soft tissue blocks | Incomplete dehydration or clearing | Optimize ethanol and xylene incubation times | Difficult sectioning, poor morphology |
| High background noise | Inadequate protease treatment or endogenous peroxidase activity | Optimize protease concentration, use Hydrogen Peroxide block [17] | Reduced signal-to-noise ratio [17] |
| Section detachment | Poor slide adhesion, insufficient drying | Use charged slides, optimize drying conditions | Tissue loss during pre-treatment steps |
Proper execution of the FFPE tissue protocol for fixation, processing, and sectioning establishes the foundation for successful RNAscope in situ hybridization analyses. Through controlled fixation parameters, optimized processing conditions, and rigorous quality assessment using housekeeping genes, researchers can reliably preserve RNA integrity within morphological context, even in tissues archived for extended periods. The guidelines presented herein enable researchers to standardize FFPE sample preparation specifically for spatial transcriptomic applications, ensuring compatibility with the sensitive detection methodology of RNAscope technology. By implementing these protocols, scientists can maximize the research utility of precious FFPE samples for both investigative studies and diagnostic development.
Within the framework of RNAscope sample preparation fixation research, the method of preparing fixed-frozen tissues is a critical cornerstone for achieving high-quality spatial transcriptomics data. Proper preparation preserves RNA integrity and tissue morphology, enabling precise localization of gene expression at the single-cell level [29]. This protocol details the use of sucrose gradients and Optimal Cutting Temperature (OCT) compound embedding, two essential procedures that mitigate ice crystal formation—a major source of RNA degradation and morphological artifacts [30] [31]. The following application note provides a standardized, detailed methodology for researchers and drug development professionals seeking reliable results from RNAscope and other advanced in situ hybridization assays on frozen tissue specimens.
The Scientist's Toolkit: Essential Research Reagent Solutions
The following table catalogues the essential materials required for the successful preparation of fixed-frozen tissue samples.
Table 1: Key Research Reagents and Materials for Fixed-Frozen Tissue Preparation
| Item | Function/Application | Key Considerations |
|---|---|---|
| Paraformaldehyde (PFA) [30] [32] | Tissue fixation; cross-links proteins to preserve tissue architecture and nucleic acids. | Always use freshly prepared 4% PFA in 1X PBS. Methanol-free formulations are recommended for optimal RNA preservation [32]. |
| Sucrose Solutions [30] [31] | Cryoprotectant; infiltrates tissue to displace water and prevent ice crystal formation during freezing. | Prepared in a graded series (e.g., 10%, 20%, 30% w/v in 1X PBS). Infiltration is complete when tissue sinks to the bottom of the container [30]. |
| OCT Embedding Medium [30] [33] | Water-soluble embedding matrix; provides structural support for cryostat sectioning. | Ensure tissue is completely surrounded and covered. Use products like Tissue-Tek OCT for consistent results. |
| SuperFrost Plus Microscope Slides [30] [15] | Tissue section adhesion. | Specifically recommended to prevent tissue loss during subsequent washing and staining steps of the RNAscope assay [30]. |
| Immedge Hydrophobic Barrier Pen [30] | Creates a hydrophobic barrier around tissue sections on slides. | Enables easy application of reagents during the RNAscope assay by forming a well around the sample [30]. |
| RNAscope Universal Pretreatment Reagents [30] | Prepares fixed-frozen sections for the RNAscope assay. | Includes Hydrogen Peroxide, Target Retrieval, and Protease Plus to permeabilize tissues and reduce background [30]. |
| Lignin | Lignin|High-Purity Reagent for Research Applications | High-purity lignin for research (RUO). Explore applications in biomedicine, 3D printing, and sustainable materials. For Research Use Only. Not for human use. |
| Anagyrine | Anagyrine, CAS:33478-03-4, MF:C15H20N2O, MW:244.33 g/mol | Chemical Reagent |
Workflow and Protocol
Comprehensive Workflow Diagram
The diagram below outlines the complete journey of tissue from dissection to being ready for the RNAscope assay.
Detailed Experimental Protocol
Part 1: Tissue Fixation and Sucrose Gradient Infiltration
This initial phase stabilizes the tissue's molecular architecture and protects it from freezing-induced damage.
Materials:
- Freshly dissected tissue
- Freshly prepared 4% Paraformaldehyde (PFA) in 1X PBS [30] [32]
- 10%, 20%, and 30% (w/v) Sucrose solutions in 1X PBS [30] [31]
- Conical tubes, orbital shaker, 4°C refrigerator or cold room
Procedure:
- Fixation: Immediately immerse the dissected tissue in a sufficient volume of 4% PFA (10-20 times the tissue volume) for 24 hours at 4°C [30]. For some tissues, vacuum infiltration can be applied to ensure complete and rapid penetration of the fixative [32].
- Wash: Rinse the fixed tissue with 1X PBS to remove excess PFA. Place the tissue on an orbital shaker and wash with 1X PBS for 20-30 minutes [31].
- Sucrose Infiltration: Transfer the tissue through a graded sucrose series for cryoprotection. Immerse the tissue in each solution at 4°C until it sinks to the bottom of the container, indicating complete infiltration.
Table 2: Sucrose Gradient Infiltration Parameters
| Solution | Incubation Endpoint | Approximate Time (Tissue-Dependent) | Temperature |
|---|---|---|---|
| 10% Sucrose | Tissue sinks to bottom | 18 hours - Overnight [30] | 4°C [30] |
| 20% Sucrose | Tissue sinks to bottom | Several hours - Overnight [31] | 4°C [30] |
| 30% Sucrose | Tissue sinks to bottom | Overnight [30] [31] | 4°C [30] |
Part 2: OCT Embedding and Freezing
This phase encapsulates the tissue in a supportive medium for cryostat sectioning.
Materials:
- Tissue processed through 30% sucrose
- Optimal Cutting Temperature (OCT) compound
- Embedding molds
- Dry ice, isopentane (pre-cooled in liquid nitrogen) or a dry ice/ethanol slurry [33] [31]
- Forceps, foil, plastic bags for storage
Procedure:
- Preparation: Blot the tissue free of extraneous sucrose solution [33]. Fill an embedding mold one-third to one-half full with OCT [33] [31].
- Orientation: Carefully orient the tissue in the mold using forceps. Maintain clearance between the tissue and the sides of the mold. Ensure the tissue is completely surrounded by OCT before final freezing to prevent detachment during sectioning [31].
- Freezing: Slowly lower the mold onto the surface of a pre-cooled isopentane bath or a dry ice-ethanol slurry. Avoid submerging the mold or allowing the OCT to directly contact the ethanol, as this can create a gel-like surface [31]. Freeze until the OCT is firm and opaque (1-2 minutes).
- Storage: Wrap the frozen block tightly in foil or place it in a labeled plastic bag. Store at -80°C in an airtight container to prevent freeze-drying [30] [33].
Part 3: Sectioning and Pretreatment for RNAscope
This final part prepares tissue sections for the RNAscope assay itself.
Materials:
- Frozen tissue block in OCT
- Cryostat, SuperFrost Plus slides [30]
- RNAscope Hydrogen Peroxide, Target Retrieval, and Protease Plus [30]
Procedure:
- Sectioning: Equilibrate the tissue block at -20°C in a cryostat for at least 1 hour. Cut sections at a thickness of 7–15 µm [30] [15]. Mount sections on SuperFrost Plus slides and air-dry for 20 minutes at -20°C [30].
- Wash: To remove OCT, wash slides in 1X PBS for 5 minutes [30].
- RNAscope Pretreatment: Follow the RNAscope Fixed-Frozen Pretreatment protocol [30]:
- Hydrogen Peroxide: Apply 2-4 drops to each section for 10 minutes at room temperature to quench endogenous peroxidases. Rinse with distilled water.
- Target Retrieval: Submerge slides in boiling 1X Target Retrieval solution for 5 minutes. Adjust time based on tissue type. Rinse in distilled water, then in 100% ethanol, and air dry.
- Create Hydrophobic Barrier: Use the Immedge pen to draw a barrier around the tissue and let it dry completely.
- Protease Digestion: Apply Protease Plus to cover the tissue sections. Incubate for 30 minutes at 40°C in a pre-warmed HybEZ oven. Rinse with distilled water [30].
Following these steps, the slides are ready for the RNAscope in situ hybridization procedure as described in the appropriate user manual.
Critical Troubleshooting and Best Practices
- Tissue Orientation: Proper orientation during embedding is crucial for sectioning the desired anatomical plane. Note that the surface placed down in the mold will be sectioned first [33].
- Freezing Artifacts: Rapid freezing using a pre-cooled isopentane bath, rather than directly in liquid nitrogen, reduces ice crystal formation, which can destroy tissue morphology and RNA integrity [31].
- RNAscope Controls: Always run RNAscope assays with control probes (e.g., positive control PPIB and negative control dapB) to validate assay conditions and RNA quality in your samples [10] [15].
- Fixation Quality: Adhere strictly to fixation parameters. Under-fixation leads to RNA degradation and poor morphology, while over-fixation reduces probe accessibility and signal intensity [10].
Within the framework of a broader thesis on RNAscope sample preparation fixation research, the processing of cultured cell samples represents a critical foundational step. The reliability and interpretability of subsequent RNA in situ hybridization (ISH) data are contingent upon the initial procedures of cell adherence, fixation, and pretreatment. This document synthesizes current methodologies and protocols to establish a standardized, robust pipeline for preparing cultured cells for advanced RNA analysis using the RNAscope platform. Proper execution of these initial steps is paramount for preserving RNA integrity and cellular morphology, thereby ensuring the spatial fidelity of gene expression analysis [10] [34].
Cell Adherence and Culture Protocols
Successful RNAscope analysis begins with the establishment of a healthy, adherent monolayer of cells. The core principle is to promote strong attachment to the growth surface, which minimizes cell loss during the multiple fluid exchange steps in the subsequent RNAscope assay.
General Protocol for Passaging Adherent Cells
Maintaining cells in the logarithmic growth phase is essential for achieving optimal health and adherence. The following general protocol is adapted from standard cell culture practices for mammalian cells [35].
- Pre-passage Assessment: Routinely monitor cell viability, which should be greater than 90% prior to subculturing. Passage cells when they are in the log phase of growth, typically between 70-80% confluence.
- Wash Step: Remove and discard the spent cell culture media. Wash the cell layer gently with a balanced salt solution without calcium and magnesium (e.g., PBS) to remove any traces of serum, which can inhibit trypsin.
- Cell Dissociation: Add a pre-warmed dissociation reagent, such as trypsin or TrypLE, to the culture vessel. Gently rock the vessel to ensure complete coverage of the cell layer.
- Incubation and Monitoring: Incubate the vessel at room temperature for approximately 2 minutes (note that incubation time varies by cell line). Observe the cells under a microscope. If less than 90% of cells are detached, tap the vessel gently or extend the incubation time in 30-second increments.
- Neutralization and Collection: Once most cells are detached, add a volume of pre-warmed complete growth medium that is at least twice the volume of the dissociation reagent to neutralize the enzyme. Pipette the medium over the cell layer surface to disperse the cells.
- Seeding New Cultures: Transfer the cell suspension to a conical tube and centrifuge at 200 x g for 5-10 minutes. Resuspend the cell pellet in a small volume of fresh growth medium, perform a cell count, and dilute the suspension to the recommended seeding density for the cell line. Plate the cells into new culture vessels.
Specialized Coating Protocols for Enhanced Adherence
Many primary cells or sensitive cell lines require coated surfaces for optimal attachment and growth. The choice of coating substrate is often cell-type specific [36].
Table 1: Coating Substrates for Different Cell Types
| Cell Type | Recommended Coating | Brief Procedure | Key Application |
|---|---|---|---|
| Embryonic Stem (ES) / Induced Pluripotent Stem (iPS) Cells | Irradiated Mouse Embryonic Fibroblasts (iMEFs) | Plate iMEFs on gelatin-coated coverslips and culture overnight before seeding stem cells [36]. | Provides a feeder layer for pluripotent stem cell maintenance. |
| ES/iPS Cells, MSCs, Neural Stem Cells (NSCs) | Defined Culture Matrix (e.g., Matrigel) | Dilute matrix 1:100 in PBS, coat surfaces for 2-3 hours at 37°C, then rinse with PBS before plating cells [36]. | Provides a complex extracellular matrix environment. |
| Mesenchymal Stem/Stromal Cells (MSCs) under serum-free conditions | Fibronectin | Dilute Fibronectin in PBS to 5 µg/mL, coat surfaces for 3 hours at room temperature or overnight at 2-8°C, then rinse with PBS [36]. | Enhances adhesion in the absence of serum adhesion factors. |
| Neural Stem Cells (NSCs) | Poly-L-ornithine & Fibronectin | Coat with Poly-L-ornithine overnight at 37°C, rinse, then coat with Fibronectin (1 µg/mL) for 3-24 hours at 37°C [36]. | Sequential coating optimal for neural cell attachment and differentiation. |
Table 2: Reagent Preparation for Coating Protocols
| Reagent | Composition / Preparation | Function |
|---|---|---|
| MEF Media | High glucose DMEM, 10% fetal bovine serum, 2 mM L-glutamine, optional 1:100 Penicillin/Streptomycin [36]. | Growth medium for feeder layer cells. |
| Poly-L-ornithine (1000X Stock) | 15 mg/mL in sterile PBS. Aliquot and store at < -20°C [36]. | Promotes initial surface attachment for neural cells. |
| Fibronectin Solution (1X) | 1 µg/mL in sterile PBS. Prepare fresh as needed [36]. | Enhances cell adhesion and spreading. |
Figure 1: Workflow for Cultured Cell Sample Preparation. The process begins with ensuring proper cell adherence, followed by critical fixation and pretreatment steps before the RNAscope in situ hybridization (ISH) assay can be performed.
Fixation and Pretreatment for RNAscope
Fixation is the most critical step for preserving RNA in situ. The goal is to rapidly stabilize cellular RNA and morphology while maintaining probe accessibility during the hybridization assay.
Fixation Protocol
The following protocol for the preparation and fixation of cells on coverslips is optimized for subsequent ICC staining and is directly applicable to RNAscope sample preparation [36].
- Wash: When cells have reached the desired density, remove the culture media from each well and wash the cells twice with PBS.
- Fix: Add 300-400 µL of 4% Formaldehyde Fixative Solution (4% paraformaldehyde in PBS) to each well.
- Incubate: Incubate for 20 minutes at room temperature.
- Wash Again: Remove the fixative and wash the cells twice with wash buffer (e.g., 0.1% BSA in PBS).
- Store: Fixed cells can be stored in PBS at 2-8°C for several days before proceeding. For long-term storage, dehydration and paraffin-embedding are recommended, though this is less common for cultured cells than for tissue specimens.
Fixation Guidelines and Optimization for RNAscope
Adherence to strict fixation protocols is non-negotiable for high-quality RNAscope results. The following guidelines are curated from ACD's technical support documents [10] [15] [37].
- Fixative and Duration: For FFPE cell pellets, fixation in fresh 10% Neutral Buffered Formalin (NBF) for 16–32 hours at room temperature is the gold standard recommended by ACD [10] [15]. Note: Do not fix at 4°C.
- Impact of Suboptimal Fixation:
- Under-fixation: Results in significant RNA loss during storage and processing. In the RNAscope assay, this can lead to poor tissue morphology and low signal due to RNA degradation [10] [37].
- Over-fixation: Leads to excessive cross-linking, which results in poor probe accessibility. This manifests as a low signal and a low signal-to-background ratio, even though tissue morphology may appear excellent [10].
- Alternative Fixative: A recently published expanded RNAscope protocol for plant tissues used 4% Formaldehyde solution with Hank’s Balanced Salt Solution (HBSS) and a surfactant (Silwet-L77) for 12-24 hours at 1-4°C, demonstrating that protocol adaptation is possible for specific applications [32].
Table 3: Fixation Parameters and Their Effects
| Parameter | Recommended Condition | Effect of Deviation |
|---|---|---|
| Fixative | 10% NBF (Neutral Buffered Formalin) [10] [15] | Other fixatives may require extensive optimization and can compromise RNA integrity. |
| Fixation Time | 16 - 32 hours [10] [15] | < 16 hrs (Under-fixation): RNA loss, poor morphology.> 32 hrs (Over-fixation): Poor probe access, low signal. |
| Temperature | Room Temperature [10] [15] | Fixation at 4°C is not recommended as it leads to suboptimal results. |
The Scientist's Toolkit: Essential Reagents and Materials
A successful RNAscope experiment in cultured cells relies on a set of core reagents and materials. The following table details these essential items and their functions.
Table 4: Research Reagent Solutions for RNAscope on Cultured Cells
| Item | Function / Explanation | Example/Reference |
|---|---|---|
| Defined Coating Substrate | Provides a surface that mimics the extracellular matrix, promoting strong cell adhesion essential for preventing loss during assay washes. | Matrigel, Fibronectin, Poly-L-ornithine [36]. |
| Neutral Buffered Formalin (10% NBF) | The gold-standard fixative. It rapidly penetrates cells to preserve morphology and immobilize RNA while minimizing degradation. | ACD's recommended fixative for FFPE samples [10] [15]. |
| RNAscope Control Probes & Slides | Critical for validating assay performance. Positive control probes (e.g., PPIB, POLR2A, UBC) verify RNA integrity, while negative controls (e.g., DapB) assess background. | ACD recommends always using control slides and probes [15]. |
| RNAscope Pretreatment Reagents | A kit containing Target Retrieval reagents and Protease (e.g., Protease Plus). These steps are optimized to reverse cross-links and permeabilize the sample for probe access. | Part of the standard RNAscope kit [32]. |
| SuperFrost Plus Microscope Slides | Positively charged slides that significantly enhance tissue section adhesion, preventing detachment during the rigorous protocol. | ACD specifically recommends these slides to avoid tissue loss [15]. |
| RNAscope Probe Set | The core of the assay; a pool of ~20 ZZ probe pairs designed to hybridize to the target RNA. This design enables massive signal amplification with high specificity. | The proprietary technology enabling single-molecule sensitivity [34]. |
| Fomentariol | Fomentariol | High-purity Fomentariol for research on diabetes. This product is For Research Use Only (RUO). Not for human or veterinary diagnostic or therapeutic use. |
| Dicentrine hydrochloride | Dicentrine hydrochloride, CAS:5742-03-0, MF:C20H22ClNO4, MW:375.8 g/mol | Chemical Reagent |
Figure 2: Troubleshooting Poor Signal. A logic flow for diagnosing and addressing the common problem of a poor signal in the RNAscope assay, often rooted in fixation issues.
The journey from a cultured cell in a flask to a robust, quantifiable RNAscope dataset is paved with meticulous attention to the protocols for adherence, fixation, and pretreatment. As detailed in this application note, the interdependence of these steps cannot be overstated: optimal coating ensures a stable monolayer survives the assay, precise fixation locks RNA in place without rendering it inaccessible, and appropriate pretreatment opens the door for specific probe hybridization. By standardizing these preparatory stages according to the guidelines and protocols herein, researchers can lay a solid foundation for the sensitive and spatially resolved gene expression analysis that the RNAscope platform provides, thereby generating reliable data that can effectively bridge laboratory findings and clinical applications.
Tissue Microarrays (TMAs) represent a pivotal high-throughput technology in pathology and biomedical research, enabling the simultaneous analysis of hundreds of tissue specimens on a single slide [38]. The technique revolutionized tissue analysis by introducing miniaturization, standardization, and parallel processing to histopathological studies [39]. Initially described by Kononen et al. in 1998, TMA technology has evolved from earlier "sausage" block methods into a precision instrument that maintains tissue integrity while dramatically increasing analytical throughput [38]. The fundamental innovation of TMAs lies not merely in technical improvements but in their capacity to systematically link clinical data to specific tissue samples arranged in a highly organized array pattern [40].
In the context of RNA biomarker research using advanced in situ hybridization techniques like RNAscope, TMAs provide an indispensable platform for validating transcriptomic findings across large patient cohorts [39] [34]. The integration of TMAs with RNAscope analysis is particularly valuable for spatial transcriptomics, allowing researchers to examine RNA expression patterns within precise histopathological contexts while maintaining tissue morphology [26]. This application note examines the critical technical considerations and core variability factors that impact TMA performance, with specific emphasis on their implications for RNAscope-based research.
TMA Design and Construction Methodologies
Core Technical Specifications
The construction of TMAs requires meticulous planning and execution to ensure representative sampling and analytical reliability. The process begins with careful selection of donor tissue blocks and their corresponding hematoxylin and eosin (H&E) stained slides, which are reviewed by pathologists to identify regions of interest [38]. The core extraction process utilizes specialized tissue microarrayers that precisely punch cylindrical cores from donor blocks and transfer them to a recipient paraffin block in a predefined grid pattern [39].
Table 1: TMA Core Size Specifications and Performance Characteristics
| Core Diameter (mm) | Surface Area (mm²) | Typical Cores per Patient | Technical Accuracy for Tumor Tissue | Advantages | Limitations |
|---|---|---|---|---|---|
| 0.6 | 0.28 | 2-4 [41] | 85.9% [41] | Higher patient density; slower donor block depletion | Lower analytical accuracy; less representative of heterogeneous tissues |
| 1.0 | 0.79 | 1-3 [41] | 94.2% [41] | Superior technical and analytical accuracy; better for small histological features | Faster donor block depletion; lower patient density per block |
| 2.0 | 3.14 | 1-2 [38] | >95% (inferred) [38] | Maximum tissue representation; highest concordance with donor tissue | Significantly limits number of patients per TMA block |
Core size selection represents a critical compromise between analytical accuracy and practical considerations. Larger core sizes (1.0-2.0mm) demonstrate superior technical accuracy, particularly for challenging tissue types like normal urothelium (80.9% for 1.0mm vs. 58.6% for 0.6mm) and carcinoma in situ (71.4% for 1.0mm vs. 63.8% for 0.6mm) [41]. The enhanced performance of larger cores is attributed to their nearly threefold greater surface area compared to 0.6mm cores, which increases the likelihood of capturing representative tissue elements [41].
TMA Construction Workflow
The following diagram illustrates the comprehensive workflow for TMA construction and analysis, highlighting key decision points and quality control checkpoints:
TMA Construction and Analysis Workflow. This diagram outlines the key steps in TMA development, from initial design to final data interpretation, highlighting critical decision points at core size selection and quality control stages.
The construction process demands precision at each stage. After array design planning, tissue cores are extracted using a microarrayer and arranged in recipient blocks with precise spatial organization to maintain sample traceability [39]. Sectioning produces thin sections (4-5μm for FFPE, 7-20μm for frozen tissues) mounted on charged slides, with multiple sections preserved for subsequent analyses [15] [26]. Quality control through H&E staining of initial sections verifies proper tissue representation before proceeding to expensive molecular analyses [41].
Special Considerations for RNAscope Analysis on TMAs
Sample Preparation and Fixation Requirements
The integration of TMAs with RNAscope technology demands strict adherence to sample preparation protocols to preserve RNA integrity and ensure valid results. Proper fixation is paramount, as both under-fixation and over-fixation significantly impact RNA quality and assay performance [10].
Table 2: Sample Preparation Guidelines for RNAscope on TMAs
| Parameter | Optimal Condition | Impact of Deviation | Recommendation |
|---|---|---|---|
| Fixative | Fresh 10% NBF [10] [15] | Alternative fixatives may require optimization | Use only fresh 10% Neutral Buffered Formalin |
| Fixation Time | 16-32 hours at room temperature [10] [15] | Under-fixation: protease over-digestion, RNA loss, poor morphologyOver-fixation: protease under-digestion, poor probe accessibility, low signal | Strictly adhere to recommended duration |
| Fixation Temperature | Room temperature [10] | Lower temperatures cause under-fixation | Never fix at 4°C |
| Tissue Processing | Standard dehydration through graded series of ethanol and xylene [15] | Inadequate processing compromises RNA integrity and tissue morphology | Follow established pathology protocols |
| Section Thickness | 5±1μm for FFPE [15] | Thicker sections may improve signal but reduce resolution | Optimize based on tissue type and assay requirements |
For tissues fixed under suboptimal conditions, the RNAscope protocol requires modification of pretreatment conditions, particularly target retrieval and protease digestion steps [10]. The use of control probes—positive controls like PPIB (cyclophilin B) and negative controls like bacterial dapB—is essential for validating assay performance on TMA sections [15]. RNAscope signals are evaluated through semi-quantitative scoring based on dots per cell rather than signal intensity, as dot count correlates directly with RNA copy numbers [15].
Addressing Technical Challenges in TMA-RNAscope Integration
The combination of TMA technology with RNAscope presents unique technical challenges. Tissue heterogeneity within individual cores may complicate RNA expression interpretation, particularly when analyzing tumors with diverse cellular populations [41]. The small core sizes standard in TMAs (0.6-2.0mm) necessitate careful selection of tumor-rich areas during array construction to ensure meaningful results [39].
RNA degradation in archival tissues represents another significant challenge. Studies demonstrate that RNAscope signals in FFPE tissues are lower than in fresh frozen tissues in an archival duration-dependent fashion, with high-expression genes like PPIB and UBC showing the most pronounced degradation [26]. This effect necessitates careful quality control using housekeeping genes (HKGs) to assess RNA integrity before proceeding with experimental assays [26].
For TMAs containing tissues with varying fixation histories, pretreatment conditions may require core-specific optimization. The commercial RNAscope platform offers flexibility in target retrieval and protease treatment to accommodate such variability [10]. Systematic testing with control slides using both positive and negative control probes is recommended when working with TMAs composed of tissues with unknown or suboptimal preparation histories [10].
Core Variability and Analytical Considerations
Impact of Core Size on Analytical Accuracy
Core size selection significantly influences both technical and analytical accuracy in TMA studies. Technical accuracy refers to the correct capture of target histologic areas from donor blocks, while analytical accuracy concerns the concordance between core measurements and the original tissue characteristics [41].
Table 3: Comparative Performance of Different TMA Core Sizes
| Performance Metric | 0.6mm Core | 1.0mm Core | Statistical Significance |
|---|---|---|---|
| Technical Accuracy: Normal Tissue | 58.6% | 80.9% | P<0.001 [41] |
| Technical Accuracy: Tumor Tissue | 85.9% | 94.2% | P<0.01 [41] |
| Technical Accuracy: CIS | 63.8% | 71.4% | P<0.05 [41] |
| Analytical Accuracy (Ki67 quantification) | Moderate (r²=0.669) | High (r²=0.979) | P=0.035 [41] |
| Recommended Cores per Patient | 3-4 | 2-3 | Context-dependent [41] |
Comparative studies demonstrate that 1.0mm cores provide significantly better analytical accuracy for quantitative assessments such as Ki67 proliferation indices and mitotic counts [41]. While increasing the number of 0.6mm cores can partially compensate for reduced technical accuracy, this approach cannot overcome the limitations in analytical accuracy [41]. The superior performance of larger cores is particularly important for applications requiring precise quantification, such as RNA transcript counting in RNAscope assays.
Strategic Approaches to Core Variability
Several strategies can mitigate the impact of core variability in TMA studies:
Multiple Sampling: Incorporating 2-3 cores per tissue specimen significantly improves representativeness, especially for heterogeneous tumors [41]. For critical applications, some researchers recommend duplicate or triplicate TMA blocks to ensure sufficient material for repeated analyses [41].
Stratified Sampling: For tissues with distinct histological zones (e.g., tumor center vs. invasive front), purposeful sampling from each region enhances comprehensive representation [38].
Pathologist Involvement: Expert pathological review during core selection is indispensable for identifying appropriate regions of interest and avoiding necrotic or poorly preserved areas [39] [41].
Control Integration: Incorporating control tissues (cell lines, tissue controls) throughout the TMA block helps monitor technical variability and assay performance across the array [41].
For RNAscope applications specifically, the inclusion of multiple housekeeping genes with varying expression levels (UBC and PPIB as high expressors; POLR2A and HPRT1 as moderate to low expressors) provides internal quality metrics for assessing RNA integrity across different TMA cores [26].
Essential Research Reagents and Solutions
Table 4: Essential Research Reagent Solutions for TMA-RNAscope Workflows
| Reagent/Category | Specific Examples | Function/Application | Technical Notes |
|---|---|---|---|
| Fixatives | 10% Neutral Buffered Formalin (NBF) [10] [15] | Tissue preservation while maintaining RNA integrity | Must be fresh; 16-32 hour fixation at RT |
| Control Probes | PPIB, POLR2A, UBC (positive); dapB (negative) [15] [26] | Assay validation and RNA quality assessment | PPIB has highest signal but shows most degradation in archived samples |
| Slide Systems | SuperFrost Plus Slides [15] [26] | Tissue section adhesion | Critical for preventing tissue loss during stringent RNAscope procedures |
| Pretreatment Reagents | Target Retrieval Reagents, Protease [10] [15] | Unmasking target RNA and permitting probe access | Requires optimization for suboptimally fixed tissues |
| Detection Systems | RNAscope Multiplex Fluorescent v2 Kit [26] | Signal amplification and detection | Compatible with multiple fluorophores (Opal 520, 570, 620, 690) |
| Imaging Systems | Vectra Polaris [26], HALO [42], QuPath [42] | Image acquisition and analysis | Automated platforms enable high-throughput TMA analysis |
Tissue Microarrays represent a powerful platform for high-throughput tissue analysis, particularly when integrated with advanced RNA detection technologies like RNAscope. Core size selection critically impacts both technical and analytical accuracy, with 1.0mm cores generally providing superior performance for most applications, especially those involving quantitative assessment of RNA expression patterns. The successful implementation of TMA-RNAscope workflows demands strict adherence to sample preparation protocols, particularly regarding fixation parameters and RNA preservation methods. By addressing the special considerations and core variability factors outlined in this application note, researchers can leverage the full potential of TMA technology to advance biomarker discovery and validation in the context of spatial transcriptomics and precision medicine.
Sample preparation is the most critical determinant of success in RNAscope in situ hybridization, a technology that enables single-molecule RNA visualization within intact cells. Within this framework, two elements are paramount: the choice of microscope slides and the management of ribonuclease (RNase) activity. This application note details the essential roles of SuperFrost Plus slides and clarifies the specific requirements for an RNase-free environment in RNAscope assays. Adherence to these guidelines ensures optimal tissue adhesion, preserves RNA integrity, and guarantees high signal-to-noise ratios, forming the bedrock of reproducible and reliable data for research and drug development.
The Scientist's Toolkit: Essential Research Reagent Solutions
The following table catalogs the essential materials required for successful RNAscope sample preparation, with a specific focus on adhesion and RNA integrity.
Table 1: Essential Research Reagents and Materials for RNAscope Sample Preparation
| Item | Function/Description | Critical Usage Notes |
|---|---|---|
| SuperFrost Plus Slides | Positively charged electrostatic coating for superior tissue section adhesion; eliminates need for protein coatings or special adhesives [43]. | Mandatory for RNAscope to prevent tissue loss during rigorous enzymatic and thermal pretreatment steps [44]. |
| RNase-Free Reagents | Reagents (e.g., PBS, water) treated to inactivate RNases, preserving target RNA from degradation. | Required during tissue handling and sectioning prior to fixation; once samples are properly fixed, further RNA degradation is minimal [10]. |
| 10% Neutral Buffered Formalin (NBF) | Gold standard fixative for RNAscope; crosslinks and preserves RNA in tissue architecture. | Must be fresh; fixation for 16–32 hours at room temperature is recommended [10] [15]. |
| ImmEdge Hydrophobic Barrier Pen | Creates a hydrophobic barrier around sections, maintaining reagent coverage and preventing drying. | The only barrier pen recommended for use throughout the RNAscope procedure [44]. |
| RNAscope Control Probes & Slides | Validate assay performance, tissue RNA quality, and pretreatment conditions (e.g., PPIB for positive, dapB for negative control) [15]. | Essential for qualifying samples, especially those with unknown or suboptimal fixation history [44]. |
| Neoeuonymine | Neoeuonymine|CAS 33510-25-7|Research Chemical | High-purity Neoeuonymine, a natural alkaloid fromTripterygium wilfordii. For Research Use Only. Not for human or veterinary use. |
| Sengosterone | Sengosterone|Phytoecdysteroid|RUO | High-purity Sengosterone, a phytoecdysteroid from Ajuga species. For Research Use Only. Not for human or veterinary diagnostic or therapeutic use. |
The Critical Role of SuperFrost Plus Slides
Mechanism and Advantages
SuperFrost Plus slides are not merely a convenience but a requirement for the RNAscope assay [44]. Their surface features a permanent positive charge that electrostatically attracts tissue sections and cytological preparations [43]. This electrostatic interaction forms a bridge that evolves into covalent bonds with formalin-fixed sections, creating an exceptionally strong attachment to the glass [43]. This mechanism provides several key advantages crucial for the demanding RNAscope workflow:
- Elimination of Tissue Loss: The strong adhesion prevents sections from detaching during high-temperature target retrieval and the extended protease digestion steps required for probe accessibility [44].
- Reduction of Background Staining: Unlike albumin-coated slides, the Plus glass does not contribute to background staining, which is critical for distinguishing specific RNA-derived signals from non-specific artifacts [43].
- Resistance to Enzymatic and Chemical Stress: The adhesion is robust enough to withstand the higher degrees of enzyme digestion sometimes needed for optimization without compromising tissue integrity [43].
Practical Workflow Integration
In practice, SuperFrost Plus slides are integrated at the sectioning stage. For fixed-frozen tissues, sections should be cut at 7–15 µm, while for fresh-frozen tissues, a thickness of 10–20 µm is recommended [15]. After sectioning, slides should be air-dried and, for FFPE samples, baked at 60°C for 1-2 hours prior to initiating the RNAscope assay [15]. The slides are available in standard white or color-coded variants (blue, green, pink, yellow) to facilitate sample tracking and organization during processing [43] [45].
Clarifying the Requirement for RNase-Free Reagents
A common point of confusion in RNAscope sample preparation is the extent to which an RNase-free environment is necessary. ACD Bio, the developer of RNAscope, provides a clear and nuanced guideline: an RNase-free environment is critical during tissue handling and sectioning prior to fixation [10]. This includes using RNase-free water, PBS, and other solutions during the dissection, washing, and embedding processes. The rationale is to prevent endogenous and exogenous RNases from degrading the target RNA before it is stabilized and protected by the fixative.
However, once tissues have been properly fixed in 10% NBF, further RNA degradation is not expected to occur, and stringent RNase-free conditions for all subsequent reagents are not strictly required [10]. The cross-linking action of formalin effectively immobilizes and protects the RNA. This distinction simplifies the protocol and reduces costs, as it means that after fixation, the assay can be performed without the need for dedicated RNase-free reagents for every solution, such as the standard buffers used in the hybridization and washing steps.
Detailed Experimental Protocol for Frozen Tissue
The protocol below, optimized for fixed frozen mouse brain sections, integrates the critical materials discussed and is adaptable for other tissues [46] [13].
Sample Preparation and Sectioning
- Perfusion and Fixation: Perfuse mice transcardially with freshly prepared, RNase-free 4% PFA in PBS. Dissect the brain and post-fix in the same fixative at 4°C overnight [46].
- Cryoprotection: Immerse the fixed tissue in 30% sucrose in RNase-free PBS at 4°C until it sinks (approximately 48 hours for brain tissue) to prevent ice crystal formation [46].
- Embedding and Storage: Snap-freeze the tissue in OCT embedding medium using dry ice or liquid nitrogen. Store the blocks in an airtight container at -80°C [46] [13].
- Sectioning: Cut 20 µm thick sections using a cryostat and mount them onto SuperFrost Plus slides. Store slides at -80°C until use [46].
RNAscope In Situ Hybridization (2-Day Protocol)
Day 1: Pretreatment and Probe Hybridization
- Slide Preparation: Warm slides to room temperature and bake at 60°C for 30 minutes in a hybridization incubator [46].
- Post-fixation and Dehydration: Post-fix slides in 4% PFA for 15 minutes at 4°C, then dehydrate through a series of ethanol baths (50%, 70%, 100%, 5 minutes each). Air dry for 5 minutes [46].
- Hydrogen Peroxide Incubation: Apply RNAscope Hydrogen Peroxide to sections for 10 minutes at room temperature to quench endogenous peroxidase activity. Wash 2x in PBS [46].
- Target Retrieval: Preheat RNAscope Target Retrieval Reagent in a steamer (>99°C). Briefly rinse slides in PBS, then steam in Target Retrieval Reagent for 3 minutes. Immediately transfer to fresh PBS, rinse, and then immerse in 100% ethanol for 3 minutes. Air dry completely [46].
- Protease Digestion: Draw a barrier around sections with an ImmEdge pen. Apply Protease Plus or Protease III and incubate at 40°C for 20 minutes in a HybEZ oven. Wash 2x in PBS. From this point forward, do not allow sections to dry out [46].
- Probe Hybridization: Dilute target probes (C1, C2, C3) as required. Apply the probe mixture to the sections and incubate at 40°C for 2 hours. Wash 2x in RNAscope Wash Buffer. Store slides in 5x SSC buffer overnight at room temperature [46].
Day 2: Signal Amplification and Development
- Amplification Steps: Perform a series of amplifications by sequentially applying and incubating AMP1 (30 min), AMP2 (30 min), and AMP3 (15 min) at 40°C, with 2x washes in Wash Buffer between each step [46].
- Fluorescent Signal Development (for Multiplex Fluorescent Assay):
- Apply HRP-C1 blocker and incubate at 40°C for 15 minutes. Wash.
- Apply TSA-plus fluorophore (e.g., diluted 1:1000) and incubate at 40°C for 30 minutes. Wash.
- Apply HRP Blocker to inactivate HRP-C1. Wash.
- Repeat steps a-c for each subsequent channel (HRP-C2 and HRP-C3) with the appropriate fluorophores [46].
- Counterstaining and Mounting: Apply DAPI for 30 seconds, remove excess, and immediately add a few drops of fluorescence-compatible, anti-fade mounting medium. Apply a coverslip, avoid air bubbles, and dry in the dark. Store at 4°C [46].
Quantitative Data and Specifications
Table 2: SuperFrost Plus Slides Specifications and Data
| Parameter | Specification / Quantitative Data | Source / Reference |
|---|---|---|
| Slide Dimensions | 25 x 75 mm, 1 mm thick | [43] |
| Color Availability | White, Blue, Green, Pink, Yellow (for coding) | [43] [45] |
| Pack Size & Pricing | 144 slides: ~$91.30; Case of 1440: ~$880.00 (White, example) | [43] |
| Key Property | Permanent positive charge; covalent bonding with fixed tissue | [43] |
| Primary Benefit in RNAscope | Prevents tissue loss during target retrieval and protease digestion | [44] |
Table 3: RNase-Free Reagent Requirements by Preparation Stage
| Sample Preparation Stage | RNase-Free Reagent Requirement | Rationale |
|---|---|---|
| Dissection & Rinsing | Mandatory (e.g., PBS, Water) | Protects RNA from degradation before fixation [10]. |
| Pre-fixation Handling | Mandatory | Prevents introduction of exogenous RNases [10]. |
| Post-fixation Processing | Not strictly required | Formalin cross-linking protects RNA [10]. |
| RNAscope Assay Buffers | Not required | Standard laboratory reagents are sufficient post-fixation [10]. |
The rigorous application of fundamental sample preparation principles is non-negotiable for achieving precise and meaningful RNAscope data. The mandatory use of SuperFrost Plus slides ensures structural integrity by preventing tissue loss, while a clear understanding of the selective need for RNase-free conditions—primarily confined to the pre-fixation phase—safeguards RNA integrity without introducing unnecessary procedural complexity. By systematically integrating these critical materials and adhering to the detailed protocols, researchers can establish a robust and reliable foundation for their RNA localization studies, thereby enhancing the validity and impact of their research in neuroscience, oncology, and drug development.
Solving Fixation Problems and Optimizing Pretreatment Conditions
Sample fixation is a foundational step in RNAscope in situ hybridization that directly determines the success or failure of gene expression analysis. Proper fixation preserves RNA integrity while maintaining tissue morphology, creating the essential balance required for accurate detection of RNA molecules within their native spatial context. Within the broader thesis on RNAscope sample preparation, understanding fixation artifacts becomes paramount for researchers, scientists, and drug development professionals who rely on precise spatial gene expression data. Suboptimal fixation manifests in characteristic staining patterns that can compromise experimental outcomes and lead to erroneous biological interpretations. This application note provides a systematic framework for diagnosing fixation-related issues through strategic control probe implementation and staining pattern analysis, enabling researchers to distinguish technical artifacts from true biological signals and ensure data reliability in both research and development pipelines.
Understanding Fixation Effects on RNA Detection
The Fundamental Fixation Balance
Fixation creates a critical balance between RNA preservation and probe accessibility. Under-fixed tissue results in insufficient cross-linking, allowing RNA to degrade or leach out during processing, ultimately manifesting as poor signal despite potentially good morphology. Conversely, over-fixed tissue creates excessive cross-links that impede probe access to target RNA sequences, resulting in diminished signal even when RNA is well-preserved [47]. The optimal fixation window depends on tissue type and size, but generally follows the recommended guideline of fresh 10% neutral-buffered formalin (NBF) for 16-32 hours [44].
Manifestation of Fixation Artifacts
The effects of improper fixation become most apparent during the protease digestion step, where the degree of tissue permeabilization must precisely match the fixation level. Over-fixed specimens resist adequate protease digestion, leading to under-digestion where probes cannot penetrate the tissue matrix, yielding weak or absent target signal despite excellent morphological preservation. Under-fixed specimens suffer from over-digestion during protease treatment, causing physical deterioration of tissue architecture with loss of cell borders and a generally "faded" appearance, coupled with significant RNA loss [22] [47]. These opposing artifacts present distinct staining patterns that experienced researchers can recognize during quality assessment.
Table: Characterizing Fixation-Related Artifacts
| Parameter | Under-Fixed/Over-Digested Tissue | Properly Fixed Tissue | Over-Fixed/Under-Digested Tissue |
|---|---|---|---|
| Tissue Morphology | Poor; faded appearance, loss of cell borders | Excellent preservation with distinct cellular boundaries | Excellent preservation with distinct cellular boundaries |
| RNA Integrity | Significant RNA loss | Well-preserved RNA | Well-preserved but inaccessible RNA |
| Probe Accessibility | Increased but non-specific | Optimal for specific binding | Severely restricted |
| Signal Outcome | High background, non-specific signal | Strong specific signal, low background | Weak or absent target signal |
| Signal-to-Noise Ratio | Low | High | Low |
The Essential Control System for Fixation Diagnosis
Control Probe Strategy and Selection
A robust control system is indispensable for diagnosing fixation issues, requiring simultaneous assessment of both positive and negative control probes on adjacent tissue sections. ACD recommends running three slides minimum per sample: the target marker panel, a positive control probe, and the negative control probe [48]. This tripartite approach enables researchers to distinguish fixation-related artifacts from true biological expression patterns and technical failures.
The negative control probe targeting the bacterial DapB gene should yield no staining in properly fixed tissue, with successful results showing a score of <1 (no staining or <1 dot/10 cells) [44] [49]. Any significant signal with DapB indicates excessive protease digestion or background staining unrelated to specific RNA detection.
Selecting the appropriate positive control probe requires matching the expression level to your target of interest. ACD provides three primary options with different expression ranges and applications:
Table: Positive Control Probe Selection Guide
| Control Probe | Expression Level (Copies/Cell) | Recommended Application | Interpretation Guidelines |
|---|---|---|---|
| UBC (Ubiquitin C) | High (>20) | High expression targets only | Score ≥3 indicates acceptable RNA quality |
| PPIB (Cyclophilin B) | Medium (10-30) | Most flexible option for general use | Score ≥2 indicates acceptable RNA quality |
| POLR2A | Low (3-15) | Low expression targets; proliferating tissues | Score ≥1-2 indicates acceptable RNA quality |
For most applications, PPIB provides the optimal balance of sensitivity and rigor as a positive control. UBC's high expression level can sometimes detect RNA even in suboptimal conditions, potentially providing false reassurance about fixation quality [50].
Systematic Workflow for Fixation Troubleshooting
The following diagnostic workflow provides a systematic approach to identify and resolve fixation-related issues in RNAscope experiments:
Comprehensive Experimental Protocols
Protocol: Control Slide Validation for Fixed-Frozen Tissues
This protocol validates fixation quality in fresh-frozen tissues using the RNAscope Multiplex Fluorescent v2 assay, adapted from established methodologies [22] [13].
Materials Required:
- SuperFrost Plus slides (Fisher Scientific, #12-550-15) [44]
- ImmEdge Hydrophobic Barrier Pen (Vector Laboratories) [44]
- RNAscope Multiplex Fluorescent Reagent Kit v2 (ACD Bio, #323100)
- Positive control probes (PPIB, UBC, or POLR2A based on target expression)
- Negative control probe (DapB, ACD Bio)
- 4% PFA in 1x PBS
- Ethanol series (50%, 70%, 100%)
- HybEZ Hybridization System [44]
Sample Preparation:
- Cryosectioning: Section fresh-frozen tissue at 7-16 μm thickness onto SuperFrost Plus slides.
- Fixation: Immerse slides in 4% PFA in 1x PBS for 2 hours at room temperature.
- Dehydration: Process through ethanol series: 5 minutes each in 50%, 70%, and two changes of 100% ethanol.
- Barrier Application: Air dry slides briefly and draw hydrophobic barrier around sections using ImmEdge pen.
Pretreatment and Hybridization:
- Hydrogen Peroxide: Apply RNAscope Hydrogen Peroxide for 10 minutes at room temperature to quench endogenous peroxidase activity.
- Protease Treatment: Apply Protease Plus for 10 minutes at room temperature.
- Probe Hybridization: Prepare control probe mixtures and hybridize for 2 hours at 40°C in HybEZ oven.
- Signal Amplification: Perform sequential AMP 1, 2, and 3 incubations per manufacturer instructions.
Validation Criteria: Successful fixation is confirmed when PPIB scores ≥2 with relatively uniform signal throughout the sample and DapB scores <1 (minimal background) [44] [49]. Tissue should maintain excellent morphology with distinct cellular boundaries.
Protocol: Pretreatment Optimization for Over-Fixed FFPE Tissues
This protocol addresses the common challenge of over-fixed FFPE tissues through systematic pretreatment optimization.
Materials Required:
- RNAscope Target Retrieval Reagents
- RNAscope Protease III or Protease Plus
- SuperFrost Plus slides
- Positive and negative control probes
- HybEZ Hybridization System
Optimization Procedure:
- Sectioning: Cut 5 μm sections from FFPE blocks and mount on SuperFrost Plus slides.
- Deparaffinization: Bake slides at 60°C for 1 hour, followed by xylene and ethanol series.
- Target Retrieval Optimization: Test increasing retrieval times (15, 20, 25 minutes) at 95-100°C.
- Protease Optimization: Test increasing protease incubation times (15, 25, 35 minutes) at 40°C.
- Control Probe Application: Run PPIB and DapB controls at each condition.
- Analysis: Score staining results and tissue morphology to identify optimal balance.
Interpretation Guidelines:
- Under-digested: Excellent morphology but weak PPIB signal → Increase protease time
- Over-digested: Poor morphology with high DapB background → Reduce protease time
- Optimal: Excellent morphology with strong PPIB signal and minimal DapB → Proceed to target probes
For automated systems like the Leica BOND RX, extend pretreatment times incrementally while maintaining constant temperatures (e.g., 20 min ER2 at 95°C + 25 min Protease at 40°C) [44] [49].
The Scientist's Toolkit: Essential Research Reagents
Table: Essential Research Reagents for Fixation Diagnosis
| Reagent/Category | Specific Product Examples | Function in Fixation Diagnosis |
|---|---|---|
| Control Probes | PPIB, UBC, POLR2A (positive); DapB (negative) | Reference standards for assessing RNA integrity and staining specificity |
| Slide System | SuperFrost Plus slides | Prevent tissue detachment during rigorous pretreatment protocols |
| Barrier Pen | ImmEdge Hydrophobic Barrier Pen | Maintain reagent coverage and prevent tissue drying during assay |
| Mounting Media | EcoMount, PERTEX, ProLong Gold | Preserve signal while maintaining morphology for accurate analysis |
| Pretreatment Reagents | RNAscope Target Retrieval, Protease Plus | Enable optimization for fixation artifacts through controlled exposure |
| Hybridization System | HybEZ Oven with Humidity Control Tray | Maintain optimal temperature and humidity for consistent hybridization |
| Image Analysis Software | QuPath, HALO, ImageJ | Provide quantitative assessment of dot counts and cellular distribution |
| 3,3,5,5-tetramethylhexanoic Acid | 3,3,5,5-tetramethylhexanoic Acid, CAS:1135681-77-4, MF:C10H20O2, MW:172.26 g/mol | Chemical Reagent |
Advanced Analysis and Interpretation Framework
Quantitative Scoring Methodology
The RNAscope assay employs a semi-quantitative scoring system that focuses on dot enumeration rather than signal intensity. Each dot represents a single RNA transcript, with dot count correlating directly with expression level [44] [48]. The standardized scoring system enables consistent interpretation across experiments and operators:
Table: RNAscope Scoring Guidelines for PPIB Control Probe
| Score | Criteria | Interpretation |
|---|---|---|
| 0 | No staining or <1 dot/10 cells | Unacceptable RNA quality or severe over-fixation |
| 1 | 1-3 dots/cell | Suboptimal, may indicate moderate over-fixation |
| 2 | 4-9 dots/cell; none or very few dot clusters | Acceptable for most targets |
| 3 | 10-15 dots/cell; <10% dots in clusters | Good RNA quality and fixation |
| 4 | >15 dots/cell; >10% dots in clusters | Excellent RNA quality and fixation |
When fewer than 5% of cells score 1 and over 95% score 0, assign a score of 0. When 5-30% of cells score 1 with over 70% scoring 0, assign a score of 0.5 [49]. These semi-quantitative assessments should be performed at 20x magnification for consistency.
Integrated Fixation Assessment Algorithm
For complex cases where fixation quality varies within a sample, implement this sequential assessment algorithm:
- Morphology Primary Assessment: Evaluate tissue architecture integrity using DAPI or hematoxylin counterstain.
- Negative Control Verification: Confirm DapB signal falls within acceptable background limits (score <1).
- Positive Control Quantification: Calculate average PPIB dots per cell across multiple representative fields.
- Homogeneity Analysis: Assess staining uniformity across tissue regions and cellular compartments.
- Correlation with Fixation History: Compare staining patterns with documented fixation parameters.
This integrated approach enables researchers to distinguish fixation artifacts from true biological heterogeneity, particularly valuable in drug development contexts where quantitative accuracy is paramount.
Optimization Workflow for Suboptimally Fixed Tissues
Within RNAscope sample preparation research, proper tissue fixation is the cornerstone for achieving reliable and interpretable results. Formalin-fixed, paraffin-embedded (FFPE) tissue specimens fixed in fresh 10% Neutral Buffered Formalin (NBF) for 16–32 hours at room temperature represent the gold standard, ensuring an optimal balance between RNA integrity, probe accessibility, and tissue morphology [10] [15]. However, in both research and drug development, scientists frequently encounter tissues with unknown or suboptimal fixation histories, such as those from legacy biobanks or collaborative studies. Such deviations from the ideal protocol can severely compromise the sensitivity and specificity of the RNAscope assay. This application note provides a detailed, practical framework for diagnosing and optimizing suboptimally fixed tissues, enabling researchers to salvage precious samples and generate high-quality spatial gene expression data.
The Impact of Suboptimal Fixation
Understanding the specific effects of fixation errors is the first step in troubleshooting. The table below summarizes the characteristics and consequences of under-fixed and over-fixed tissues.
Table: Consequences of Suboptimal Tissue Fixation on RNAscope Assay Performance
| Fixation Type | Impact on Protease Digestion | Impact on RNA & Morphology | Final Assay Outcome |
|---|---|---|---|
| Under-fixation | Over-digestion [10] [37] | Significant RNA loss and poor tissue morphology [10] [37] | Low signal [10] [37] |
| Over-fixation | Under-digestion [10] | Poor probe accessibility; excellent tissue morphology preserved [10] | Low signal and poor signal-to-background ratio [10] |
The underlying mechanism is rooted in the cross-linking nature of formalin-based fixatives. Under-fixation fails to adequately preserve and protect RNA, making it vulnerable to degradation during subsequent processing and to over-digestion by the protease pretreatment, which is necessary for probe access [10] [37]. Conversely, over-fixation creates an excessively dense network of cross-links, making the target RNA inaccessible to the probes even with standard protease treatment, leading to low signal despite pristine morphology [10].
Diagnostic and Optimization Workflow
A systematic approach is essential for successfully optimizing suboptimally fixed tissues. The following workflow guides you from initial assessment to a finalized protocol.
Diagram: A systematic workflow for diagnosing and optimizing suboptimally fixed tissues for the RNAscope assay.
Initial Quality Control with Probes and Controls
Before optimizing, establish a baseline with control probes. This is a non-negotiable first step [15] [17].
- Essential Controls: Always run your sample alongside, or in combination with, the recommended control probes.
- Positive Control: A housekeeping gene like PPIB (Cyclophilin B), POLR2A, or UBC. Successful staining typically requires a PPIB/POLR2A score of ≥2 or a UBC score of ≥3 [15] [17].
- Negative Control: The bacterial dapB gene, which should yield a score of <1, indicating minimal background and non-specific binding [15] [17].
- Interpretation: A low signal with the positive control probe indicates an issue with the sample or pretreatment conditions. Compare the control results with the assessment of tissue morphology to diagnose the likely type of fixation error.
Optimization of Pretreatment Conditions
The primary lever for optimizing suboptimally fixed tissues is the pretreatment protocol, specifically the protease digestion step [10] [37]. The following table provides a detailed methodology for this optimization.
Table: Detailed Experimental Protocol for Pretreatment Optimization
| Optimization Step | Parameter | Recommended Action | Rationale & Technical Notes |
|---|---|---|---|
| 1. Diagnosis | Control Probes | Run RNAscope with PPIB and dapB controls on the test tissue [10] [15]. | Establishes a baseline for RNA integrity and assay performance. |
| 2. Morphology Assessment | Tissue Integrity | Evaluate H&E stained consecutive section for morphology [14]. | Poor morphology suggests under-fixation; excellent morphology with low signal suggests over-fixation [10]. |
| 3. Protease Optimization | Digestion Time | Suspected Under-fixation: Reduce protease treatment time [10]. Suspected Over-fixation: Increase protease treatment time [10]. | Protease digestion must be balanced to unmask RNA targets without destroying the tissue or RNA. |
| 4. Systematic Testing | Experimental Design | Test a range of protease times (e.g., 10, 20, 30 minutes) using control slides and probes [10]. | ACD recommends testing tissues alongside control slides using positive and negative control probes [10]. |
The Scientist's Toolkit: Essential Reagents and Materials
Successful optimization relies on using the correct reagents. The following table lists key solutions required for the workflow described in this note.
Table: Key Research Reagent Solutions for RNAscope Optimization
| Reagent / Material | Function / Application | Examples / Notes |
|---|---|---|
| RNAscope Control Slides | Verify assay workflow conditions [15]. | Human Hela Cell Pellet (Cat. #310045) or Mouse 3T3 Cell Pellet (Cat. #310023) [15]. |
| Positive Control Probes | Assess RNA integrity and optimize pretreatment [15] [17]. | Probes for housekeeping genes (e.g., PPIB, POLR2A, UBC). |
| Negative Control Probes | Determine background and nonspecific signal [15] [17]. | Probe for the bacterial dapB gene. |
| RNAscope Protease Reagents | Key enzyme for optimization; permeabilizes membranes and unmasks RNA targets [17]. | Protease Plus is commonly used; other types (Protease III, IV) are available for different samples [17]. |
| RNAscope Target Retrieval | Reverses cross-linking from fixation to provide probe access [17]. | Used with heating for FFPE samples; not required for fresh-frozen tissue [17]. |
| SuperFrost Plus Slides | Prevents tissue loss during rigorous processing steps [15]. | Critical for ensuring tissue adhesion, especially during heat treatment [15] [51]. |
Optimizing suboptimally fixed tissues for RNAscope is a systematic and achievable process. By leveraging control probes to diagnose the problem and methodically adjusting the protease digestion step, researchers can successfully recover data from valuable tissue resources that would otherwise be unusable. This workflow empowers scientists and drug development professionals to maintain the integrity of their spatial biology studies, even when working with non-ideal samples, thereby advancing research within the broader context of RNAscope fixation science.
Addressing No Signal, Weak Signal, and Poor Morphology
In the context of advancing RNAscope sample preparation fixation research, achieving reliable and interpretable results hinges on a fundamental, yet often overlooked, process: tissue fixation. Proper fixation preserves tissue architecture (morphology) and protects the target RNA molecules in their native spatial context. When assays fail—manifesting as no signal, weak signal, or poor morphology—the root cause can almost invariably be traced back to the initial sample preparation and fixation steps. This application note provides a detailed, evidence-based guide to diagnosing and resolving these common challenges, ensuring that your RNAscope assays deliver on the promise of single-molecule sensitivity and superb cellular resolution. The proprietary RNAscope technology, with its "double Z" probe design and signal amplification, is highly robust, but its performance is fundamentally dependent on the quality of the starting sample [52]. This document synthesizes the latest protocols and troubleshooting guidelines to empower researchers and drug development professionals to optimize their spatial biology workflows.
Diagnosis: Linking Symptoms to Root Causes in Fixation
Effective troubleshooting requires a systematic approach where specific symptoms point to likely root causes. The table below summarizes the primary failure modes, their underlying fixation-related causes, and the corresponding morphological and signal manifestations.
Table 1: Diagnostic Guide to Common RNAscope Issues Related to Sample Preparation
| Observed Symptom | Primary Root Cause | Impact on Assay & Morphology |
|---|---|---|
| No or Very Weak Signal | Over-fixation (>32 hours in formalin) [10] | Excessive crosslinking masks RNA, preventing probe access. Protease under-digestion occurs, leading to poor signal despite excellent morphology [10]. |
| Weak Signal & High Background | Under-fixation (<16 hours in formalin) [10] | Incomplete tissue preservation fails to protect RNA. Protease over-digestion degrades RNA and compromises tissue structure [10]. |
| Poor Tissue Morphology | Under-fixation or Over-digestion [10] [22] | Inadequate crosslinking or excessive protease treatment leads to loss of cellular detail, holes, or tissue detachment. |
| Heterogeneous Staining | Variable Fixation (e.g., delayed fixation, uneven penetration) [10] | Delayed fixation can degrade RNA, producing lower or no signal in parts of the tissue [10]. |
| Signal Fading in Fresh Frozen Tissue | Extended storage after perfusion or sectioning [22] | RNA integrity degrades over time, especially in non-FFPE samples. Protocols recommend performing the assay as soon as possible after sample preparation [22]. |
The following diagram illustrates the logical relationship between fixation quality, protease digestion, and the ultimate assay outcome. This decision tree can guide the initial diagnosis.
Optimized Protocols for Routine and Challenging Samples
Gold-Standard Fixation Protocol for FFPE Tissues
For formalin-fixed, paraffin-embedded (FFPE) tissues, ACD explicitly recommends fixation in fresh 10% Neutral Buffered Formalin (NBF) for 16–32 hours at room temperature [10] [15]. Tissues should be trimmed to a thickness of 3-4 mm to ensure adequate fixative penetration [15]. Deviations from this protocol are a primary source of assay failure. Do not fix at 4°C, as this leads to under-fixation. Following fixation, tissues must be dehydrated through a graded series of ethanol and xylene before being infiltrated with paraffin at a temperature not exceeding 60°C [15]. For sectioning, use 5 μm ± 1 μm thick sections mounted on SuperFrost Plus slides to prevent tissue loss [25] [15].
Protocol for Fresh-Frozen Tissues
For fresh-frozen tissues, the fixation process occurs after cryosectioning. The following workflow, adapted from a validated protocol, is tailored for mouse brain but applicable to other tissues [22].
Table 2: Essential Protocol for Fresh-Frozen Tissue Preparation and Fixation
| Step | Procedure | Critical Parameters |
|---|---|---|
| 1. Perfusion & Embedding | Perfuse with 1x PBS. Embed tissue in OCT. Freeze using 2-methylbutane surrounded by dry ice. | Perfusion rate: ~7 ml/min. PBS volume (ml) ≈ mouse weight (g). [22] |
| 2. Sectioning | Section on a cryostat at 16 μm thickness. | Collect sections on SuperFrost Plus slides. Store at -80°C. [22] |
| 3. Fixation | Immerse slides in 4% PFA in 1x PBS. Incubate for 2 hours at room temperature. | A minimum of 15 minutes is required, but 2 hours is recommended. [22] |
| 4. Dehydration | Dehydrate through a series of 50%, 70%, and two changes of 100% ethanol, 5 min each at RT. | Slides can be stored in 100% EtOH at -20°C for up to 1 week, though this is not recommended. [22] |
Optimization Strategies for Non-Ideal or Sub-Optimally Fixed Samples
When sample preparation history is unknown or sub-optimal, the pretreatment steps of Target Retrieval and Protease digestion require optimization. The recommended workflow is to first qualify the sample using control probes and slides [25]. The following diagram outlines this systematic optimization workflow.
For automated platforms like the Leica BOND RX, specific optimization parameters exist. The standard pretreatment is 15 minutes Epitope Retrieval 2 (ER2) at 95°C and 15 minutes Protease at 40°C. For over-fixed tissues, extended pretreatment is recommended: increase ER2 time in 5-minute increments and Protease time in 10-minute increments (e.g., 20 min ER2/25 min Protease, then 25 min ER2/35 min Protease) while keeping temperatures constant [25].
The Scientist's Toolkit: Essential Reagents and Controls
The following table details the critical reagents and materials required for a successful RNAscope assay, as emphasized in official documentation [25] [22] [15].
Table 3: Key Research Reagent Solutions for RNAscope Assays
| Item | Function & Importance | Specific Recommendation |
|---|---|---|
| Fixative | Preserves tissue morphology and RNA integrity. | Fresh 10% NBF for FFPE; 4% PFA in 1x PBS for fresh-frozen. Avoid old or used fixative. [10] [22] |
| Microscope Slides | Prevents tissue detachment during stringent assay steps. | Superfrost Plus slides are mandatory. Other types may cause tissue loss. [25] [15] |
| Hydrophobic Barrier Pen | Creates a well to contain reagents and prevent drying. | ImmEdge Hydrophobic Barrier Pen is the only pen certified for use throughout the RNAscope procedure. [25] |
| Control Probes & Slides | Validates assay performance, reagent functionality, and sample RNA quality. | Use species-specific control slides (e.g., Human Hela, Mouse 3T3) with positive control probes (PPIB, POLR2A, UBC) and the negative control probe (dapB). [25] [15] |
| Protease | Permeabilizes tissue to allow probe access. | Protease Plus is used in many manual assays. Condition must be optimized based on fixation. [22] |
| Mounting Media | Preserves staining for microscopy. | Xylene-based media (e.g., CytoSeal XYL) for Brown assays; EcoMount or PERTEX for Red and multiplex assays. [25] |
| HybEZ Oven | Maintains optimum humidity and temperature during hybridization. | Required for manual RNAscope hybridization steps to ensure consistent and reliable results. [25] [22] |
Mastering sample preparation and fixation is not merely a preliminary step but the cornerstone of generating publication-quality, reliable data with RNAscope technology. By understanding the direct link between fixation quality, protease digestion, and assay output, researchers can proactively prevent issues or systematically troubleshoot them. Adherence to the recommended protocols for fixation, the mandatory use of controls for validation, and the careful optimization of pretreatment conditions for challenging samples will significantly enhance experimental outcomes. This rigorous approach ensures that spatial gene expression data is both sensitive and specific, thereby advancing research in drug development, biomarker discovery, and basic biological understanding.
Validating Fixation Quality and Comparing with Gold Standards
Within the broader context of RNAscope sample preparation and fixation research, robust quality assessment (QA) is the cornerstone of generating reliable, interpretable in situ hybridization data. The integrity of RNA targets within tissue specimens is profoundly influenced by pre-analytical variables, including fixation time, processing methods, and storage conditions [10] [37]. Success with any RNAscope assay begins with good and consistent quality control practices to distinguish true biological signal from technical artifacts [50] [53]. This application note details a comprehensive QA strategy employing a panel of control probes—PPIB, POLR2A, UBC, and dapB—to provide researchers and drug development professionals with a rigorous framework for assessing sample quality, RNA integrity, and technical performance. This multi-level verification is essential for validating findings in preclinical studies across various animal models and human tissues [53].
Control Probe Profiles and Selection Criteria
A strategic approach to control probe selection is necessary to accurately qualify samples and the assay itself. The expression level of the target gene of interest should guide the choice of an appropriate positive control probe [50].
Table 1: RNAscope Positive Control Probe Characteristics and Selection Guide
| Control Probe | Target Gene | Expression Level (Copies/Cell) | Primary Recommendation |
|---|---|---|---|
| PPIB | Cyclophilin B | Medium (10-30) | The most flexible option for most tissues; provides a rigorous control for sample quality and technical performance [50]. |
| POLR2A | DNA-directed RNA polymerase II | Low (3-15) | Use with low expression targets; an alternative to PPIB for proliferating tissues like tumors [50]. |
| UBC | Ubiquitin C | High (>20) | Use with high expression targets only; not recommended for low-expression targets as it can lead to false-negative interpretations [50]. |
| dapB | Bacterial gene from B. subtilis | N/A | Universal negative control; confirms minimal background and specific staining [50] [15]. |
For the negative control, ACD's universal negative control probe targets the dapB gene from Bacillus subtilis, a soil bacterium not present in mammalian tissues [50]. This probe is essential for verifying that observed signals are specific to the target RNA and not due to non-specific background staining or inadequate sample preparation. Alternative negative controls, such as sense or scrambled probes, are available but are generally discouraged as sense strands may occasionally be transcribed, leading to ambiguous results [50].
Experimental Protocol for Quality Assessment
Sample Preparation and Fixation Essentials
Proper sample preparation is the most critical factor influencing RNAscope assay outcomes. Suboptimal preparation is the most common reason for subpar results [37]. The following protocol is recommended for formalin-fixed, paraffin-embedded (FFPE) tissues:
- Tissue Fixation: Fix tissue specimens (blocked to 3-4 mm thickness) in fresh 10% Neutral Buffered Formalin (NBF) for 16–32 hours at room temperature. Do not fix at 4°C [10].
- Processing: Dehydrate in a graded series of ethanol and xylene, followed by infiltration with melted paraffin held at no more than 60°C [15] [37].
- Sectioning: Cut embedded tissue into 5 ± 1 μm sections using a microtome and mount on Superfrost Plus Slides [15] [37].
- Slide Storage: Air-dry slides overnight at room temperature. For near-term use, slides can be baked at 60°C for 1-2 hours prior to the RNAscope assay. Analyze specimens within 3 months of sectioning when stored at room temperature with desiccant [15].
Under-fixation (<16 hours) results in protease over-digestion during pretreatment, leading to RNA loss and poor morphology. Over-fixation (>32 hours) causes protease under-digestion, resulting in poor probe accessibility, low signal, and suboptimal signal-to-background ratio, even while tissue morphology may appear excellent [10].
Recommended Workflow for Sample Qualification
When sample preparation conditions are unknown or deviate from recommended guidelines, qualifying samples using control probes before running target-specific experiments is strongly advised [44]. The following workflow ensures systematic quality assessment:
Figure 1: A flowchart illustrating the recommended workflow for qualifying samples of unknown or non-standard fixation prior to target-specific experiments.
- Run Control Probes: Perform the RNAscope assay on test sample sections using the positive control probe PPIB (recommended for its medium expression level) and the negative control probe dapB simultaneously or on adjacent slides [15] [44].
- Interpret and Score Staining: Evaluate the staining results using semi-quantitative scoring guidelines. Focus on the number of punctate dots per cell, as this correlates directly with RNA copy number, rather than signal intensity [15] [44].
- Acceptance Criteria: A sample is qualified for target-specific probing if it meets the following criteria:
- Troubleshoot with Pretreatment: If signals are low (PPIB <2) or background is high (dapB ≥1), adjust pretreatment conditions. This often involves optimizing the epitope retrieval time (in 5-minute increments) and/or protease treatment time (in 10-minute increments) to optimally expose target RNA while preserving tissue integrity [53] [44].
Interpretation and Scoring of Control Results
Accurate interpretation of control probe results is fundamental for assay validation. The RNAscope assay uses a semi-quantitative scoring system based on the number of punctate dots per cell, where each dot represents a single RNA molecule [53] [1].
Table 2: RNAscope Semi-Quantitative Scoring Guidelines for Control Probes
| Score | Criteria (Dots per Cell) | Interpretation for Sample Qualification |
|---|---|---|
| 0 | No staining or <1 dot/10 cells | Unacceptable for PPIB (failed sample). Required for dapB. |
| 1 | 1-3 dots/cell | Unacceptable for PPIB (suboptimal sample). |
| 2 | 4-9 dots/cell; very few dot clusters | Minimum acceptable score for PPIB [15] [44]. |
| 3 | 10-15 dots/cell; <10% dots in clusters | Good positive control signal. |
| 4 | >15 dots/cell; >10% dots in clusters | Strong positive control signal. |
For a successful experiment, staining with the positive control PPIB should yield a score of ≥2, while the negative control dapB should yield a score of <1 [15] [44]. If using UBC as a positive control, a score of ≥3 is required for the sample to be considered qualified [15] [44]. Samples failing these criteria require pretreatment optimization or should be excluded from target analysis due to poor RNA quality or inappropriate fixation.
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Research Reagent Solutions for RNAscope Quality Assessment
| Item | Function/Description | Example/Catalog Reference |
|---|---|---|
| Positive Control Probes | Species-specific probes for housekeeping genes (PPIB, POLR2A, UBC) to assess RNA integrity and assay technique. | RNAscope Positive Control Probes (Human, Mouse, Rat) [50] [54]. |
| Negative Control Probe | Probe targeting the bacterial dapB gene to confirm absence of non-specific background staining. | RNAscope Negative Control Probe (dapB) [50] [15]. |
| Control Slides | Pre-made slides with fixed cell pellets to verify the technical workflow is performed correctly. | Human HeLa (Cat. No. 310045) or Mouse 3T3 (Cat. No. 310023) Control Slides [15] [44]. |
| Hydrophobic Barrier Pen | Creates a hydrophobic barrier around the tissue section to prevent reagent spread and slide drying. | ImmEdge Hydrophobic Barrier Pen (Vector Labs) [44]. |
| Superfrost Plus Slides | Microscope slides with enhanced tissue adhesion to prevent detachment during the rigorous assay procedure. | Fisherbrand Superfrost Plus Microscope Slides [15] [44]. |
| RNAscope Pretreatment Kits | Reagents for deparaffinization, target retrieval, and protease digestion to optimally permeabilize tissue for probe access. | RNAscope Target Retrieval Reagents, Protease Plus [53] [32]. |
The accurate assessment of gene expression is a cornerstone of molecular biology and clinical diagnostics, directly influencing research outcomes and patient treatment decisions. For years, quantitative real-time PCR (qPCR) and immunohistochemistry (IHC) have served as foundational techniques for quantifying RNA and protein expression, respectively. However, these methods have inherent limitations; qPCR requires nucleic acid extraction, losing crucial spatial information, while IHC depends on antibody specificity and may not directly reflect RNA levels [2]. The introduction of RNA in situ hybridization (ISH) technologies, particularly the RNAscope assay, represents a significant advancement by enabling the visualization and quantification of RNA within the context of intact tissue architecture.
This application note synthesizes systematic review evidence to evaluate the concordance between RNAscope and established gold standard methods. Furthermore, it provides detailed protocols to guide researchers in generating robust, reproducible data that bridges molecular quantification with morphological context, thereby supporting applications from basic research to drug development.
Systematic Review of Concordance Evidence
A 2021 systematic review provides a comprehensive evaluation of RNAscope's performance in the clinical diagnostic field compared to established methods [2]. The review, which included 27 retrospective studies, found that RNAscope is a highly sensitive and specific method with a high concordance rate when compared to techniques that measure similar molecules (i.e., RNA).
Table 1: Concordance Rates Between RNAscope and Other Techniques
| Comparison Method | Concordance Rate (CR) with RNAscope | Notes |
|---|---|---|
| qPCR / qRT-PCR | 81.8% - 100% | High concordance when measuring the same analyte (RNA) [2]. |
| DNA ISH | 81.8% - 100% | High concordance for gene detection [2]. |
| IHC | 58.7% - 95.3% | Lower concordance expected as techniques measure different molecules (RNA vs. protein); discrepancies can arise from post-transcriptional regulation [2]. |
Specific studies substantiate these high concordance rates. For instance, in a study focusing on HER2 status in invasive breast carcinoma, RNAscope demonstrated a 97.3% concordance with qPCR in cases where FISH results were unequivocal. Notably, RNAscope proved superior to qPCR in resolving cases with intratumoral heterogeneity or equivocal FISH results, highlighting its advantage in providing spatial context [55].
A more recent 2025 study on breast cancer biomarkers further supports the strong correlation between mRNA measurement by qRT-PCR and protein detection by IHC. The study reported an overall concordance of 95.9% for ESR1/ER and 100% for ERBB2/HER2, though concordance for PGR/PR was lower at 79.3% [56]. Another 2025 study confirmed that molecular biology approaches for HER2 determination (using both DNA and RNA) showed completely identical results with IHC in control samples, underscoring the legitimacy of these methods [57].
Principles of the RNAscope Technology
The high sensitivity and specificity of the RNAscope assay are rooted in its unique proprietary signal amplification and background suppression mechanics.
Underlying Principle
The core of the technology is the use of a double "Z" probe design. Each "Z" probe consists of three elements:
- A lower region that hybridizes to the target RNA sequence.
- A linker sequence (spacer).
- A tail region that binds to pre-amplifier molecules [2].
This design is critical because the assay requires two "Z" probes to bind side-by-side on the target RNA for the pre-amplifier to attach and initiate the signal amplification cascade. This double-Z requirement is the foundation of the assay's high specificity, making off-target binding and background noise highly unlikely [2].
Signal Amplification Workflow
The workflow involves a series of sequential hybridization steps that build a large signal complex on each target RNA molecule:
- Hybridization: Pairs of "Z" probes bind to the target RNA.
- Pre-amplification: A pre-amplifier molecule binds to the tails of the "Z" probe pair.
- Amplification: Multiple amplifier molecules bind to the pre-amplifier.
- Labeling: Finally, many labeled probes (chromogenic or fluorescent) conjugate to the amplifiers [2].
This cascade results in an amplification of up to 8,000-fold for each target RNA molecule, which is visualized as a distinct dot under a microscope. Each dot corresponds to a single RNA molecule, enabling direct quantification [2].
Detailed Experimental Protocols
Proper sample preparation is critical for the success of the RNAscope assay. The following protocols are optimized for the most common sample types.
Protocol A: FFPE Tissue Sample Preparation and Staining
This protocol is designed for formalin-fixed, paraffin-embedded tissues, the most common sample type in pathology.
Sample Fixation and Embedding (Critical Step)
- Fixation: Fix tissue samples in FRESH 10% Neutral Buffered Formalin (NBF) for 16–32 hours at room temperature. Do not fix at 4°C [10] [15].
- Impact of Improper Fixation:
- Under-fixation: Leads to protease over-digestion during pretreatment, resulting in RNA loss and poor tissue morphology [10] [15].
- Over-fixation: Leads to protease under-digestion, causing poor probe accessibility, low signal, and a low signal-to-background ratio, though tissue morphology may remain excellent [10] [15].
- Embedding: Process fixed tissues through a graded series of ethanol and xylene, then infiltrate with paraffin. The tissue block should be cut to a thickness of 3-4 mm [15].
Sectioning and Slide Preparation
- Cut 5 μm ± 1 μm sections using a microtome [15].
- Mount sections on Fisher Scientific SuperFrost Plus Slides to prevent tissue loss [15].
- Air-dry slides and bake at 60°C for 1-2 hours prior to initiating the RNAscope assay [15].
RNAscope Assay Workflow (Manual or Automated)
- Deparaffinization and Dehydration: Use xylene and ethanol series.
- Hydrogen Peroxide Incubation: Incubate with RNAscope Hydrogen Peroxide for 10 minutes at room temperature to quench endogenous peroxidase activity. Wash in distilled water [22].
- Target Retrieval: Expose slides to target retrieval reagents in a steamer or water bath. The conditions may require optimization based on fixation. Always start with the manufacturer's recommended guidelines [15].
- Protease Digestion: Incubate slides with Protease Plus for 10-30 minutes at room temperature. Protease treatment time is a key optimization variable; over-fixed tissues may require longer digestion [22] [15].
- Probe Hybridization: Apply the target-specific probe mixture and incubate for 2 hours at 40°C in a HybEZ oven or similar hybridization system [22].
- Signal Amplification: Perform a series of amplifier incubations (AMP 1, AMP 2, AMP 3) as per the kit instructions, with wash steps in between [22].
- Signal Detection: Develop the signal using chromogenic or fluorescent substrates compatible with your detection system.
- Counterstaining and Mounting: Counterstain with hematoxylin (chromogenic) or DAPI (fluorescent). Apply coverslips with a suitable mounting medium [22].
Protocol B: Fresh-Frozen Tissue Preparation and Staining
This protocol is tailored for sensitive tissues where RNA preservation is a priority, such as brain tissue.
Tissue Freezing and Sectioning
- Embed the fresh tissue in OCT compound in a mold.
- Freeze by immersing the mold in a metal beaker filled with 2-methylbutane, surrounded by dry ice in methanol [22].
- Store the block at -80°C.
- Section on a cryostat at 16 μm thickness and mount on SuperFrost Plus slides. Store slides at -80°C until use [22].
Sample Fixation and Dehydration
- Immerse slides in 4% PFA in 1x PBS for a minimum of 15 minutes, but optimally 2 hours at room temperature [22].
- Wash twice in 1x PBS.
- Dehydrate the tissue by immersing slides in a series of ethanol solutions:
- 50% EtOH in DDW for 5 min at RT.
- 70% EtOH in DDW for 5 min at RT.
- 100% EtOH in DDW for 5 min at RT (twice) [22].
- Let slides dry, then draw a hydrophobic barrier around the sections.
Tissue Pretreatment and Hybridization
- Hydrogen Peroxide: Apply Hâ‚‚Oâ‚‚ for 10 minutes at RT. Wash in DDW [22].
- Protease Treatment: Apply Protease Plus for 10 minutes at RT. Wash in 1x PBS [22].
- Probe Hybridization: Prepare probe master mix. Apply to slides and incubate for 2 hours at 40°C in a pre-warmed HybEZ oven [22].
- Amplification and Detection: Follow the same amplification and development steps as for FFPE tissues (Steps 6-8 from Protocol A). For fluorescent detection, Opal fluorophores can be used at a dilution of 1:750 in TSA buffer [22].
The Scientist's Toolkit: Essential Reagents and Controls
Table 2: Key Research Reagent Solutions for RNAscope
| Reagent / Solution | Function / Purpose | Examples / Notes |
|---|---|---|
| 10% NBF (Neutral Buffered Formin) | Standard fixative for FFPE tissues; preserves RNA and morphology. | Must be fresh; fixation time is critical (16-32 hrs) [10]. |
| 4% PFA (Paraformaldehyde) | Fixative for fresh-frozen tissues and cells. | Can be made with RNase-free PBS for pre-fixation steps [10] [22]. |
| Protease Plus / Protease III | Enzyme for digesting proteins and making target RNA accessible. | Digestion time is a key optimization variable [22] [15]. |
| Target Retrieval Reagents | Unmask target RNA sequences cross-linked by fixatives. | Required for FFPE tissues; conditions may need optimization [15]. |
| Positive Control Probes | Validate assay success and RNA integrity. | Probes for housekeeping genes (e.g., PPIB, POLR2A, UBC) [2] [15]. |
| Negative Control Probe | Assess background and non-specific binding. | Probe for bacterial gene dapB [2] [15]. |
| Multiplex Fluorescent v2 Kit | Enable simultaneous detection of multiple RNA targets. | Contains HRP blockers and amplifiers for multiplexing [22]. |
| Opal Fluorophores | Fluorescent labels for signal detection in multiplex assays. | Used with TSA buffer for high-sensitivity detection [22]. |
The body of evidence from systematic reviews and original research consistently demonstrates that the RNAscope assay exhibits high concordance with molecular techniques like qPCR and DNA ISH, while providing invaluable spatial resolution that these methods lack. Its application is particularly powerful for resolving equivocal cases, analyzing tumor heterogeneity, and validating biomarkers in drug development. The provided detailed protocols for FFPE and fresh-frozen tissues serve as a foundational guide for generating robust and reliable data. By integrating quantitative power with morphological context, RNAscope establishes itself as an indispensable tool for researchers and drug development professionals, complementing and enhancing the capabilities of traditional gold-standard methods.
The reliability of any RNAscope assay is fundamentally rooted in proper sample preparation. Fixation quality directly determines the success of subsequent digital quantification, forming a critical link in the research workflow [10]. Under-fixation leads to protease over-digestion and RNA degradation, while over-fixation reduces probe accessibility, resulting in low signal-to-noise ratios [10]. For formalin-fixed, paraffin-embedded (FFPE) tissues, ACD recommends fixation in fresh 10% Neutral Buffered Formalin (NBF) for 16–32 hours at room temperature to optimally preserve RNA integrity and morphology [10] [15]. This precise fixation creates the foundation for accurate, software-assisted quantification, enabling researchers to extract meaningful, reproducible gene expression data from tissue context.
The HALO Image Analysis Platform
The HALO image analysis platform, developed through a partnership between ACD (a Bio-Techne brand) and Indica Labs, is a digital pathology solution designed specifically for quantitative RNA in situ hybridization analysis [58] [59]. It brings objective, accurate quantification to RNAscope assays, enabling a new generation of applications based on single-cell analysis while preserving crucial histopathological context [58]. The platform is engineered for high-throughput, quantitative tissue analysis, offering rapid whole-slide processing and interactive data exploration [60]. Its user-friendly interface allows image analysts with no prior experience to master basic functions within hours, significantly reducing the learning curve associated with digital image analysis [60].
Key Analysis Modules for RNAscope Assays
HALO utilizes a modular approach, providing purpose-built analysis modules that eliminate the need to build algorithms from scratch [60]. The table below summarizes the key modules relevant to RNAscope and in situ hybridization analysis:
Table 1: Key HALO Analysis Modules for RNAscope and In Situ Hybridization
| Module Name | Primary Application | Key Capabilities |
|---|---|---|
| ISH Module [58] | Single-plex and multiplex ISH quantification | Quantitative RNAscope spot counting; brightfield and fluorescent ISH analysis; cell-by-cell expression profiles |
| FISH Module [58] | Fluorescent in situ hybridization | Analysis of unlimited fluorescently-labeled DNA/RNA ISH probes; spot counting and area measurement per cell |
| ISH-IHC Module [58] | RNA-protein co-detection | Simultaneous analysis of ISH probes and IHC biomarkers; cell-by-cell co-expression data |
| Spatial Analysis Module [58] | Spatial relationship investigation | Proximity analysis, nearest neighbor analysis, infiltration analysis, and density heatmap generation |
| Tissue Classifier Add-on [61] [60] | Multi-tissue segmentation | Separation of multiple tissue classes using a learn-by-example approach for region-specific analysis |
| Highplex FL Module [60] | Highly multiplex fluorescence | Analysis of an unlimited number of fluorescent biomarkers with AI-based nuclear and membrane segmentation |
Supported Assays and Technical Compatibility
The HALO platform supports all RNAscope manual and automated assays, including the RNAscope 2.5 HD BROWN and RED kits, RNAscope LS Multiplex assays, BaseScope assays, and miRNAscope assays [58] [59]. This broad compatibility ensures researchers can apply consistent quantification methods across different assay formats. The platform maintains extensive file format interoperability, supporting proprietary slide formats from major manufacturers (Nikon, Hamamatsu, Aperio, Zeiss, Leica, Ventana, Philips) as well as non-proprietary formats (JPG, TIF, OME.TIFF) [60]. This flexibility allows integration into diverse laboratory workflows without requiring extensive file conversion steps.
RNAscope Quantification Workflow and Data Outputs
Integrated Analysis Workflow
The following diagram illustrates the comprehensive workflow for RNAscope sample preparation through HALO software analysis, highlighting the critical role of proper fixation:
Figure 1: Integrated workflow from sample preparation to digital quantification with HALO
Quantitative Data Outputs and Analysis Capabilities
HALO provides multiple analytical outputs for RNAscope data interpretation, ranging from single-cell data to spatial distribution patterns. The platform generates automatic H-scores and histograms that bin cells according to their expression levels (ACD scores 0-4), calculating an overall H-score from 0-400 using the formula: H-score = Σ (ACD score × percentage of cells per bin) [18]. This quantitative approach enables precise characterization of gene expression heterogeneity within tissue samples.
The platform's cell-by-cell expression profiles allow researchers to mine millions of cells while maintaining an interactive link between numerical data and visual representation in the tissue context [58] [60]. Users can sort and filter cell data, then immediately locate corresponding cells in the image markup, facilitating visual validation of analysis results. The spatial analysis module extends this capability further by providing heat mapping, proximity analysis, and infiltration analysis to characterize the tumor microenvironment and cellular interactions [58].
Table 2: RNAscope Data Analysis Scenarios and Corresponding HALO Outputs
| Expression Scenario | Description | Recommended HALO Analysis Method |
|---|---|---|
| Homogeneous Expression [18] | Uniform target expression in a cell population | Average dots per cell across entire population |
| Heterogeneous Expression [18] | Variable expression levels within a cell type | H-score calculation and expression histogram profiling |
| Subpopulation Expression [18] | Target expressed in specific cell subset | Region-specific analysis with Tissue Classifier |
| Cellular Co-expression [18] | Two targets expressed in same cells | Dual-positive quantification with ISH-IHC module |
| Rare Cell Expression [18] | Target in scarce cell population | Cell count percentage over expression level focus |
| Spatial Relationships [58] | Cellular interactions and positioning | Proximity and infiltration analysis with Spatial module |
Experimental Protocol: Fresh-Frozen Tissue RNAscope with HALO Analysis
Sample Preparation and Fixation
The following protocol is adapted for fresh-frozen mouse brain tissue but can be applied to other central nervous system areas with appropriate modifications [22].
Materials:
- 1x PBS (using autoclaved Hâ‚‚O)
- OCT Compound
- 2-methylbutane
- SuperFrost Plus slides
- 4% PFA in 1x PBS
- Fresh 50%, 70%, 100% Ethanol
- RNAscope Multiplex Fluorescent Reagent Kit v2
- Target probes and control probes
- HybEZ Hybridization System
- Opal fluorophores
- ProLong Gold Antifade Reagent with DAPI
Tissue Freezing and Sectioning:
- Perfuse mouse with 1x PBS at approximately 7ml/min perfusion rate.
- Immediately place harvested brain into OCT mold, cover with OCT, and freeze in a metal beaker filled with 2-methylbutane, surrounded by dry ice in methanol.
- Store brain at -80°C overnight.
- Section on cryostat at 16 μm thickness onto SuperFrost Plus slides.
- Store slides at -80°C overnight.
Fixation and Dehydration:
- Immerse slides in 4% PFA in 1x PBS for 2 hours at room temperature.
- Wash twice in 1x PBS.
- Dehydrate through graded ethanol series: 5 minutes each in 50% EtOH, 70% EtOH, and two changes of 100% EtOH.
- If necessary, store slides in 100% EtOH at -20°C for up to one week (not recommended for optimal results).
- Let slides air dry briefly and draw a hydrophobic barrier around sections.
RNAscope Assay Procedure
Tissue Pretreatment:
- Apply RNAscope Hydrogen Peroxide and incubate for 10 minutes at room temperature.
- Wash in DDW for 2 minutes (twice).
- Apply Protease Plus and incubate for 10 minutes at room temperature.
- During incubation, warm probes at 40°C for 10 minutes, then equilibrate to room temperature.
- Wash in 1x PBS for 2 minutes (twice).
Probe Hybridization and Amplification:
- Prepare probe master mix with C1:C2:C3 ratio of 50:1:1.
- Apply probe mix to slides and incubate in HybEZ oven at 40°C for 2 hours.
- Wash in 1x wash buffer for 2 minutes (twice).
- Perform sequential amplification:
- Apply AMP1, incubate 30 minutes at 40°C, wash
- Apply AMP2, incubate 30 minutes at 40°C, wash
- Apply AMP3, incubate 15 minutes at 40°C, wash
- Prepare Opal fluorophore secondaries at 1:750 dilution in TSA buffer.
Signal Development and Mounting:
- For each channel (C1, C2, C3):
- Apply appropriate HRP blocker, incubate 15 minutes at 40°C, wash
- Apply diluted fluorophore, incubate 30 minutes at 40°C, wash
- Apply HRP blocker, incubate 15 minutes at 40°C, wash
- Apply DAPI counterstain for 30 seconds to 5 minutes at room temperature.
- Apply ProLong Gold Antifade Reagent and coverslip with 1.5 thickness coverslips.
- Let slides dry overnight in dark at 4°C before imaging.
HALO Analysis Procedure
The following diagram outlines the core steps for analyzing RNAscope images using the HALO platform:
Figure 2: HALO software workflow for RNAscope image analysis
Analysis Steps:
- Image Loading and Quality Control: Load scanned RNAscope images and visually assess staining quality using positive and negative controls as reference.
- Module Selection: Choose appropriate analysis module based on assay type (ISH for brightfield, FISH for fluorescent, ISH-IHC for co-detection).
- Segmentation Optimization: Use pre-trained AI networks for nuclear and membrane segmentation with real-time tuning for parameter optimization.
- Phenotype Definition: Assign channel combinations using intuitive grid interface to identify cell types of interest.
- Analysis Execution: Run analysis on single slides or queue batch analysis for high-throughput processing.
- Data Exploration: Investigate results with interactive markup images, toggle cell populations, and explore spatial relationships.
- Export and Visualization: Customize export of summary data, cell-by-cell data, or publication-quality figures.
Essential Research Reagent Solutions
Successful implementation of RNAscope assays with HALO quantification requires specific research reagents and materials. The table below details essential components and their functions within the experimental workflow:
Table 3: Essential Research Reagents for RNAscope Assays with HALO Analysis
| Reagent/Material | Function in Workflow | Compatibility Notes |
|---|---|---|
| SuperFrost Plus Slides [22] [15] | Tissue adhesion and section support | Recommended for all tissue types to prevent tissue loss |
| 10% NBF or 4% PFA [10] [22] | Tissue fixation and RNA preservation | 10% NBF optimal for FFPE; 4% PFA for frozen sections |
| RNAscope Multiplex Fluorescent Kit v2 [22] | Signal amplification and detection | Compatible with HALO FISH and ISH-IHC modules |
| Protease Plus [22] | Tissue permeabilization for probe access | Requires optimization for fixation conditions |
| Positive Control Probes (PPIB, POLR2A, UBC) [15] | RNA quality and assay performance verification | Essential for validating experimental conditions |
| Negative Control Probes (dapB) [15] | Background staining assessment | Critical for establishing specificity thresholds |
| Opal Fluorophores [22] | Multiplex signal detection | Compatible with HALO fluorescence modules |
| DAPI [22] | Nuclear counterstain | Essential for cellular segmentation in HALO |
The integration of optimized RNAscope sample preparation with HALO software quantification creates a powerful pipeline for spatial gene expression analysis. The critical importance of proper fixation—using fresh 10% NBF for 16-32 hours for FFPE tissues or 4% PFA for fresh-frozen tissues—cannot be overstated, as it fundamentally determines the quality and reliability of subsequent digital quantification [10] [22]. HALO's purpose-built modules for in situ hybridization provide researchers with objective, reproducible tools for quantifying gene expression at single-cell resolution, enabling sophisticated analysis of expression heterogeneity, cellular co-expression patterns, and spatial relationships within the tissue microenvironment [58] [18]. This integrated approach from sample preparation through digital quantification advances the rigor and reproducibility of spatial transcriptomics research, supporting drug development and discovery with quantitative data derived directly from morphological context.
Conclusion
Proper sample preparation fixation is the cornerstone of successful RNAscope assays, directly influencing RNA integrity, probe accessibility, and ultimately, the reliability of gene expression data. Adherence to standardized fixation protocols using fresh 10% NBF for 16-32 hours establishes the foundation, while systematic troubleshooting and validation using control probes ensure assay quality. The high concordance of RNAscope with molecular techniques like qPCR positions it as a powerful tool for spatial gene expression analysis in research and emerging clinical applications. Future directions include broader adoption in clinical diagnostics through rigorous validation studies and continued refinement of multiplexing capabilities, solidifying RNAscope's role in advancing precision medicine and biomarker discovery.
References
- 1. : A Novel in Situ RNA Analysis Platform for Formalin-Fixed... RNAscope [https://pmc.ncbi.nlm.nih.gov/articles/PMC3338343/]
- 2. Evaluation of the Suitability of RNAscope as a Technique to Measure Gene Expression in Clinical Diagnostics: A Systematic Review - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8710359/]
- 3. RNAscope Enables Spatial Genomic ISH, Spatial Tissue Staining | ACDbio [https://acdbio.com/science/how-it-works]
- 4. 神ç»çŽ¯è·¯åº”用(35)丨RNAscope技术简介åŠåŽŸä½æ‚交实验方法 [https://view.inews.qq.com/a/20251124A02A9N00]
- 5. RNASCOPE®检测的实验原ç†æ˜¯ä»€ä¹ˆï¼Ÿ [https://acdbio.com/cn/science/how-it-works]
- 6. bio-techne.com/resources/protocols-troubleshooting/using-cellprofiler... [https://www.bio-techne.com/resources/protocols-troubleshooting/using-cellprofiler-analyze-rnascope-basescope-data]
- 7. BOND-RX 全自动ã€å¤šé‡IHC æŸ“è‰²å¹³å° [https://www.leicabiosystems.com/zh-cn/ihc-ish/ihc-ish-instruments/bond-rx/]
- 8. 使用RNA 原ä½æ‚交测定对å°é¼ è„‘ä¸çš„èƒ°å²›ç´ å—体A å’ŒB 进行 ... [https://www.jove.com/cn/t/68677/precise-visualization-insulin-receptors-b-murine-brain-with-an-rna]
- 9. 一ç§ç»„织å°é—试剂ã€å¤šæŒ‡æ ‡rna-蛋白共染方法åŠåº”用 [https://patents.google.com/patent/CN119932161A/zh]
- 10. Sample Preparation | In Situ Hybridization, RNA-ISH [https://acdbio.com/support-solution-category/sample-preparation]
- 11. RNAscope in situ hybridization (ISH) FAQs [https://www.bio-techne.com/reagents/rnascope-ish-technology/getting-started/faqs]
- 12. FFPE Sample Preparation and Pretreatment: For the RNAscopeâ„¢ 2.5 Assay | Bio-Techne [https://www.bio-techne.com/resources/protocols-troubleshooting/ffpe-sample-preparation-pretreatment-rnascope-2.5-assay]
- 13. Quantitative Analysis of Gene Expression in RNAscope ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9901451/]
- 14. Identification of Optimal Decalcification Method and Tissue ... [https://www.mdpi.com/2304-6767/13/11/538]
- 15. RNAscope Assay Technical Support and FAQ [https://acdbio.com/technical-support/getting-started]
- 16. Effect of formalin-fixation and paraffin-embedded tissue ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11185121/]
- 17. Learn How RNAscope Technology Works [https://www.bio-techne.com/reagents/rnascope-ish-technology/get-started]
- 18. A Guide for RNAscopeâ„¢ Data Analysis [https://www.bio-techne.com/reagents/rnascope-ish-technology/rnascope/guide-rnascope-data-analysis]
- 19. FFPE | In Situ Hybridization, RNA-ISH [https://acdbio.com/innovation/ffpe]
- 20. FFPE vs Frozen Tissue Samples [https://www.biochain.com/blog/ffpe-vs-frozen-tissue-samples/]
- 21. Fresh frozen vs FFPE samples for next-generation ... [https://www.lexogen.com/blog/fresh-frozen-vs-ffpe-samples-for-next-generation-sequencing-studies/]
- 22. Fresh Frozen RNAScope Protocol [https://www.protocols.io/view/fresh-frozen-rnascope-protocol-hfnab3maf.html]
- 23. Fixed frozen tissue | In Situ Hybridization, RNA-ISH [https://acdbio.com/sample-compatibility/fixed-frozen-tissue]
- 24. Comparison of Fresh Frozen Tissue With Formalin-Fixed ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7083147/]
- 25. RNAscope Troubleshooting Guide and FAQ [https://acdbio.com/technical-support/solutions]
- 26. RNAscope Multiplex FISH Signal Assessment in FFPE and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11755420/]
- 27. What are FFPE samples and how are they prepared? [https://www.lexogen.com/blog/what-are-ffpe-samples-and-how-are-they-prepared/]
- 28. RNAscope in situ hybridization confirms mRNA integrity ... [https://www.oncotarget.com/article/21851/text/]
- 29. Advances in spatial transcriptomics and its application ... [https://www.nature.com/articles/s41413-025-00429-w]
- 30. Fixed Frozen Tissue Sample Preparation- RNAscope 2.5 ... [https://www.bio-techne.com/resources/protocols-troubleshooting/fixed-frozen-tissue-sample-preparation-rnascope2.5-chromogenic-assay]
- 31. Cryopreservation and Sample Embedding – insitutech [https://insitutech.wordpress.com/2023/10/04/cryopreservation-and-sample-embedding/]
- 32. Expanded RNAscopeâ„¢ In Situ Hybridization Assay [https://www.protocols.io/view/expanded-rnascope-in-situ-hybridization-assay-dp8p5rvn.html]
- 33. How to Submit Tissues for Embedding | Research Histology Services [https://rhslab.pitt.edu/drop-info/how-submit-tissues-embedding]
- 34. RNAscope: A Novel in Situ RNA Analysis Platform for ... [https://www.sciencedirect.com/science/article/pii/S1525157811002571]
- 35. Adherent Cell Culture Protocol [https://www.thermofisher.com/us/en/home/references/gibco-cell-culture-basics/cell-culture-protocols/subculturing-adherent-cells.html]
- 36. Protocol for the Preparation and Fluorescent ICC Staining ... [https://www.rndsystems.com/resources/protocols/protocol-preparation-and-fluorescent-icc-staining-stem-cells-coverslips]
- 37. In Situ Hybridization Troubleshooting | ACD [https://acdbio.com/ebook/troubleshooting/sample-preparation-and-optimization]
- 38. Demystified … Tissue microarray technology - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC1187321/]
- 39. Tumor Tissue Microarrays (TMAs): Importance, ... [https://blog.crownbio.com/tumor-tissue-microarrays-cancer-research]
- 40. Tissue Microarrays: Methods and Protocols [https://link.springer.com/book/10.1007/978-1-60761-806-5]
- 41. Larger core size has superior technical and analytical ... [https://www.nature.com/articles/labinvest2016151]
- 42. Immune Biomarkers on Tissue Microarray Cores Support ... [https://www.sciencedirect.com/science/article/pii/S0023683725000017]
- 43. SUPERFROST® PLUS - Adhesion Slide [https://www.emsdiasum.com/superfrost-plus-adhesion-slide?srsltid=AfmBOooNJMeA_6vxubqdemEBtGoj9Luc7dTYiWj1Yyn7J9xs9OJowe6-]
- 44. RNAscope Troubleshooting Guide and FAQ [https://acdbio.com/technical-support/solutions/faq]
- 45. Superfrost Plus and ColorFrost Plus Microslides [https://www.weberscientific.com/superfrost-plus-and-colorfrost-plus-microslides?srsltid=AfmBOopeTaHzEUndgZXwUw3J9G3ggEmYgKY0tactu9nwQ4ARL3qUTHGy]
- 46. RNA In situ hybridization (ISH) using RNAscope® [https://www.protocols.io/view/rna-in-situ-hybridization-ish-using-rnascope-dydu7s6w.html]
- 47. Unexpected Staining Patterns: ISH Staining | ACD [https://acdbio.com/ebook/troubleshooting/unexpected-staining-patterns]
- 48. RNAscope Assay Visualization and Interpretation [https://acdbio.com/support-solution-category/rnascope-assay-visualization-and-interpretation]
- 49. RNAscope ISH Troubleshooting [https://www.bio-techne.com/resources/protocols-troubleshooting/rnascope-ish-troubleshooting]
- 50. Control Slides and Control Probes for RNAscope [https://acdbio.com/control-slides-and-control-probes-rnascope]
- 51. Frontiers | Combining RNAscope and immunohistochemistry to... [https://www.frontiersin.org/journals/molecular-neuroscience/articles/10.3389/fnmol.2023.1225847/full]
- 52. 2025 Jan 14 - Unlock the role of spatial profiling in ... [https://acdbio.com/2025-jan-14-unlock-role-spatial-profiling-detecting-diverse-mrna-markers-and-short-targets]
- 53. RNAscope ISH Reference Guide [https://www.bio-techne.com/reagents/rnascope-ish-technology/rnascope/rnascope-ish-reference-guide]
- 54. Control Probes: Manual Multiplex | In Situ Hybridization, ... [https://acdbio.com/control-probes-manual-multiplex]
- 55. Automated Quantitative RNA in Situ Hybridization for ... [https://www.sciencedirect.com/science/article/pii/S1525157812003157]
- 56. Assessment of ESR1, PGR, ERBB2, and MKI67 mRNA in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12261032/]
- 57. Molecular Biological Determination of HER2 Status Using ... [https://www.mdpi.com/1422-0067/26/5/2148]
- 58. HALO® image analysis platform | In Situ Hybridization ... [https://acdbio.com/halo-image-analysis-platform]
- 59. HALO® image analysis platform | In Situ Hybridization ... [https://acdbio.com/halo-image-analysis-platform-new]
- 60. HALO | Quantitative Image Analysis for Pathology [https://indicalab.com/halo/]
- 61. Getting Started with RNAscope™ Image Analysis in HALO® [https://acdbio.com/2023-mar-28-getting-started-rnascope%E2%84%A2-image-analysis-halo%C2%AE]