Wnt Signaling in Early Embryogenesis: From Molecular Mechanisms to Therapeutic Targets
This article provides a comprehensive analysis of the Wnt signaling pathway's critical functions during early embryogenesis.
Wnt Signaling in Early Embryogenesis: From Molecular Mechanisms to Therapeutic Targets
Abstract
This article provides a comprehensive analysis of the Wnt signaling pathway's critical functions during early embryogenesis. It explores the foundational biology of canonical and non-canonical Wnt pathways, their roles in cell fate determination, pluripotency, and body axis patterning. For a research and drug development audience, we examine advanced methodological approaches for studying Wnt signaling, address common experimental challenges and optimization strategies, and validate findings through cross-species comparisons and clinical implications. The synthesis of current research highlights Wnt signaling as a pivotal regulator of development and a promising therapeutic target, with future directions focusing on overcoming technical barriers for clinical translation.
Core Mechanisms of Wnt Signaling in Embryonic Development
Wnt signaling represents a cornerstone of cellular communication, governing fundamental processes in embryonic development and tissue homeostasis. The term "Wnt" originated as a merger of two homologous proteins: Drosophila Wingless (Wg) and mouse Int-1, ultimately coined by Nusse et al. in 1991 [1]. This evolutionarily conserved pathway regulates cell fate determination, proliferation, migration, and polarity across species from diploblastic cnidarians to mammals [2] [3]. The Wnt family comprises 19 highly conserved glycoproteins in humans that function as secreted signaling molecules [1] [3]. These proteins undergo extensive post-translational modifications including glycosylation and palmitoylation by the Porcupine protein, which are essential for their secretion and function [1] [4]. Wnt distribution occurs through various mechanisms—free diffusion, restricted diffusion, and active transport—forming concentration gradients that provide positional information during embryogenesis [5].
The broader thesis context of Wnt signaling in early embryogenesis research reveals its indispensable roles in axial patterning, gastrulation, neural specification, and organ formation [2] [5]. During mammalian embryogenesis, Wnt signaling specifies pattern formation and regulates the maintenance and differentiation of stem cells both in vivo and in vitro [2]. Disruption of Wnt signaling during embryonic development results in severe abnormalities, including spina bifida and other birth defects, while dysregulation in adults contributes to various pathologies including cancer, skeletal disorders, and metabolic diseases [1] [4]. This technical guide provides a comprehensive overview of the canonical and non-canonical Wnt pathways, emphasizing their mechanisms, regulatory networks, and experimental approaches relevant to research in early embryogenesis.
Pathway Mechanisms and Molecular Components
Canonical Wnt/β-catenin Pathway
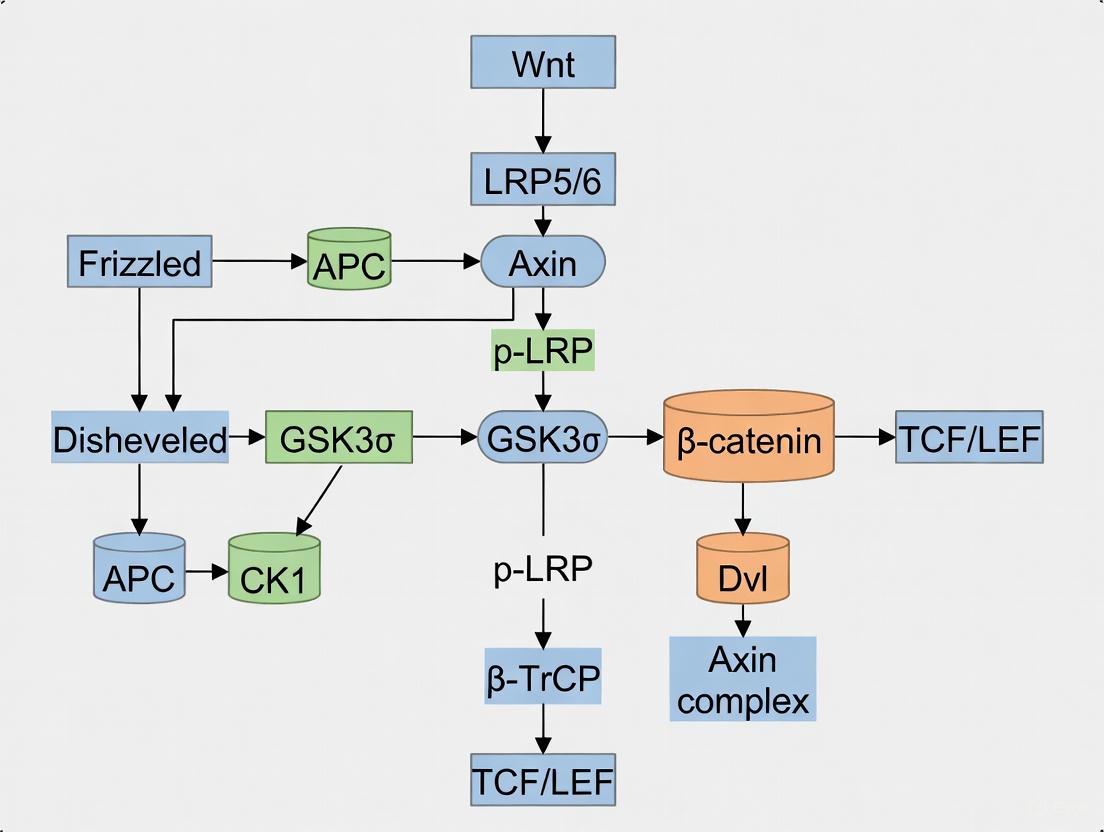
The canonical Wnt pathway, also known as the Wnt/β-catenin pathway, centers on the regulation of β-catenin stability and nuclear translocation. In the absence of Wnt ligand, a destruction complex composed of Axin, Adenomatous Polyposis Coli (APC), Glycogen Synthase Kinase 3β (GSK3β), Casein Kinase 1α (CK1α), and β-transducin repeat containing protein (β-TrCP) constitutively targets β-catenin for proteasomal degradation [1] [3] [6]. This complex facilitates the phosphorylation of β-catenin by GSK3β, leading to its ubiquitination and subsequent degradation [1]. When canonical Wnt ligands (including Wnt1, Wnt2, Wnt3, Wnt3a, Wnt8a, Wnt8b, Wnt10a, and Wnt10b) bind to Frizzled (Fz) receptors and LRP5/6 co-receptors, Dishevelled (Dsh/Dvl) is recruited to the membrane [1] [6]. This recruitment inhibits the destruction complex, allowing β-catenin to accumulate in the cytoplasm and translocate to the nucleus [1] [3]. Within the nucleus, β-catenin associates with T-cell factor (TCF)/lymphoid enhancer factor (LEF) transcription factors to activate target genes involved in cell proliferation, survival, and differentiation [1] [6] [4].
Table 1: Core Components of the Canonical Wnt Pathway
| Component | Function | Representative Members |
|---|---|---|
| Ligands | Activate pathway by binding receptors | Wnt1, Wnt2, Wnt3, Wnt3a, Wnt8a, Wnt8b, Wnt10a, Wnt10b [1] [6] |
| Receptors | Bind Wnt ligands initiate signaling | Frizzled (Fz) family [1] [3] |
| Co-receptors | Essential for signal transduction | LRP5/6 [1] [3] |
| Intracellular Transducers | Relay signal from membrane | Dishevelled (Dsh/Dvl), β-catenin [1] [3] |
| Transcription Factors | Regulate target gene expression | TCF/LEF family [1] [6] |
| Target Genes | Execute cellular responses | MYC, CCND1 (Cyclin D1), WISP2 [7] |
Non-Canonical Wnt Pathways
Non-canonical Wnt pathways operate independently of β-catenin and LRP5/6 co-receptors, diversifying into two major branches: the Wnt/Planar Cell Polarity (PCP) pathway and the Wnt/Ca²⺠pathway [3] [4]. Non-canonical Wnt ligands include Wnt4, Wnt5a, Wnt5b, Wnt6, Wnt7a, Wnt7b, and Wnt11 [1] [6]. These ligands bind to Frizzled receptors along with alternative co-receptors such as ROR1/2, RYK, and CTHRC1 [1]. The diversity of receptor/co-receptor/ligand combinations creates context-specific signaling outcomes depending on cell type and receptor availability [1].
The Wnt/PCP pathway regulates coordinated cellular polarization and migration within the plane of epithelial sheets [3] [4]. Upon activation, Frizzled recruits Dishevelled, which then interacts with Dishevelled-associated activator of morphogenesis 1 (DAAM1) [3]. DAAM1 activates the small GTPase Rho through guanine exchange factors, leading to activation of Rho-associated kinase (ROCK)—a key regulator of cytoskeletal dynamics [3]. Simultaneously, Dishevelled can form complexes with Rac1, leading to Jun N-terminal kinase (JNK) activation and actin polymerization [3].
The Wnt/Ca²⺠pathway triggers the release of intracellular calcium from the endoplasmic reticulum [3] [4]. This pathway involves Frizzled receptors along with co-receptors Knypek and Ror2 [4]. Activation leads to stimulation of G-proteins, phospholipase C (PLC), and protein kinase C (PKC) [4]. The resulting increase in intracellular calcium activates calcium/calmodulin-dependent protein phosphatase calcineurin, which dephosphorylates the transcription factor NF-AT (Nuclear Factor of Activated T-cells), promoting its nuclear accumulation and subsequent regulation of target genes [3] [4].
Table 2: Core Components of Non-Canonical Wnt Pathways
| Component | Function | Representative Members |
|---|---|---|
| Ligands | Activate β-catenin-independent signaling | Wnt4, Wnt5a, Wnt5b, Wnt6, Wnt7a, Wnt7b, Wnt11 [1] [6] |
| Receptors | Bind non-canonical Wnt ligands | Frizzled (Fz) family [1] [3] |
| Co-receptors | Mediate alternative signaling | ROR1/2, RYK, CTHRC1, Knypek [1] [4] |
| PCP Effectors | Regulate cytoskeleton and polarity | DAAM1, Rho, Rac, ROCK, JNK [3] |
| Calcium Effectors | Mediate calcium signaling | PLC, PKC, Calcineurin, NF-AT [3] [4] |
Pathway Regulation and Cross-Talk
Extracellular and Intracellular Regulation
Wnt signaling is tightly regulated at multiple levels to ensure precise spatiotemporal control. Extracellular regulation occurs through various secreted antagonists that sequester Wnt ligands or block receptors [3] [6]. These include Secreted Frizzled-Related Proteins (sFRPs), Wnt Inhibitory Factor (WIF), and Dickkopf (DKK) family proteins [3] [6]. While sFRPs and WIF1 bind directly to Wnt ligands, preventing their interaction with Frizzled receptors, Dickkopf proteins specifically inhibit canonical signaling by binding to LRP5/6 co-receptors [6]. Conversely, R-spondin proteins enhance Wnt signaling by binding to LGR4/5/6 receptors and preventing Frizzled ubiquitination and degradation [6].
Intracellularly, multiple mechanisms fine-tune Wnt signaling responses. The destruction complex maintains low β-catenin levels in unstimulated cells [1]. Additionally, nuclear regulators such as ICAT and duplin directly interact with β-catenin to prevent its association with TCF/LEF transcription factors [4]. Recent findings also reveal intricate cross-talk between Wnt pathways and other signaling networks. For instance, non-canonical Wnt signaling interacts with the Hippo-YAP/TAZ pathway, forming an integrated signaling axis that regulates biological functions traditionally attributed to non-canonical Wnt signaling [1]. Furthermore, non-canonical signaling can inhibit canonical pathway activation through multiple mechanisms, including increased β-catenin degradation via GSK3β-independent mechanisms and enhanced secretion of canonical Wnt inhibitors such as DKK1 [1].
Integrated Wnt Signaling Model
The traditional binary classification of Wnt signaling has been challenged by proposals for an integrated Wnt signaling model that acknowledges the complexity of pathway cross-talk [1]. The emerging paradigm suggests that the cellular context—including specific receptor availability, cytoplasmic components, and nuclear mediators—determines the signaling outcome rather than a simple ligand-receptor coupling [1]. This perspective is particularly relevant in early embryogenesis, where Wnt signaling pleiotropy enables diverse context-dependent functions including mitogenic stimulation, cell fate specification, and differentiation [3].
Quantitative Data and Experimental Approaches
Key Quantitative Measurements
Advanced imaging and computational approaches have yielded quantitative insights into Wnt pathway dynamics. A study investigating β-catenin spatial dynamics in HEK293T cells developed a novel 3D confocal quantitation protocol to measure temporal and spatial changes in β-catenin concentrations following pathway perturbations [8]. The researchers acquired spatial data from two cellular compartments (nucleus and cytosol-membrane) and quantified target protein concentrations after treatments with cycloheximide (protein synthesis inhibitor) or Wnt3A (pathway activator) [8].
Table 3: Quantitative β-catenin Dynamics in HEK293T Cells After Perturbation [8]
| Treatment | Time Point | β-catenin Change (Whole Cell) | Compartmental Dynamics |
|---|---|---|---|
| Cycloheximide | 0-4 hours | Decrease at constant rate | Similar decrease rate in both nuclear and cytosol-membrane compartments |
| Wnt3A | 0-1 hour | Initial increase | Faster increase in nuclear compartment |
| Wnt3A | 1-4 hours | Continued increase | Balanced increase across compartments |
This study demonstrated that with Wnt3A stimulation, the total cellular β-catenin rises throughout the cell, but the increase occurs initially faster in the nuclear compartment during the first hour [8]. When protein synthesis was inhibited with cycloheximide, β-catenin decreased at similar rates in both compartments, suggesting that diffusional transport is rapid compared to β-catenin degradation in the cytosol [8]. Computational modeling revealed that a two-compartment model with active transport mechanisms best reproduced the experimental data, highlighting the importance of spatial organization in Wnt signaling [8].
Experimental Protocols
3D Confocal Quantification of β-catenin Dynamics
The following methodology was adapted from the HEK293T study to quantify spatial and temporal protein dynamics in Wnt signaling [8]:
Cell Culture and Treatment:
- Culture HEK293T cells in appropriate medium under standard conditions.
- For perturbation experiments: apply either Wnt3A ligand (e.g., 100-200ng/mL) to activate signaling or cycloheximide (e.g., 10-100μg/mL) to inhibit protein biosynthesis.
- Include untreated controls at each time point.
- Incubate for predetermined time points (e.g., 0, 1, 2, 4 hours).
Cell Staining and Fixation:
- Fix cells with paraformaldehyde (e.g., 4% for 15 minutes).
- Permeabilize with Triton X-100 (e.g., 0.1% for 10 minutes).
- Block with serum-based blocking buffer.
- Incubate with primary antibodies: anti-β-catenin, anti-N-cadherin (cell boundary marker).
- Incubate with fluorophore-conjugated secondary antibodies.
- Counterstain nuclei with DAPI.
Image Acquisition and Calibration:
- Use confocal microscopy with consistent settings across samples.
- Include InSpeck microspheres (0.3% rated) as intensity calibration standards to enable quantitative comparisons between samples and time points.
- Acquire z-stacks to generate 3D volume data.
Image Analysis and Quantification:
- Use image processing software to identify cellular compartments based on marker signals (DAPI for nuclei, N-cadherin for cell boundary).
- Measure β-catenin intensity in each compartment.
- Normalize intensities using calibration standards.
- Perform statistical analysis across multiple cells and replicates.
This protocol enables quantitative analysis of protein localization and abundance changes in response to pathway perturbations, providing spatial and temporal resolution essential for understanding signaling dynamics.
Gene Expression Analysis in Pathological Contexts
To investigate Wnt pathway involvement in disease contexts, such as tumorigenesis, the following QPCR-based approach can be employed [7]:
Sample Collection and Preparation:
- Obtain tissue samples (e.g., pituitary tumors) during surgical procedures.
- Microdissect to separate tumoral from non-tumoral tissues.
- Snap-freeze in liquid nitrogen and store at -70°C.
- Isolate total RNA using TRIzol reagent.
- Assess RNA integrity by spectrophotometry (260/280nm ratio) and agarose gel electrophoresis.
- Synthesize cDNA using reverse transcription kits.
Gene Expression Profiling:
- Design or select TaqMan assays for genes of interest covering:
- Canonical pathway components (WNT ligands, receptors, destruction complex members, target genes)
- Non-canonical pathway components (PCP and Ca²⺠pathway members)
- Endogenous controls (GUSβ, TBP, PGK1)
- Perform quantitative PCR with appropriate cycling conditions.
- Calculate gene expression using efficiency-corrected methods.
- Determine fold changes relative to control tissues.
Data Analysis:
- Use hierarchical clustering to identify expression patterns.
- Perform statistical comparisons between sample groups.
- Correlate expression patterns with clinical outcomes.
This approach revealed that most Wnt pathway components are not mis-expressed in pituitary tumors, contrasting with other tumors like colorectal cancer and craniopharyngioma where Wnt signaling plays established roles [7].
Research Tools and Reagent Solutions
Table 4: Essential Research Reagents for Wnt Signaling Studies
| Reagent/Category | Specific Examples | Research Application | Function |
|---|---|---|---|
| Cell Lines | HEK293T [8] | Pathway mechanism studies | Responsive to Wnt3A stimulation; no known Wnt pathway mutations |
| Wnt Ligands | Recombinant Wnt3A [8] | Canonical pathway activation | Binds Fz/LRP5/6 receptors to stabilize β-catenin |
| Pathway Inhibitors | Cycloheximide [8], IWP-2 [5] | Block protein synthesis or Wnt production | Inhibit global translation or Porcupine-mediated Wnt processing |
| Antibodies | anti-β-catenin, anti-N-cadherin [8] | Protein localization and quantification | Visualize and quantify target proteins in cellular compartments |
| Fluorescent Markers | DAPI [8] | Nuclear staining | Demarcate nuclear compartment for spatial analysis |
| Calibration Standards | InSpeck microspheres [8] | Intensity calibration | Enable quantitative comparisons between samples |
| Gene Expression Assays | TaqMan assays [7] | Expression profiling | Quantify transcript levels of pathway components |
Visualizing Wnt Signaling Pathways
Canonical Wnt/β-catenin Pathway
Non-Canonical Wnt Pathways
Experimental Workflow for Spatial Analysis
The canonical and non-canonical Wnt pathways represent sophisticated signaling networks that orchestrate critical processes in early embryogenesis and maintain tissue homeostasis throughout life. While the canonical pathway primarily regulates gene expression through β-catenin stabilization and nuclear translocation, the non-canonical pathways control cytoskeletal organization, cell polarity, and calcium-mediated signaling. The emerging paradigm of integrated Wnt signaling acknowledges the extensive cross-talk between these pathways and their context-dependent functions. Advanced experimental approaches, including 3D quantitative imaging and spatial modeling, continue to reveal new insights into the dynamic regulation of Wnt signaling. For researchers and drug development professionals, understanding these intricate mechanisms provides fertile ground for developing novel therapeutic strategies targeting Wnt-related pathologies while harnessing its regenerative potential.
The Wnt signaling pathway is a highly conserved, crucial system that governs fundamental aspects of embryonic development, including body axis patterning, cell fate specification, proliferation, and migration [9] [10]. Its function is paramount during early embryogenesis, directing processes from the establishment of the primary body axis to the formation of numerous tissues and organs [11]. The pathway's name is a portmanteau of the Drosophila segment polarity gene Wingless (Wg) and the vertebrate proto-oncogene Int-1, reflecting its evolutionary conservation and diverse functional roles [9] [12]. The intricate orchestration of Wnt signaling is achieved through a complex interplay between its core molecular components: the Wnt ligands, their cell surface receptors, and a suite of intracellular transducers that propagate the signal, ultimately leading to specific nuclear responses and changes in gene expression [9] [13]. This guide provides an in-depth technical overview of these key components, framing them within the context of embryogenesis research.
The Wnt Ligand Family
Wnt ligands constitute a large family of secreted, lipid-modified glycoproteins that are approximately 350-400 amino acids in length [9]. In humans, this family comprises 19 members, each playing distinct yet sometimes overlapping roles during development and homeostasis [9] [14]. A defining characteristic of all Wnts is a conserved palmitoleoylation event at a single cysteine residue. This modification, mediated by the Porcupine (PORCN) enzyme in the endoplasmic reticulum, is essential for the ligand's secretion via the Wntless (WLS) transporter and for its subsequent ability to bind to Frizzled receptors [9] [11]. Wnt proteins also undergo glycosylation, which further ensures their proper secretion and function [9].
While the functional classification of Wnt ligands can be context-dependent, they are often categorized based on their propensity to activate different downstream signaling branches. The table below summarizes the primary Wnt ligands found in Homo sapiens and their predominant signaling pathways.
Table 1: Major Wnt Ligands in Humans and Their Primary Signaling Pathways
| Wnt Ligand | Primary Signaling Pathway | Key Roles in Early Embryogenesis |
|---|---|---|
| WNT1 | Canonical [15] | Central nervous system development [9] |
| WNT2 | Canonical [15] | |
| WNT3 | Canonical [15] | Primitive streak formation, somiteogenesis [9] |
| WNT3A | Canonical [15] | |
| WNT4 | Non-canonical & Canonical [15] | Kidney, reproductive tract development [9] |
| WNT5A | Non-canonical [15] [12] | Limb bud patterning, cell migration [9] |
| WNT5B | Non-canonical [15] | |
| WNT6 | Non-canonical & Canonical [15] | |
| WNT7A | Non-canonical & Canonical [15] | Limb patterning, dorsoventral axis specification [9] |
| WNT7B | Non-canonical & Canonical [15] | |
| WNT8A | Canonical [15] | |
| WNT8B | Non-canonical & Canonical [15] | |
| WNT9A | Non-canonical & Canonical [15] | |
| WNT9B | Non-canonical & Canonical [15] | |
| WNT10A | Canonical [15] | |
| WNT10B | Canonical [15] | |
| WNT11 | Non-canonical [15] [12] | Cell movements during gastrulation [14] |
| WNT16 | Non-canonical & Canonical [15] |
Receptors and Co-receptors
The initiation of Wnt signaling occurs at the plasma membrane through the binding of a Wnt ligand to a receptor complex. The core of this complex is formed by receptors from the Frizzled (FZD) family, often in conjunction with various co-receptors that determine the specificity of the downstream signaling cascade [9] [11].
Frizzled (FZD) Receptors
Frizzled receptors are a family of G-protein coupled receptor (GPCR)-like proteins that span the plasma membrane seven times [9] [10]. In humans, there are 10 FZD genes (FZD1-FZD10) [14]. The extracellular N-terminal domain of FZD receptors features a characteristic cysteine-rich domain (CRD) that is responsible for direct binding to the Wnt ligand [9] [11]. The specific combination of Wnt ligand and the FZD receptor it engages with is a primary factor in channeling the signal into the canonical or a non-canonical pathway.
Co-receptors
Co-receptors are essential partners that work alongside FZD receptors to transduce the Wnt signal effectively.
- LRP5/6: The Low-density lipoprotein receptor-related proteins 5 and 6 are single-pass transmembrane proteins that act as the primary co-receptors for the canonical Wnt/β-catenin pathway [13] [10]. Their interaction with Wnt ligands and FZD receptors is crucial for initiating the intracellular events that lead to β-catenin stabilization.
- ROR1/ROR2 and RYK: For the non-canonical pathways, different co-receptors are employed. The tyrosine-protein kinase transmembrane receptors ROR1 and ROR2, as well as the RYK (Receptor-like tyrosine kinase), are key co-receptors for the Planar Cell Polarity (PCP) pathway [9] [15] [11]. These receptors help activate signaling cascades that regulate cytoskeletal organization and cell polarity.
Table 2: Primary Wnt Receptors and Co-receptors and Their Pathway Associations
| Receptor / Co-receptor | Family | Primary Signaling Pathway | Function |
|---|---|---|---|
| FZD1-10 | 7-transmembrane GPCR | Canonical & Non-canonical [15] | Primary receptor for Wnt ligands [9] |
| LRP5/6 | Single-pass transmembrane | Canonical [15] | Co-receptor for β-catenin pathway [13] |
| ROR1 | Receptor tyrosine kinase | Non-canonical [15] | Co-receptor for PCP pathway [11] |
| ROR2 | Receptor tyrosine kinase | Non-canonical [15] | Co-receptor for PCP pathway [11] |
| RYK | Receptor tyrosine kinase | Non-canonical [15] | Co-receptor for PCP and Wnt/Ca2+ pathways [16] |
Intracellular Signal Transduction
Upon ligand-receptor binding, the Wnt signal is propagated inside the cell by a network of intracellular transducers. The central cytoplasmic node for all Wnt signaling branches is the Dishevelled (DVL/Dsh) protein.
The Central Mediator: Dishevelled (DVL)
Dishevelled is a multi-domain, cytoplasmic phosphoprotein that is directly recruited to the plasma membrane by the activated Frizzled receptor [9] [10]. It serves as a molecular hub, directing the signal into different pathways based on its specific protein domains [9]:
- DIX Domain: Essential for canonical pathway signaling. It facilitates interactions with the β-catenin destruction complex [11].
- PDZ Domain: Involved in both canonical and non-canonical pathways. It mediates protein-protein interactions with FZD and downstream effectors [11].
- DEP Domain: Primarily associated with non-canonical signaling, including the PCP and Wnt/Ca2+ pathways [11].
The Canonical Pathway: β-Catenin and the Destruction Complex
The hallmark of the canonical pathway is the regulation of the transcriptional co-activator β-catenin.
- "Off" State (No Wnt ligand): In the absence of a Wnt signal, cytoplasmic β-catenin is targeted for proteasomal degradation by a multi-protein "destruction complex." This complex includes the scaffold proteins Axin and Adenomatous Polyposis Coli (APC), and the kinases Glycogen Synthase Kinase 3β (GSK3β) and Casein Kinase 1α (CK1α). These kinases sequentially phosphorylate β-catenin, marking it for ubiquitination by β-TrCP and subsequent degradation [13] [14]. This keeps cytoplasmic β-catenin levels low and prevents target gene transcription.
- "On" State (Wnt ligand present): Binding of a canonical Wnt (e.g., WNT3A) to FZD and LRP5/6 recruits DVL and Axin to the plasma membrane. This disrupts the destruction complex, preventing β-catenin phosphorylation and degradation. Stabilized β-catenin accumulates in the cytoplasm and translocates into the nucleus. There, it binds to transcription factors of the TCF/LEF (T-cell factor/lymphoid enhancer factor) family, displacing transcriptional repressors and activating the expression of target genes (e.g., c-MYC, CYCLIN D1) that drive cell proliferation and fate decisions [9] [13] [11].
Non-Canonical Pathway Transducers
The non-canonical pathways operate independently of β-catenin/TCF/LEF-mediated transcription.
- Planar Cell Polarity (PCP) Pathway: This pathway regulates cytoskeletal organization and cell polarity. Activated DVL, via its PDZ domain, forms a complex with DAAM1 (Dishevelled-associated activator of morphogenesis 1). DAAM1 activates the small GTPase RhoA, which in turn activates ROCK (Rho-associated kinase). In a parallel branch, DVL activates the small GTPase Rac, which then activates JNK (Jun N-terminal kinase). These cascades ultimately lead to actin cytoskeleton remodeling, which is critical for convergent extension movements during gastrulation [9] [16] [3].
- Wnt/Ca2+ Pathway: Activation of this pathway (e.g., by WNT5A) leads to an increase in intracellular calcium ions (Ca2+). DVL, through its PDZ and DEP domains, can activate Phospholipase C (PLC) via G-proteins. PLC generates inositol 1,4,5-trisphosphate (IP3), triggering the release of Ca2+ from the endoplasmic reticulum. The elevated Ca2+ activates enzymes like Protein Kinase C (PKC) and Ca2+/calmodulin-dependent kinase II (CaMKII), which can influence transcription through factors like NFAT (Nuclear Factor of Activated T-cells), regulating processes such as cell adhesion and migration [16] [14].
Visualization of Wnt Signaling Pathways
The following diagrams, generated using DOT language, illustrate the core components and signal flow of the canonical and non-canonical Wnt pathways.
Canonical Wnt/β-catenin Pathway
Non-Canonical Wnt Pathways
Experimental Analysis of Key Components
Studying the role of Wnt pathway components in embryogenesis requires a multifaceted experimental approach. The following section details key methodologies for analyzing the expression, localization, and functional involvement of ligands, receptors, and transducers.
Protocol: In Situ Hybridization for Mapping Wnt Ligand Expression in Embryo
Objective: To spatially localize the expression patterns of specific Wnt ligand mRNAs (e.g., Wnt3a, Wnt5a) in early-stage mouse embryos (e.g., E8.5-E12.5) [10].
Tissue Fixation and Sectioning:
- Dissect embryos in cold PBS and fix in 4% paraformaldehyde (PFA) in PBS for 12-24 hours at 4°C.
- Dehydrate through an ethanol series, clear in xylene, and embed in paraffin wax.
- Section the embedded embryos into 5-7 µm thick slices using a microtome and mount on positively charged glass slides.
Riboprobe Synthesis and Labeling:
- Clone a specific fragment of the target Wnt cDNA (e.g., 500-1000 bp) into a plasmid with opposing RNA polymerase promoters (e.g., T7, SP6).
- Linearize the plasmid and perform in vitro transcription with the appropriate RNA polymerase in the presence of digoxigenin (DIG)-labeled UTP to generate antisense (experimental) and sense (control) riboprobes.
Hybridization and Detection:
- Deparaffinize and rehydrate the sections. Treat with proteinase K (1-10 µg/mL) for permeabilization.
- Pre-hybridize sections with a hybridization buffer containing formamide, salts, and blocking agents (e.g., yeast tRNA, salmon sperm DNA) for 1-2 hours at 55-65°C.
- Hybridize with the DIG-labeled riboprobe (0.5-1.0 ng/µL) in hybridization buffer overnight at 55-65°C in a humidified chamber.
Washing and Signal Development:
- Perform stringent washes with SSC buffers (e.g., 2x SSC, 0.2x SSC) to remove non-specifically bound probe.
- Block non-specific binding sites with a blocking reagent (e.g., 2% normal sheep serum, 2% BSA).
- Incubate with an alkaline phosphatase (AP)-conjugated anti-DIG antibody for 1-2 hours.
- Wash thoroughly and incubate with the colorimetric AP substrates NBT/BCIP in the dark until a purple-blue precipitate forms. Stop the reaction with TE buffer.
- Counterstain with a nuclear fast red or eosin, dehydrate, clear, and mount with a permanent mounting medium.
Imaging and Analysis:
- Image sections using a bright-field microscope. The expression pattern of the target Wnt ligand will be visualized by the localized NBT/BCIP precipitate.
Protocol: Immunofluorescence for Localizing Intracellular Transducers
Objective: To visualize the subcellular localization and relative abundance of key intracellular transducers (e.g., β-catenin, DVL) in embryonic tissues or stem cell models of embryogenesis.
Sample Preparation:
- For embryos: Fix in 4% PFA, cryoprotect in 15-30% sucrose, embed in OCT compound, and section on a cryostat (8-12 µm thickness).
- For cultured cells: Grow on glass coverslips, treat with recombinant Wnt ligands (e.g., WNT3A) or inhibitors, and fix with 4% PFA.
Permeabilization and Blocking:
- Permeabilize samples with 0.1-0.5% Triton X-100 in PBS for 10-15 minutes.
- Block with 5-10% normal serum (from the species of the secondary antibody) and 1-5% BSA in PBS for 1 hour at room temperature to prevent non-specific antibody binding.
Antibody Incubation:
- Incubate samples with primary antibodies diluted in blocking buffer overnight at 4°C.
- Examples: Mouse anti-β-catenin (to assess nuclear accumulation), Rabbit anti-Dishevelled (to assess membrane recruitment), Rabbit anti-Axin.
- The next day, wash samples 3-4 times with PBS containing 0.05% Tween-20 (PBST).
- Incubate with fluorophore-conjugated secondary antibodies (e.g., Alexa Fluor 488, 568) diluted in blocking buffer for 1 hour at room temperature, protected from light.
- Incubate samples with primary antibodies diluted in blocking buffer overnight at 4°C.
Counterstaining and Mounting:
- Incubate with DAPI (4',6-diamidino-2-phenylindole) for 5-10 minutes to stain nuclei.
- Wash extensively with PBS and mount coverslips onto glass slides using an anti-fade mounting medium (e.g., ProLong Gold).
Imaging and Analysis:
- Image using a confocal or epifluorescence microscope. Analyze images for changes in protein localization (e.g., β-catenin nuclear/cytoplasmic ratio, DVL puncta formation at the membrane).
Protocol: Functional Knockdown using siRNA/shRNA
Objective: To assess the functional requirement of a specific Wnt component (e.g., FZD receptor, DVL) in a developmental process using in vitro models.
Design and Selection of RNAi Constructs:
- Design 2-3 different siRNA oligonucleotides or shRNA-encoding plasmids targeting distinct regions of the mRNA of the gene of interest (e.g., FZD5, DVL1). Include a non-targeting scrambled sequence as a negative control.
Cell Transfection/Transduction:
- Culture relevant cells (e.g., mouse embryonic stem cells, primary mesenchymal cells).
- For siRNA: Transfect cells at 40-60% confluency using a lipofection or electroporation reagent according to the manufacturer's protocol.
- For shRNA: Transduce cells with lentiviral particles encoding the shRNA and select with puromycin (1-5 µg/mL) for 3-5 days to generate a stable knockdown pool.
Validation of Knockdown:
- 48-96 hours post-transfection (or after selection), harvest cells.
- Validate knockdown efficiency via quantitative RT-PCR (qRT-PCR) to measure mRNA levels and/or western blotting to assess protein depletion.
Phenotypic Analysis:
- Perform functional assays relevant to embryogenesis:
- Cell Migration/Invasion: Use Transwell or scratch/wound healing assays to model cell movements.
- Gene Expression Profiling: Analyze the expression of downstream Wnt target genes (e.g., Axin2, Sp5) by qRT-PCR.
- Cytoskeletal Analysis: Stain for F-actin with phalloidin and analyze cell morphology and polarity.
- Perform functional assays relevant to embryogenesis:
The Scientist's Toolkit: Essential Research Reagents
The following table catalogs key reagents and tools essential for investigating the Wnt signaling pathway in a research setting.
Table 3: Key Research Reagents for Wnt Pathway Analysis
| Reagent / Tool | Category | Primary Function in Research | Example Application |
|---|---|---|---|
| Recombinant Wnt Proteins | Ligand | Activate Wnt signaling pathways [11] | Treatment of stem cells to direct differentiation; study of pathway activation in cell culture. |
| IWP-2 / LGK974 | Small Molecule Inhibitor | Inhibit Porcupine (PORCN), blocking Wnt ligand secretion and all downstream signaling [11] | Determine if a phenotypic effect is Wnt-dependent; create Wnt-free conditions. |
| XAV-939 / IWR-1 | Small Molecule Inhibitor | Stabilize the Axin/APC/GSK3β destruction complex, promoting β-catenin degradation (Canonical pathway specific) [11] | Probe the specific role of canonical signaling in a biological process. |
| Anti-β-catenin Antibodies | Antibody | Detect total and active (non-phospho) β-catenin protein levels and localization [14] | Immunofluorescence (nuclear vs. cytoplasmic), Western blot analysis. |
| Anti-FZD Antibodies | Antibody | Detect Frizzled receptor expression and localization [14] | Flow cytometry, immunoprecipitation, blocking receptor function. |
| Dkk-1 | Secreted Antagonist | Binds LRP5/6 co-receptors, specifically inhibiting the canonical Wnt pathway [13] [14] | Selectively block canonical signaling without affecting non-canonical pathways. |
| TOPFlash/FOPFlash | Reporter Assay | Luciferase-based reporters for β-catenin/TCF transcriptional activity [10] | Quantify the activity of the canonical Wnt pathway in cell-based screens. |
| siRNA/shRNA Libraries | Functional Genomics | Knockdown expression of specific Wnt pathway genes [11] | Perform loss-of-function screens to identify essential pathway components. |
| KWKLFKKGIGAVLKV | KWKLFKKGIGAVLKV Cationic Antimicrobial Peptide | Research-grade cationic helical peptide "KWKLFKKGIGAVLKV" for antimicrobial mechanism studies. For Research Use Only. Not for human, veterinary, or household use. | Bench Chemicals |
| Bmeda | BMEDA (N,N-bis(2-mercaptoethyl)-N',N'-diethylenediamine) | BMEDA is a chelating agent for Rhenium-186 in liposomal nanoliposome research (e.g., 186RNL for glioma). For Research Use Only. Not for human or veterinary diagnostic or therapeutic use. | Bench Chemicals |
The Wnt/β-catenin signaling pathway constitutes an evolutionarily conserved system that plays a pivotal role in early embryonic development, tissue homeostasis, and cell fate determination. At the core of this pathway lies β-catenin, a dual-function protein that serves as both a transcriptional co-activator and a component of cell-cell adhesion complexes. This technical review comprehensively examines the molecular mechanisms governing β-catenin dynamics, from its regulation by the cytoplasmic destruction complex to its nuclear translocation and transcriptional functions. Special emphasis is placed on the pathway's critical functions during early embryogenesis, including its established roles in maintaining pluripotency, regulating cell lineage specification, and facilitating implantation processes. Additionally, this review integrates recent advances in our understanding of nuclear translocation mechanisms and discusses emerging therapeutic strategies targeting β-catenin dynamics in disease contexts, particularly cancer. The information presented herein provides researchers and drug development professionals with a detailed framework for understanding β-catenin regulation and its implications in both developmental biology and pathogenesis.
The Wnt/β-catenin pathway, also known as the canonical Wnt pathway, represents a fundamental signaling cascade that is highly conserved across metazoan species, from Drosophila to humans [9]. The name "Wnt" originates from the fusion of two terms: the Drosophila segment polarity gene "Wingless" and the murine proto-oncogene "integration 1" [9] [17]. This pathway functions as a master regulator of various physiological processes, with particularly crucial functions during early embryonic development. The mammalian pre-implantation period constitutes one of the most critical and unique phases during early embryonic developmental processes, involving a precise transition from a single-cell zygote to the blastocyst stage through a series of meticulously regulated events [18]. These processes initiate cell lineage specification and differentiation into the inner cell mass (ICM) and trophectoderm (TE), which are fundamentally regulated by the activation of several intracellular signaling cascades, with Wnt signaling playing a predominant role [18].
Wnt signaling is primarily categorized into two distinct branches: the canonical pathway (β-catenin-dependent) and non-canonical pathways (β-catenin-independent) [18] [19]. The canonical Wnt/β-catenin pathway is distinguished by its reliance on the stabilization and nuclear translocation of β-catenin, which subsequently acts as a transcriptional co-activator for T-cell factor/lymphoid enhancer factor (TCF/LEF) family transcription factors [18] [19] [20]. During early embryogenesis, spatially defined and well-controlled Wnt signaling orchestrates normal embryonic development in a process that initiates at fertilization and continues throughout organism formation [18]. The pathway plays an indispensable role in maintaining pluripotency both in vivo and in vitro in human and mouse embryonic stem cell (ESC) cultures, primarily through regulation of core pluripotency factors such as Oct4, Nanog, Sox2, and Klf4 [18] [17].
Table 1: Key Components of the Wnt/β-Catenin Signaling Pathway
| Component Category | Key Elements | Primary Functions |
|---|---|---|
| Extracellular Signals | Wnt proteins (Wnt1, Wnt3a, Wnt2, etc.) | Secreted ligands that initiate pathway activation by binding to receptors [19] [17] |
| Membrane Receptors | Frizzled (FZD), LRP5/6 | Seven-transmembrane receptors and co-receptors that transduce Wnt signals [19] [20] |
| Cytoplasmic Destruction Complex | APC, Axin, GSK-3β, CK1α | Phosphorylates β-catenin, targeting it for degradation in the absence of Wnt signaling [19] [20] [21] |
| Signal Transducers | Dvl (Dishevelled) | Recruited upon receptor activation; disrupts the destruction complex [19] [20] |
| Nuclear Components | β-catenin, TCF/LEF | Transcriptional co-activator complex that regulates target gene expression [18] [19] [20] |
Molecular Composition of the Destruction Complex
Architecture and Regulation
The β-catenin destruction complex represents a multi-protein machinery that maintains precise control over cytoplasmic β-catenin levels in the absence of Wnt signaling. This complex consists of several core components: adenomatous polyposis coli (APC), Axin, glycogen synthase kinase 3β (GSK-3β), and casein kinase 1α (CK1α) [19] [20] [21]. Under unstimulated conditions (Wnt-OFF state), this destruction complex facilitates the phosphorylation and subsequent degradation of β-catenin, thereby preventing its accumulation and nuclear translocation [19] [20].
Axin functions as a critical scaffolding protein within the destruction complex, containing binding domains that facilitate interactions with other complex components [19]. The regulator of G protein signaling (RGS) domain at the N-terminus of Axin specifically interacts with the APC protein, while the DIX domain at the C-terminus enables interaction with Dishevelled (Dvl) and also contributes to Axin oligomerization [19]. APC, a large multi-domain protein, serves as a scaffold in conjunction with Axin, promoting the sequential phosphorylation of β-catenin by the kinase components [19]. The destruction complex operates through a precisely coordinated phosphorylation cascade: CK1α initially phosphorylates β-catenin at serine 45 (Ser45), which serves as a priming phosphorylation event that enables subsequent phosphorylation by GSK-3β at threonine 41 (Thr41), serine 37 (Ser37), and serine 33 (Ser33) [19] [22].
Table 2: Phosphorylation Events in β-Catenin Regulation
| Residue | Kinase | Functional Consequence |
|---|---|---|
| Ser45 | CK1α | Priming phosphorylation that enables subsequent GSK-3β-mediated phosphorylation [19] [22] |
| Thr41, Ser37, Ser33 | GSK-3β | Creates recognition site for β-TrCP E3 ubiquitin ligase; targets β-catenin for proteasomal degradation [19] [22] |
| Ser552 | AKT | Promotes dissociation from cell-cell contacts and enhances nuclear accumulation; independent of destruction complex [9] |
This multi-site phosphorylation creates a recognition motif for the E3 ubiquitin ligase β-TrCP (SCFβ-TrCP), which subsequently ubiquitinates β-catenin, marking it for proteasomal degradation [19] [20]. This meticulous regulatory mechanism ensures that cytoplasmic β-catenin levels remain low in the absence of Wnt signaling, thereby preventing inappropriate activation of target genes.
Wnt-Mediated Disassembly Mechanism
Upon Wnt ligand binding to the Frizzled receptor and LRP5/6 co-receptor, a series of intracellular events leads to the inhibition of the destruction complex [19] [20]. The transmembrane receptor complex recruits Dishevelled (Dvl) to the plasma membrane, which becomes activated through sequential phosphorylation, poly-ubiquitination, and polymerization [20]. Activated Dvl subsequently recruits Axin and the destruction complex to the plasma membrane through interactions with the intracellular domains of Frizzled and LRP5/6 [19] [20]. The phosphorylation of LRP5/6 at multiple PPPSPxS motifs creates docking sites for Axin, effectively sequestering the scaffolding protein away from the cytoplasmic destruction complex [19]. This redistribution and membrane localization of Axin disrupts the integrity and functionality of the destruction complex, thereby preventing β-catenin phosphorylation and degradation [19] [20]. Consequently, β-catenin accumulates in the cytoplasm and subsequently translocates to the nucleus to initiate transcriptional programs essential for embryonic development.
Diagram 1: Wnt/β-Catenin Pathway Regulation - illustrating the key molecular events in the destruction complex during Wnt OFF and ON states
Cytoplasmic Stabilization and Nuclear Translocation Mechanisms
Cytoplasmic Accumulation and Transport
Following disruption of the destruction complex, β-catenin accumulates in the cytoplasm through a tightly regulated process. The stability of β-catenin is significantly enhanced as it evades phosphorylation-mediated ubiquitination and proteasomal degradation [19] [20]. This stabilized β-catenin then undergoes active transport toward the nucleus, a process recently discovered to involve specific motor proteins and adaptor complexes [22]. Recent research has revealed that the intraflagellar transport A complex (IFT-A) associates with Kinesin-2 to facilitate the nuclear translocation of β-catenin upon Wnt pathway activation [22]. IFT-A, traditionally known for its role in ciliogenesis, forms a complex with β-catenin via its N-terminal region, specifically through interaction with amino acid residues 24-79 in mammals (equivalent to Arm34-87 in Drosophila) [22]. This interaction is essential for the efficient nuclear translocation of β-catenin, as loss of function in either IFT-A components or Kinesin-2 results in impaired Wnt signaling and developmental defects despite normal cytoplasmic stabilization of β-catenin [22].
The molecular mechanism involves direct binding between IFT140 and the N-terminal region of β-catenin, which serves as a recognition site for the transport machinery [22]. Kinesin-2 interacts with IFT140 through Kap3 (kinesin-associated protein 3) and functions as the molecular motor that transports the IFT-A/β-catenin complex along cytoplasmic microtubules toward the nuclear envelope [22]. This active transport system ensures the efficient delivery of β-catenin to the nucleus, where it can execute its transcriptional functions. The critical nature of this process is demonstrated by experimental evidence showing that expression of a small N-terminal β-catenin peptide (β-catenin24-79) acts as a competitive inhibitor by binding to IFT140, thereby interfering with nuclear translocation of endogenous full-length β-catenin and attenuating Wnt signaling output [22].
Nuclear Import and Transcriptional Activation
Upon reaching the nuclear envelope, β-catenin translocates into the nucleus through the nuclear pore complex via mechanisms that involve Rac1 and other import factors [20]. Once inside the nucleus, β-catenin forms a transcriptional activation complex with T-cell factor/lymphoid enhancer factor (TCF/LEF) transcription factors [18] [19] [20]. This complex displaces transcriptional repressors, such as Groucho, that are bound to TCF/LEF in the absence of Wnt signaling [20] [17]. The formation of the β-catenin/TCF transcriptional complex recruits additional co-activators, including B-cell lymphoma 9 (BCL9), Pygopus, and CREB-binding protein (CBP)/p300, which possess histone acetyltransferase activity that modifies chromatin structure to facilitate transcription [19] [9].
The β-catenin/TCF complex activates the expression of numerous target genes that regulate fundamental processes during early embryogenesis, including cell proliferation, differentiation, and stem cell maintenance [18] [19]. Key target genes include c-Myc, Cyclin D1, Axin2, and CD44, many of which are associated with cell cycle progression and proliferation [19] [21]. Importantly, the specific transcriptional output of β-catenin is context-dependent and varies between cell types and developmental stages, influenced by the availability of co-factors and the epigenetic landscape [18] [9]. During early embryonic development, β-catenin also interacts with other transcription factors beyond TCF/LEF, including Oct4 and Yamanaka factors, to maintain pluripotency in embryonic stem cells [18]. This interaction enhances the expression of core pluripotency factors in a TCF-dependent manner, establishing a regulatory network that supports the pluripotent state in the inner cell mass of developing blastocysts [18].
Experimental Methodologies for Studying β-Catenin Dynamics
Live-Cell Imaging and Localization Assays
Advanced imaging techniques have revolutionized the study of β-catenin dynamics in living cells, enabling researchers to monitor its localization and movement in real time. The HiBiT Protein Tagging System represents a cutting-edge approach that allows for endogenous tagging of β-catenin with a small, luminescent tag [21]. When combined with LgBiT and Nano-Glo Live Cell substrate, the luminescent signal serves as a sensitive proxy for β-catenin abundance and subcellular location [21]. This methodology enables researchers to visualize β-catenin dynamics dynamically in live cells at single-cell resolution without the need for overexpression, fixation, or staining procedures that can introduce artifacts [21]. Experimental protocols typically involve:
- Endogenous Tagging: CRISPR/Cas9-mediated insertion of the HiBiT tag into the CTNNB1 gene locus, ensuring physiological expression levels and regulation [21].
- Live-Cell Imaging: Treated cells are imaged using systems such as the GloMax Galaxy Bioluminescence Imager, which captures spatial and temporal information over extended time courses [21].
- Quantitative Analysis: Image processing software quantifies the redistribution of β-catenin from cytoplasm to nucleus following pathway activation or inhibition [21].
This approach has demonstrated that in untreated cells, β-catenin remains predominantly cytoplasmic, while after treatment with GSK-3β inhibitors such as AZD2858, cells show clear nuclear accumulation of β-catenin over a five-hour time course [21]. The capability to track these dynamics in real time provides valuable insights for validating the mechanism of action of pathway modulators and identifying potential off-target effects early in the drug discovery process [21].
Reporter Assays and High-Throughput Screening
Luciferase-based reporter assays represent a cornerstone methodology for quantifying Wnt/β-catenin pathway activity and identifying regulatory compounds [21]. These assays typically utilize a β-catenin-responsive luciferase reporter construct containing multiple TCF/LEF binding sites upstream of a minimal promoter driving firefly or NanoLuc luciferase expression [21]. Standardized protocols include:
- Cell Line Development: Stable integration of the reporter construct into relevant cell lines (e.g., HEK293, HT-29, or primary stem cells) to ensure consistent expression and response [21].
- Compound Screening: High-throughput screening of chemical libraries using automated plate readers to measure Relative Luminescence Units (RLU) as a proxy for pathway activity [21].
- Validation Studies: Follow-up experiments using pathway activators (e.g., CHIR99021, Wnt3a conditioned medium) or inhibitors (e.g., iCRT compounds, XAV939) to confirm specificity [21].
In a landmark study, researchers employed a β-catenin-responsive luciferase reporter to screen nearly 15,000 compounds, leading to the discovery of the iCRT class of inhibitors that disrupt β-catenin/TCF interactions [21]. These compounds demonstrated selective toxicity toward colon cancer cells with constitutive Wnt activity and reduced tumor growth in mouse models, highlighting the therapeutic potential of targeting nuclear β-catenin function [21]. While reporter assays provide population-level data on pathway activity, they are often combined with complementary approaches such as immunofluorescence, subcellular fractionation, and quantitative PCR to obtain a comprehensive understanding of β-catenin dynamics across molecular, cellular, and functional levels.
Diagram 2: Experimental Workflow for Studying β-Catenin Dynamics - outlining key methodological approaches
Research Reagent Solutions for β-Catenin Studies
Table 3: Essential Research Reagents for Investigating β-Catenin Dynamics
| Reagent Category | Specific Examples | Research Applications | Key Features |
|---|---|---|---|
| Pathway Activators | CHIR99021, AZD2858, Wnt3a conditioned medium, 6-Bio | GSK-3β inhibition; stabilizes β-catenin by preventing phosphorylation [18] [21] [23] | Chemical inhibitors provide temporal control; recombinant proteins offer physiological activation |
| Pathway Inhibitors | iCRT3, iCRT5, iCRT14, XAV939, GSK3787 | Disrupt β-catenin/TCF interactions; tankyrase inhibition stabilizes Axin [18] [21] | Target different pathway nodes; useful for mechanism studies and therapeutic development |
| Live-Cell Imaging Tools | HiBiT Protein Tagging System, Nano-Glo Live Cell Substrate, HaloTag-β-catenin fusions | Real-time tracking of β-catenin localization and dynamics [21] | Enable endogenous tagging; minimal perturbation to native protein function |
| Reporter Systems | TCF/LEF Luciferase Reporters (BAR, TOPFlash), GFP Reporters | Quantitative measurement of pathway activity; high-throughput screening [21] | Sensitive readouts; compatible with automated screening platforms |
| Antibodies | Phospho-specific β-catenin (Ser33/37/Thr41, Ser45), Total β-catenin, Non-phospho β-catenin (Active) | Immunofluorescence, Western blot, immunohistochemistry for detection and localization [22] [23] | Distinguish activation states; validate subcellular localization |
| Genetic Tools | CRISPR/Cas9 editing constructs, siRNA/shRNA libraries, Dominant-negative peptides (β-catenin24-79) | Gene knockout, knockdown, and functional interference studies [22] | Enable mechanistic studies; assess requirement of specific pathway components |
β-Catenin in Embryonic Development and Therapeutic Implications
Roles in Early Embryogenesis
β-catenin dynamics play indispensable roles during early embryonic development, particularly in the critical stages surrounding blastocyst formation and implantation. During mammalian pre-implantation development, Wnt/β-catenin signaling is instrumental in maintaining pluripotency and regulating cell lineage specification [18]. The pathway is active from the earliest stages of development, with β-catenin detected from the 2-cell stage through to the blastocyst stage in mouse embryos [18]. The functional importance of this pathway is evidenced by experiments demonstrating that enhanced activation of Wnt signaling through GSK-3β inhibition (using compounds such as 6-Bio or CHIR99021) increases the inner cell mass (ICM) cell proliferation index and leads to improved quality and yield of bovine blastocysts [18] [23].
The trophectoderm (TE), which forms the outer cell layer of the blastocyst, exhibits particularly important dependence on proper β-catenin regulation [23]. Successful development of mammalian embryos relies on appropriate TE formation and function, as this tissue maintains blastocyst structure, forms the placenta to enable nutrient exchange between mother and embryo, and facilitates implantation [23]. Recent research has demonstrated that β-catenin accumulation in TE cells promotes their migratory and invasive capacities, which are essential for successful implantation [18] [23]. This function is mediated through the upregulation of key target genes such as c-Myc and PPARδ (peroxisome proliferator-activated receptor delta) [18]. The interplay between β-catenin and PPARδ establishes a regulatory network that coordinates cell proliferation and invasive capabilities during early development, with PPARδ strongly coupling with c-Myc expression in both ICM and trophoblast stem cells under elevated Wnt conditions [18].
Dysregulation in Disease and Therapeutic Targeting
Dysregulation of β-catenin dynamics contributes significantly to various human diseases, most notably cancer [19] [21] [17]. Aberrant activation of the Wnt/β-catenin pathway occurs through multiple mechanisms, including mutations in key pathway components such as APC, AXIN, CTNNB1 (encoding β-catenin), and other elements [19] [21]. In colorectal cancer, approximately 90% of cases involve mutations that disrupt normal β-catenin regulation, most commonly through loss-of-function mutations in APC or stabilizing mutations in CTNNB1 that prevent β-catenin phosphorylation and degradation [21]. These alterations lead to constitutive β-catenin signaling that drives uncontrolled proliferation and tumor progression [21] [24].
The clinical importance of targeting β-catenin dynamics is demonstrated by ongoing efforts to develop therapeutic interventions [19] [21] [17]. Several classes of inhibitors have been identified, including:
- Small molecule inhibitors that disrupt β-catenin/TCF interactions (iCRT compounds) [21]
- Tankyrase inhibitors (XAV939) that stabilize Axin and promote β-catenin degradation [19]
- Porcupine inhibitors that prevent Wnt secretion and ligand-mediated pathway activation [19]
- Antisense oligonucleotides that target pathway components [17]
- Peptide inhibitors that interfere with β-catenin nuclear translocation (β-catenin24-79) [22]
The β-catenin24-79 peptide represents a particularly promising therapeutic approach, as it acts as a dominant-negative inhibitor by competitively binding to IFT140 and preventing nuclear translocation of full-length β-catenin [22]. This mechanism effectively attenuates Wnt/β-catenin signaling output even in contexts with stabilized β-catenin, such as those caused by APC or CTNNB1 mutations [22]. While most of these therapeutic strategies are still in preclinical development, they offer considerable promise for targeting Wnt-driven cancers and other diseases characterized by aberrant β-catenin signaling.
The intricate regulation of β-catenin dynamics, from its controlled degradation by the cytoplasmic destruction complex to its regulated nuclear translocation and transcriptional functions, represents a cornerstone of embryonic development and tissue homeostasis. The molecular mechanisms governing these processes continue to be elucidated through advanced experimental approaches that enable real-time visualization and precise manipulation of pathway components. The essential functions of β-catenin during early embryogenesis—particularly in pluripotency maintenance, cell lineage specification, and implantation processes—highlight its fundamental importance in developmental biology. Furthermore, the frequent dysregulation of β-catenin dynamics in human diseases, especially cancer, underscores the therapeutic potential of targeting this pathway. Ongoing research continues to refine our understanding of β-catenin regulation and to develop innovative strategies for modulating its activity in pathological contexts, offering promising avenues for future therapeutic interventions.
TCF/LEF Transcription Factors as Gatekeepers of Gene Expression
Within the canonical Wnt signaling pathway, T-cell factor/lymphoid enhancer factor (TCF/LEF) proteins function as the ultimate nuclear effectors, translating β-catenin signals into precise transcriptional programs that govern early embryogenesis. This whitepaper delineates the sophisticated molecular architecture of TCF/LEF transcription factors, their context-dependent regulation through alternative splicing and post-translational modifications, and their non-redundant functions in developmental processes such as nephron formation and axis patterning. We further present quantitative data on their DNA-binding properties, detailed experimental protocols for assessing Wnt pathway activity at single-cell resolution, and emerging therapeutic strategies targeting TCF/LEF regulatory kinases for cancer and fibrotic diseases. The integral role of these factors as binary molecular switches—mediating either repression or activation of Wnt target genes—establishes them as critical gatekeepers of gene expression during embryonic development and in stem cell niches.
The Wnt signaling pathway is a fundamental molecular pathway governing cell fate decisions, tissue patterning, and stem cell maintenance during embryonic development [25] [5]. At the core of the canonical Wnt/β-catenin pathway are TCF/LEF transcription factors, which serve as the major endpoint mediators, translating the influx of β-catenin signals into discrete gene expression programs [26] [27]. The discovery of TCF/LEF genes as nuclear Wnt pathway components in the 1990s was a pivotal breakthrough that plugged a critical knowledge gap in understanding how nuclear β-catenin could regulate target genes despite lacking a DNA-binding domain [28]. The subsequent establishment of a model wherein Wnt-regulated β-catenin partners with DNA-binding TCF/LEF proteins on specific genomic sequences, known as Wnt Response Elements (WREs), provided the mechanistic link between extracellular signals and transcriptional outcomes [28].
TCF/LEF factors function as bimodal transcriptional switches that actively repress target genes in the absence of Wnt signaling and activate them when Wnt signaling is active [28]. This review examines the structure-function relationships of TCF/LEF proteins, their complex regulation, and their indispensable roles in early embryogenesis, with particular emphasis on their function as molecular gatekeepers of gene expression. We also explore experimental approaches for investigating TCF/LEF activity and the therapeutic potential of modulating their function in disease contexts.
Molecular Architecture of TCF/LEF Proteins
Domain Organization and Functional Motifs
TCF/LEF proteins possess a modular architecture consisting of highly conserved domains that enable their function as context-dependent transcription factors. Vertebrates have four TCF/LEF family members (TCF7, LEF1, TCF7L1, and TCF7L2) that arose through gene duplication, allowing for functional specialization beyond the single TCF/LEF ortholog found in invertebrates [26] [28].
Table 1: Functional Domains of TCF/LEF Transcription Factors
| Domain | Function | Conservation |
|---|---|---|
| N-terminal β-catenin binding domain | Recruits β-catenin for transcriptional activation; contains conserved motifs essential for the interaction | High across all vertebrate TCF/LEF members |
| Control region | Mediates transcriptional repression; contains binding sites for Groucho/TLE corepressors | Variable due to alternative splicing |
| DNA-binding HMG domain | Sequence-specific DNA binding to 5′-SCTTTGATS-3′ consensus; induces DNA bending | Extremely high; single amino acid changes can disrupt binding |
| C-clamp (CRARF domain) | Secondary DNA-binding domain recognizing GC-rich helper sites; not present in all isoforms | Limited to specific isoforms; absent in some family members |
The DNA-binding domain comprises a high-mobility group (HMG) box and a small peptide motif of basic residues that together recognize a specific DNA sequence (5′-SCTTTGATS-3′) with nanomolar affinity [26]. This HMG domain not only confers sequence specificity but also enforces a sharp bend in the DNA helix between 90° and 127°, likely facilitating the assembly of multi-protein complexes at regulatory elements [26].
The C-clamp domain, located carboxy-terminal to the HMG domain, is enriched in basic, cysteine, and aromatic residues and serves as a secondary DNA-binding domain with specificity for GC-rich "helper" sites [26]. This domain is not present in all TCF/LEF family members, with its inclusion or exclusion contributing to functional diversification and target gene specificity [26].
Structural Basis for Bimodal Transcriptional Regulation
TCF/LEF proteins function as binary switches that determine whether Wnt target genes are activated or repressed. This bimodal functionality is encoded within their structural domains:
Repression State: In the absence of Wnt signaling, TCF/LEF proteins interact with transcriptional corepressors from the Groucho/transducin-like enhancer of split (Gro/TLE) family via their control region, actively suppressing target gene expression [26] [28]. Additional corepressors include myeloid translocation gene-related 1 (Mtgr1) and corepressor of Pan (Coop) [26].
Activation State: Upon Wnt pathway activation, stabilized β-catenin translocates to the nucleus and binds to the N-terminal domain of TCF/LEF, displacing corepressors and recruiting coactivators to initiate transcription [26] [28]. The C-terminal transactivation domains of β-catenin then interact with additional transcriptional machinery to drive gene expression.
Diagram: Bimodal transcriptional regulation by TCF/LEF proteins. In the absence of Wnt signaling (OFF state), TCF/LEF associates with corepressors to silence target genes. When Wnt signaling is active (ON state), β-catenin translocates to the nucleus, binds TCF/LEF, displaces corepressors, and recruits coactivators to initiate transcription.
TCF/LEF in Early Embryogenesis
Roles in Nephron Development and Axis Patterning
During embryonic development, TCF/LEF transcription factors play indispensable roles in multiple morphogenetic processes. In kidney development, nephron formation depends critically on Wnt signaling, with two specific ligands—Wnt9b and Wnt4—required for nephron differentiation [25]. The canonical Wnt/β-catenin pathway acts downstream of these ligands in metanephric mesenchymal progenitor cells, where forced activation triggers progression toward proto-epithelial aggregates, while selective antagonism inhibits differentiation [25]. Titration of pathway activity appears central for proper coordination of differentiation and morphogenesis, with transient activation of the pathway during epithelial differentiation [25].
Beyond organogenesis, TCF/LEF factors are essential for fundamental patterning processes. They contribute to anterior-posterior patterning of the developing central nervous system, neural crest development, and the initial induction of the dorsal body axis [28]. The vertebrate embryo utilizes different TCF/LEF family members to achieve spatial and temporal specificity in these diverse developmental contexts, with partial functional redundancy but also unique, non-overlapping functions [26] [28].
Isoform Diversity and Context-Dependent Functions
Functional diversification of TCF/LEF proteins occurs through several mechanisms:
Gene-specific specializations: The four mammalian TCF/LEF paralogs have evolved distinct expression patterns and functions, with genetic evidence indicating only partial redundancy [26] [28].
Alternative splicing: Extensive alternative splicing generates isoforms with different domain compositions, particularly in TCF7 and TCF7L2 genes. These include isoforms that lack the β-catenin binding domain (functioning as constitutive repressors) and isoforms with varying C-terminal domains that influence DNA-binding specificity and cofactor interactions [26] [28].
Post-translational modifications: Phosphorylation by kinases such as TNIK (TRAF2 and NCK-interacting kinase) and HIPK2 (homeodomain-interacting protein kinase 2) dynamically regulates DNA-binding affinity and co-factor interactions [27]. The ubiquitin-proteasome system also contributes to regulation, particularly through UBR5-mediated clearance of Groucho/TLE corepressors [27].
Table 2: TCF/LEF Family Members in Vertebrates
| Gene Name | Common Aliases | Key Functions in Development | Notable Isoforms |
|---|---|---|---|
| TCF7 | TCF1 | T-cell development, Wnt pathway repression | Dominant-negative isoforms lacking β-catenin binding domain |
| LEF1 | TCF1α | Hair follicle development, neural crest specification | Multiple isoforms with varying transactivation potential |
| TCF7L1 | TCF3 | Pluripotency maintenance, neural patterning | Often functions as a transcriptional repressor |
| TCF7L2 | TCF4 | Central nervous system development, energy metabolism | Numerous splice variants with different C-terminal |
Experimental Analysis of TCF/LEF Function
Single-Cell RNA and DNA Sequencing for Pathway Activity Assessment
Advanced methodologies now enable simultaneous analysis of pathway activity and genetic mutations at single-cell resolution. A microfluidic-based approach allows differential extraction of RNA and DNA from individual cells through a two-stage lysis protocol [29]:
Protocol: Single-Cell RNA and DNA Extraction for Wnt Pathway Analysis
Cell Capture and Lysis:
- Capture individual calcein-stained cells in picoliter-volume traps within a microfluidic chip.
- Apply first lysis buffer (0.5× TBE with 0.5% Triton X-100) to lyse plasma membranes while keeping nuclear membranes intact.
- Collect cytoplasmic contents (including RNA) from separate outlets for off-chip cDNA synthesis and amplification.
- Apply second lysis buffer (0.5× TBE with 0.5% Triton X-100 and Proteinase K) to lyse nuclear membranes and release DNA.
- Perform on-chip whole genome amplification (WGA) at 30°C followed by heat inactivation at 60°C.
Pathway Activity Modeling:
- Construct a Bayesian network model representing the Wnt transcriptional program with three node types: transcription complex (TC), Wnt target genes, and expression level measurements.
- Calibrate the model using RNA-seq data from samples with known Wnt activity status (e.g., LS180 and SW1222 as Wnt-active; RKO as Wnt-inactive).
- Apply the calibrated model to single-cell RNA-seq data by entering measured expression values and using Bayesian reasoning to infer the probability of transcription complex activity.
Validation and Application:
- Verify model performance using control cell lines with established Wnt pathway status.
- Analyze single cells from experimental conditions to infer pathway activity states despite intercellular heterogeneity.
This approach successfully classified all seven analyzed single LS174T cells as Wnt-active and six RKO cells as Wnt-inactive, demonstrating robust pathway activity inference at single-cell resolution [29].
Diagram: Experimental workflow for single-cell analysis of Wnt pathway activity and genotype. Cells are captured in a microfluidic device, followed by sequential lysis to separate cytoplasmic RNA and nuclear DNA. Sequencing and Bayesian modeling enable correlation of pathway activity with mutational status.
Research Reagent Solutions
Table 3: Essential Research Reagents for TCF/LEF and Wnt Pathway Studies
| Reagent/Category | Specific Examples | Function/Application |
|---|---|---|
| TCF/LEF Antibodies | Anti-Prep1, Anti-Pbx1 | Chromatin immunoprecipitation (ChIP); protein localization |
| Wnt Pathway Modulators | CHIR (Wnt agonist), IWP-2 (Wnt antagonist) | Titration of pathway activity in progenitor cells |
| Microfluidic Systems | Custom picoliter-volume trap chips | Single-cell capture and processing |
| Lysis Buffers | Triton X-100 based buffers with Proteinase K | Differential extraction of RNA and DNA |
| Bayesian Model Components | 34 Wnt target gene panel | Inference of pathway activity from expression data |
Therapeutic Targeting and Future Perspectives
TCF/LEF Regulation as a Therapeutic Strategy
Direct targeting of TCF/LEF transcription factors has emerged as a promising therapeutic approach for Wnt-driven pathologies. Challenges in directly targeting the intrinsically disordered β-catenin binding domains have prompted innovative strategies:
Kinase inhibition: Targeting upstream regulatory kinases such as TNIK (TRAF2 and NCK-interacting kinase) offers an indirect approach to modulate TCF/LEF activity. The TNIK inhibitor INS018_055 has successfully completed Phase II clinical trials for idiopathic pulmonary fibrosis, demonstrating statistically significant attenuation of lung function decline over 52 weeks [27].
Advanced modalities: Proteolysis targeting chimeras (PROTACs) and AI-designed protein scaffolds show promise for precise interaction with TCF/LEF proteins, potentially overcoming the limitations of conventional small-molecule approaches [27].
This therapeutic strategy represents a significant advance over broad Wnt pathway inhibition, which often carries substantial toxicity due to the pathway's fundamental roles in tissue homeostasis [27]. By targeting downstream regulatory nodes, these approaches aim to fine-tune rather than globally inhibit Wnt signaling, preserving physiological functions while correcting pathological activation.
Future Research Directions
The field continues to evolve with several emerging research priorities:
Enhanceosome dynamics: Recent evidence indicates that the Wnt enhanceosome is pre-assembled in a poised state before signal activation, enabling rapid transcriptional responses [27]. Understanding the composition and regulation of these enhanceosomes will provide deeper insights into context-specific Wnt responses.
Chromatin interactions: The interplay between TCF/LEF proteins and chromatin remodeling complexes creates a dynamic regulatory landscape that influences target gene accessibility [27]. Mapping these interactions in different developmental contexts remains a active area of investigation.
Synthetic biology approaches: Advances in computational protein design and synthetic biology may enable engineering of bespoke TCF/LEF modulators with unprecedented specificity, potentially heralding a new era of personalized molecular therapies [27].
TCF/LEF transcription factors serve as essential gatekeepers of gene expression in the Wnt signaling pathway, integrating multiple regulatory inputs to determine transcriptional outcomes during embryonic development. Their sophisticated domain architecture, isoform diversity, and context-dependent regulation enable precise control of fundamental processes including nephron formation, axis patterning, and stem cell maintenance. Experimental approaches such as single-cell RNA/DNA sequencing coupled with Bayesian modeling provide powerful tools to dissect their functions in heterogeneous cellular populations. Emerging therapeutic strategies that target TCF/LEF regulatory networks offer promising avenues for treating Wnt-driven diseases while minimizing toxicity. As our understanding of these transcriptional gatekeepers continues to deepen, so too will our ability to harness their regulatory potential for both basic research and clinical applications.
Wnt's Role in Cell Fate Specification and Pluripotency Maintenance
The Wnt signaling pathway is a complex, evolutionarily conserved network that functions as a master regulator of embryonic development, stem cell maintenance, and tissue homeostasis [5] [30]. This pathway coordinates critical cellular processes including proliferation, migration, cell fate specification, and the maintenance of pluripotency [31]. The name "Wnt" derives from the integration of two Drosophila phenotypes: wingless and int [5]. In mammalian systems, the pathway has generally been divided into the canonical (β-catenin-dependent) and non-canonical (β-catenin-independent) branches, with the canonical pathway being particularly implicated in cell fate decisions [5] [30]. Within early embryogenesis, WNT signaling activity is involved in the regulation of many cellular functions, including the maintenance of pluripotency and the induction of differentiation toward specific tissue lineages [31]. This whitepaper examines the molecular mechanisms through which Wnt signaling governs cell fate specification and pluripotency maintenance, with particular emphasis on implications for therapeutic development.
Molecular Mechanisms of Canonical Wnt Signaling
Core Pathway Components
The canonical Wnt/β-catenin pathway initiates when Wnt ligands bind to Frizzled receptors and their co-receptors, Low-Density Lipoprotein Receptor-Related Protein 5/6 (LRP5/6) on the cell surface [32]. This interaction triggers a intracellular signaling cascade that prevents the phosphorylation and degradation of β-catenin [30]. In the absence of Wnt signaling, cytoplasmic β-catenin is phosphorylated by a destruction complex consisting of glycogen synthase kinase 3β (GSK3β), casein kinase I (CK I), Axin, and adenomatous polyposis coli (APC) [33] [30]. This phosphorylation event targets β-catenin for ubiquitin-mediated proteasomal degradation [30]. When Wnt ligands activate the pathway, they disrupt this destruction complex through the recruitment of cytosolic Disheveled (Dvl) protein, allowing β-catenin to accumulate in the cytoplasm and subsequently translocate to the nucleus [30]. Within the nucleus, β-catenin partners with T-cell factor/lymphoid enhancer-binding factor (TCF/LEF) transcription factors to activate the expression of target genes governing cell fate decisions [32] [30].
Pathway Regulation and Feedback Loops
The Wnt signaling pathway is carefully controlled by multiple regulatory mechanisms, including feedback loops involving pathway agonists and antagonists [31]. Key negative regulators include RNF43 and ZNRF3, which are E3 ubiquitin ligases that promote Wnt receptor endocytosis and degradation, thereby dampening pathway activity [33]. Additionally, the Wnt target gene Axin2 functions as a negative feedback regulator by enhancing β-catenin degradation [33]. The availability of intracellular pools of active β-catenin and cross-regulation by β-catenin-independent pathways further fine-tune signaling output [31]. Recent research has revealed that the distribution of Wnt ligands occurs through various mechanisms, including free diffusion, restricted diffusion, active transport, and via filopodia extending to adjacent cells [5].
Wnt Signaling Pathway Diagram
Canonical Wnt/β-catenin Signaling Pathway
Wnt Signaling in Pluripotency States
Naive versus Primed Pluripotency
The role of Wnt signaling in pluripotent stem cells demonstrates remarkable context-dependency, particularly when comparing "naive" and "primed" states of pluripotency [34]. Mouse embryonic stem cells (mESCs) represent a naive pluripotent state similar to the pre-implantation inner cell mass, and in these cells, Wnt/β-catenin signaling supports self-renewal and prevents differentiation [34]. In contrast, human ESCs (hESCs) and mouse epiblast stem cells (EpiSCs) exist in a primed pluripotent state resembling the post-implantation epiblast, where Wnt signaling promotes differentiation toward definitive endoderm and mesoderm lineages [34]. This fundamental difference explains the seemingly contradictory effects of Wnt activation observed in different stem cell systems and underscores the importance of cellular context in determining Wnt functional outcomes.
Table 1: Characteristics of Pluripotent Stem Cell States
| Characteristic | Mouse ESC (Naive) | Human ESC (Primed) | Mouse EpiSC (Primed) |
|---|---|---|---|
| Representative State | Pre-implantation | Post-implantation | Post-implantation |
| Culture Conditions for Self-renewal | Serum + LIF; Wnt3a/GSK3i | Fgf2 + Activin A | Fgf2 + Activin A |
| Effect of Wnt Activation | Self-renewal | Differentiation | Differentiation |
| Key Pluripotency Factors | High Klf4, Rex1, Stella | Low Klf4, Rex1, Stella | Low Klf4, Rex1, Stella |
| Developmental Potential | Broad (including germline) | Limited | Limited |
Molecular Mechanisms of Pluripotency Regulation
In naive mESCs, Wnt/β-catenin signaling stabilizes the pluripotency network through interactions with core transcription factors including Oct4, Sox2, and Nanog [34]. β-catenin can enhance Oct-4 activity and reinforce pluripotency through a TCF-independent mechanism [35]. Additionally, Wnt signaling inhibits differentiation toward the epiblast stem cell state, thereby maintaining the naive pluripotent condition [34]. The transition from naive to primed pluripotency involves significant changes in gene expression profiles, including downregulation of naive markers such as Klf4, Rex1, and Stella, and upregulation of primed markers including Sox17, Eomes, and Fgf5 [34]. These molecular rearrangements fundamentally alter how cells interpret Wnt signals, shifting the pathway's function from maintaining pluripotency to driving lineage specification.
Wnt Signaling in Cell Fate Specification
Early Embryonic Lineage Decisions
During early embryogenesis, Wnt signaling plays pivotal roles in tissue lineage differentiation [31]. In mouse embryos, Wnt/β-catenin signaling is absolutely required for the generation of mesendoderm lineage, which gives rise to cardiovascular tissues and other mesodermal and endodermal derivatives [35]. However, this pathway is dispensable for neuroectoderm differentiation, indicating its specific function in mesendodermal fate specification [35]. Studies in gastrulating mouse embryos have revealed that p120-catenin regulates WNT signaling and epithelial-mesenchymal transition (EMT), with mutants showing expanded expression of the primitive streak marker Brachyury (T) and defects in mesoderm migration [5]. These findings highlight Wnt's crucial role in orchestrating the complex morphogenetic events during early development.
Tissue-Specific Differentiation
Beyond early lineage decisions, Wnt signaling continues to influence cell fate specification across various tissue contexts. In the gastrointestinal system, Wnt maintains the pool of undifferentiated intestinal progenitor cells and controls the maturation and correct positioning of specialized cell types such as Paneth cells [5]. In osteogenesis, Wnt/β-catenin signaling promotes the differentiation of mesenchymal stem cells (MSCs) into osteoblasts while simultaneously suppressing their differentiation into adipocytes or chondrocytes [32]. This pathway also regulates dopaminergic differentiation in neural crest-derived mesenchymal stem cells, with Wnt activation directing cells toward a dopaminergic fate [5]. The diversity of these outcomes demonstrates how Wnt signaling is integrated with tissue-specific factors to generate appropriate cellular responses in different contexts.
Table 2: Wnt Signaling in Cell Fate Decisions
| Biological Context | Wnt Role | Key Targets/Effectors | Experimental Evidence |
|---|---|---|---|
| Cardiomyocyte Differentiation | Required | Mesendoderm specification | β-catenin knockout hPSCs fail to generate cardiomyocytes [35] |
| Neuroectoderm Differentiation | Dispensable | Not required | β-catenin knockout hPSCs differentiate normally to neuroectoderm [35] |
| Osteoblast Differentiation | Promotes | Runx2, OPG/RANKL ratio | LG-HMF, cerium oxide nanoparticles stimulate Wnt-induced osteogenesis [32] |
| Dopaminergic Neuron Differentiation | Promotes | Nurr-1, MAP2 | Wnt agonist CHIR enhances dopaminergic markers in TM-MSCs [5] |
| Intestinal Progenitor Maintenance | Sustains | Proliferation genes | Prevents differentiation; controls Paneth cell positioning [5] |
Experimental Approaches and Research Tools
Key Methodologies in Wnt Research
The investigation of Wnt signaling in pluripotency and cell fate specification employs diverse experimental approaches. CRISPR-Cas9 genome editing has enabled the generation of β-catenin knockout human pluripotent stem cells, demonstrating extremely high knockout efficiency (up to 25%) and providing definitive evidence for Wnt pathway requirements in specific differentiation processes [35]. Small molecule agonists and antagonists offer precise temporal control of Wnt signaling, allowing researchers to define critical windows for pathway activity during differentiation protocols. The synchronization system called Retention Using Selective Hook (RUSH) has been utilized to study Wnt trafficking from endoplasmic reticulum to Golgi, plasma membrane, and filopodia in real time, revealing that Wnt-containing vesicles are associated with filopodia extending to adjacent cells [5]. Additionally, sophisticated in vitro culture systems have been developed to model the effects of Wnt signaling on early embryonic development, such as porcine embryo cultures that demonstrate IGF-1 promotion of trophectoderm cell proliferation through Wnt/β-catenin pathway activation [23].
Research Reagent Solutions
Table 3: Essential Research Reagents for Wnt Signaling Studies
| Reagent/Category | Specific Examples | Function/Application | Experimental Notes |
|---|---|---|---|
| Wnt Agonists | CHIR99021, Wnt3a, LG-HMF, 6% Sr-MSNs | Activates canonical Wnt signaling by inhibiting GSK-3β or providing Wnt ligands | CHIR99021 (typically 3μM) used to direct hPSCs toward mesendoderm lineages [5] [35] |
| Wnt Antagonists | IWP-2, XAV939, Dkk-1 | Inhibits Wnt signaling by targeting Porcupine or stabilizing destruction complex | IWP-2 (typically 3μM) used to confirm Wnt-dependent effects [5] |
| Pathway Reporters | TCF/LEF-GFP, BAT-GAL, Axin2-luciferase | Visualize and quantify Wnt pathway activity in live or fixed cells | Axin2 serves as both target and feedback regulator [33] |
| Genome Editing Tools | CRISPR-Cas9, sgRNA systems | Create knockout models of Wnt pathway components | Achieved 25% β-catenin knockout efficiency in hPSCs [35] |
| Signaling Pathway Cross-talk Inhibitors | Picropodophyllin (IGF-1R inhibitor) | Investigate interaction between Wnt and other pathways | Rescued by Wnt activator CHIR99021 in porcine embryos [23] |
Experimental Workflow Diagram
Experimental Approach for Wnt Studies
Therapeutic Implications and Future Directions
The pivotal role of Wnt signaling in cell fate specification and pluripotency maintenance has significant implications for regenerative medicine and disease treatment. In osteoporosis research, both natural and pharmacological modulators of the canonical Wnt pathway have demonstrated potential to stimulate osteogenesis and inhibit osteoclastogenesis [32]. Conversely, in colorectal cancer, where hyperactive Wnt signaling drives tumor progression, inhibitors targeting various pathway components are under development as therapeutic agents [30]. The dual role of Wnt pathway modulators—with stimulators enhancing tissue formation and inhibitors potentially counteracting pathological overactivation—highlights the pathway's potential for guiding targeted therapies [32]. Future research directions include developing more precise temporal and spatial control of Wnt signaling, understanding pathway crosstalk with other signaling networks, and identifying genetic markers for personalized treatment approaches across various diseases.
Wnt signaling represents a master regulatory pathway that exerts profound influence over cell fate decisions and pluripotency states in context-dependent manners. Through the canonical β-catenin-mediated pathway and various non-canonical branches, Wnt signaling integrates extracellular cues to direct transcriptional programs that determine whether cells maintain pluripotency or commit to specific lineages. The experimental approaches outlined herein, including CRISPR-Cas9 genome editing, small molecule modulation, and advanced imaging techniques, continue to refine our understanding of this complex signaling network. As research advances, the therapeutic targeting of Wnt signaling holds promise for addressing diverse conditions ranging from degenerative diseases to cancer, ultimately translating fundamental insights in early embryogenesis into clinical applications.
Research Models and Techniques for Studying Embryonic Wnt Signaling
The Wnt signaling pathway is an evolutionarily conserved cell-to-cell coordination mechanism that is highly critical for a variety of physiological processes including stem cell regeneration, proliferation, division, migration, and cell fate determination [36]. In mammalian embryonic development, Wnt activation is indispensable for establishing the primitive streak, the embryonic structure through which cells migrate to form mesoderm and endoderm during gastrulation [37]. This fundamental developmental process can be recapitulated in vitro using human pluripotent stem cells (hPSCs), including both embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs), when differentiated as three-dimensional aggregates known as embryoid bodies (EBs) [37] [38].
Research has demonstrated that EBs display an unexpected degree of self-organization, forming anteroposterior polarity and primitive streak-like regions dependent on local activation of the Wnt pathway [37] [38]. This in vitro model system provides a powerful platform for studying early human embryogenesis, particularly aspects that are not ethically accessible for in vivo investigation [39]. The ability to visualize and manipulate Wnt signaling in EBs has revealed remarkable similarities to in vivo gastrulation processes, offering unprecedented opportunities for developmental biology research, disease modeling, and regenerative medicine applications [37] [40].
Core Concepts: Embryoid Bodies as Models of Early Development
Self-Organization and Polarization in EBs
Embryoid bodies, when left to differentiate without external guidance, were traditionally thought to form tissues in a disorganized fashion [37] [39]. However, seminal research using Wnt reporter cell lines has demonstrated that EBs undergo spontaneous polarization through self-reinforcing Wnt signaling mechanisms [37]. Using Axin2LacZ/+ and 7xTCF-eGFP Wnt reporter systems, researchers observed that after 3 days of differentiation, Wnt pathway activation becomes visible as one or two highly localized spots on each EB that gradually expand over time [37].
This polarized Wnt activation establishes anteroposterior polarity and creates a primitive streak-like region where cells undergo epithelial-to-mesenchymal transition (EMT) and differentiate into mesendodermal progenitors [37] [38]. The dependence of this process on endogenous Wnt signaling was confirmed through inhibition experiments using extracellular Wnt antagonists such as Dkk1 and Fz8CRD, which delayed or prevented reporter activation altogether [37]. This demonstrates that EBs possess an inherent capacity to recapitulate the initial stages of body axis formation, providing a valuable model for studying this fundamental developmental process.
Wnt-Mediated Germ Layer Specification
The localized activation of Wnt signaling in EBs directly controls germ layer specification by promoting mesendodermal differentiation while suppressing neuroectodermal fates [37]. When GFP-positive cells from 7xTCF-eGFP embryoid bodies were separated from GFP-negative cells after 4 days of differentiation, molecular analysis revealed striking differences in gene expression profiles:
- Mesendodermal markers including Brachyury, FoxA2, Mixl1, and Flk1 were enriched 2- to 10-fold in Wnt-active (GFP-positive) cells [37]
- Neurectodermal markers Pax6 and Sox1 were repressed in the Wnt-active population [37]
- The anterior marker Otx2 showed little change but was excluded from Wnt-active regions [37]
Experimental manipulation of Wnt signaling consistently alters germ layer specification in EBs. The addition of exogenous Wnt3a protein "posteriorizes" embryoid bodies, resulting in predominantly mesendodermal differentiation, while inhibiting Wnt signaling promotes anterior character and neurectodermal differentiation [37]. This establishes Wnt signaling as a master regulator of germ layer patterning in both embryonic development and in vitro EB models.
Technical Approaches: Controlling and Monitoring Wnt Signaling
Research Reagent Solutions for Wnt Pathway Manipulation
Table 1: Essential Research Reagents for Wnt Pathway Studies in hPSCs and EBs
| Reagent Category | Specific Examples | Function/Application | Key Findings Using Reagent |
|---|---|---|---|
| Wnt Reporters | Axin2LacZ/+, 7xTCF-eGFP, TCF-GFP | Visualize and monitor Wnt pathway activity in live cells | Revealed heterogeneous Wnt activity in hESC cultures and polarization in EBs [37] [41] |
| Wnt Agonists | Wnt3a protein, CHIR99021 (GSK3β inhibitor) | Activate Wnt/β-catenin signaling pathway | Posteriorizes EBs, promotes mesendodermal differentiation [37] [39] |
| Wnt Antagonists | Dkk1 protein, Fz8CRD, IWP2 | Inhibit Wnt signaling at extracellular or secretion level | Promotes anterior character, neurectodermal differentiation [37] [41] |
| Cell Culture Additives | Knockout Serum Replacement (KOSR), Lysophosphatidic acid (LPA) | Modulate Wnt signaling effects in culture media | KOSR blocks Wnt-mediated differentiation in defined media [41] |
Quantitative Data on Wnt Manipulation in Differentiation
Table 2: Quantitative Effects of Wnt Pathway Manipulation on Differentiation Outcomes
| Experimental Condition | Target Markers | Fold Change vs Control | Differentiation Outcome |
|---|---|---|---|
| Wnt3a Treatment | Brachyury, FoxA2, Flk1 | Accelerated and increased induction | Predominantly mesendodermal [37] |
| Wnt3a Treatment | Otx2, Pax6, Sox1 | Repressed | Suppressed neuroectodermal [37] |
| Dkk1 Treatment | Brachyury, FoxA2, Flk1 | Delayed induction | Impaired mesendodermal [37] |
| Dkk1 Treatment | Otx2, Pax6, Sox1 | Promoted expression | Enhanced neuroectodermal [37] |
| Wnthigh hESCs | Brachyury, Mixl1, Goosecoid | >10x higher | Primitive streak/mesoderm [41] |
| Wnthigh hESCs | CXCR4, Sox17 | 4x higher | Endodermal propensity [41] |
| Wntlow hESCs | Pax6 | Higher expression | Neuroectodermal propensity [41] |
Monitoring and Visualization Techniques
The establishment of reliable Wnt reporter systems has been fundamental to advancing our understanding of Wnt dynamics in EBs. Both transiently transfected and stably integrated reporters have been utilized, with the latter proving more effective for detecting endogenous Wnt pathway activity in hPSCs [41]. These tools have revealed that hPSC cultures are heterogeneous with respect to Wnt signaling, containing mixtures of cells with different levels of pathway activity that correlate with distinct differentiation propensities [41].
Immunostaining for Wnt target genes and lineage markers provides spatial resolution of patterning within EBs. For example, Brachyury and FoxA2 are consistently expressed in Wnt-responsive domains, while Otx2 is expressed throughout the EB except in regions with active Wnt signaling [37]. Flow cytometry enables quantification and isolation of subpopulations based on Wnt activity, allowing researchers to directly compare the molecular and functional characteristics of Wnthigh and Wntlow cells [41]. Gene expression profiling through real-time PCR and RNA sequencing further elucidates the transcriptional networks downstream of Wnt activation during EB differentiation [37] [40].
Key Experimental Protocols
Micropatterned Differentiation for Quantitative Propensity Screening
Micropatterned differentiation provides a standardized platform for quantitatively assessing the differentiation propensity of hPSC lines by confining cells to defined circular geometries [42]. This approach allows for high-throughput screening of lineage specification efficiency and has been validated for robust evaluation of multiple hPSC lines [42].
Protocol Overview:
- Plate hPSCs on micropatterned surfaces with defined circular colonies
- Induce differentiation using BMP4 for peri-gastrulation-like patterning or specific factors for definitive endoderm specification
- After 48 hours, fix cells and perform immunostaining for key markers (SOX2, T for BMP4 induction; SOX17, FOXA2 for endoderm)
- Quantify differentiation efficiency by measuring the proportion of cells expressing lineage markers
- Classify cell lines based on their differentiation propensity
Application: This protocol identified the VUB04 hESC line as having poor definitive endoderm differentiation efficiency due to endogenous suppression of WNT signaling, demonstrating how this method can reveal line-specific differentiation biases [42].
Bulk Cell Density Modulation for Lineage Specification
Modulating the bulk cell density (BCD), defined as cell number per culture volume, provides a straightforward method to control anteroposterior patterning of primitive streak-like priming in hPSCs [39]. This approach takes advantage of paracrine signaling gradients that form at different cell densities.
Protocol Overview:
- Initiate differentiation with equal cell numbers per well
- Modulate BCD by adjusting culture medium volume during the first 24 hours of CHIR99021 treatment
- Use BCD in conjunction with specific CHIR99021 concentrations to direct lineage specification:
- Low BCD (0.33 × 10^6 cells/ml) with 7.5 μM CHIR: precardiac mesoderm
- Intermediate BCD (0.5 × 10^6 cells/ml) with 10-12.5 μM CHIR: mixed outcomes
- High BCD (1 × 10^6 cells/ml) with 15 μM CHIR: definitive endoderm
- After 24 hours, replace medium with appropriate subsequent differentiation conditions
Key Findings: BCD modulation only during the first 24 hours results in distinct gene expression patterns equivalent to specific cell fates along a primitive streak-like anteroposterior axis [39]. This effect is mediated through distinct, time-dependent medium conditioning involving TGFβ superfamily members and Nodal signaling antagonists LEFTY1 and CER1 [39].
Figure 1: Bulk Cell Density Modulation Workflow: Schematic representation of how BCD and CHIR99021 concentration interact to determine lineage specification outcomes within the first 24 hours of differentiation.
Hematopoietic Differentiation via Embryoid Body Formation
EB-based protocols provide a valuable model for studying hematopoietic development, with Wnt signaling playing a crucial role in endothelial and hematopoietic stem and progenitor cell (HSPC) emergence [40].
Protocol Overview:
- Generate EBs from hiPSCs using established EB generation protocols
- Monitor activation of Wnt signaling pathway through gene expression (e.g., RSPO3)
- Assess emergence of endothelial and hematopoietic lineages over time
- Validate through real-time quantitative PCR for key markers:
- SOX17 and CDH5 for endothelial development
- RUNX1, GATA2, and HOXA9 for HSPC specification
- Confirm population emergence through flow cytometry analysis
Key Findings: Activation of the Wnt pathway induces emergence of endothelial cells and facilitates specification of HSPCs [40]. The robust upregulation of key genes over time validates progressive development of endothelial and hematopoietic lineages, although response variability between experiments suggests need for further protocol optimization [40].
Critical Factors and Optimization Strategies
Culture Conditions and Media Composition
The effects of Wnt signaling on hPSCs are highly dependent on culture conditions and media composition [41]. Research has demonstrated that the same Wnt manipulation can produce opposite effects in different culture systems:
- In standard hESC conditions (feeder cells + serum replacement), Wnt3a enhances self-renewal and expansion of undifferentiated hESCs [41]
- In defined media (N2B27 and mTeSR1 with Matrigel), Wnt3a causes rapid differentiation with loss of pluripotency markers and gain of differentiation markers [41]
The key component responsible for these differential effects was identified as knockout serum replacement (KOSR), which contains factors that block Wnt-mediated differentiation without interfering with Wnt signal transduction [41]. Lysophosphatidic acid (LPA) present in KOSR was found to be a critical factor maintaining pluripotency in the presence of Wnt activation [41].
Inter-Line Variability and Endogenous Wnt Suppression
Significant variability in differentiation efficiency exists between individual hPSC lines, often related to differences in endogenous Wnt signaling activity [42]. Some hESC lines exhibit endogenous suppression of WNT signaling that prevents the switch from self-renewal to definitive endoderm specification [42].
Strategies to overcome differentiation blocks:
- Higher WNT stimulation through increased CHIR99021 concentration or additional Wnt agonists
- Inhibition of PI3K/AKT signaling to redirect Activin/NODAL pathway activity toward endoderm specification
- Pre-screening lines for Wnt activity using micropatterned differentiation or reporter systems
- Selecting lines with appropriate endogenous Wnt levels for specific differentiation applications
Research has shown that redirecting the activity of the Activin/NODAL pathway by WNT signaling toward mediating definitive endoderm fate specification represents a vulnerable spot in the differentiation process, as disruption of this redirection can result in poor hPSC specification [42].
Figure 2: Wnt/β-catenin Signaling Pathway in hPSCs: Simplified representation of the canonical Wnt pathway showing key steps from ligand binding to target gene activation, highlighting points of experimental manipulation.
Practical Considerations for Experimental Reproducibility
Several practical factors significantly impact the reproducibility of EB differentiation studies:
- Aggregate size control: Equal-sized aggregates should be generated to avoid differentiation bias originating from differentially sized EBs [42]
- Temporal precision: The first 24 hours of differentiation are critical for lineage determination [39]
- Bioreactor systems: Agitation culture systems can improve reproducibility by enhancing nutrient exchange and paracrine factor distribution [39]
- Platform independence: BCD effects remain consistent across 2D monolayer and 3D aggregate cultures [39]
Human pluripotent stem cells and embryoid bodies provide invaluable in vitro models for studying the role of Wnt signaling in early embryogenesis. The inherent self-organizing capacity of EBs, coupled with advanced techniques for monitoring and manipulating Wnt pathway activity, has revealed remarkable conservation of developmental mechanisms between in vivo embryogenesis and in vitro differentiation systems. The controlled modulation of Wnt signaling through pharmacological agents, genetic tools, and culture parameter adjustments enables precise control over germ layer specification and spatial patterning in these models.
Continued refinement of EB differentiation protocols, including standardized micropatterning approaches, optimized bulk cell density parameters, and defined culture conditions, will enhance the reproducibility and reliability of these systems. Furthermore, recognizing and accounting for inter-line variability in endogenous Wnt signaling will improve the efficiency of directed differentiation applications. As research progresses, these Wnt-controlled in vitro models will continue to provide fundamental insights into human development and offer powerful platforms for disease modeling and regenerative medicine applications.
The integration of CRISPR-Cas9 genome editing with lineage tracing technologies represents a transformative approach for investigating complex biological processes, particularly the role of signaling pathways in early embryogenesis. The Wnt signaling pathway is a quintessential model for studying these advanced genetic tools, as it governs critical developmental events including body axis patterning, cell fate determination, and tissue morphogenesis [43] [44]. CRISPR-Cas9 enables precise manipulation of Wnt pathway components, while modern lineage tracing methods powered by CRISPR allow researchers to reconstruct developmental lineages with unprecedented resolution by recording cellular ancestry in mutable genetic barcodes [45]. This technical guide explores the convergence of these technologies, providing detailed methodologies, data analysis frameworks, and practical resources for researchers investigating Wnt signaling in embryonic development.
The fundamental importance of Wnt signaling in embryogenesis is well-established, with roles spanning from anterior-posterior axis formation to craniofacial development [46] [43]. However, traditional approaches to studying this pathway faced significant limitations: incomplete genetic perturbation and static snapshots of dynamic processes. The advent of CRISPR-based screening and lineage tracing has overcome these hurdles, allowing for systematic functional genomics and real-time tracking of cell fate decisions. These approaches are particularly valuable for deciphering how Wnt signaling orchestrates complex morphogenetic events during gastrulation and neural development, processes where temporal-spatial regulation is critical [46] [43].
Core Technologies and Methodologies
CRISPR-Cas9 for Functional Genomics of Wnt Signaling
CRISPR-Cas9 knockout screening has emerged as a powerful method for identifying novel regulators of the canonical Wnt pathway. This approach typically involves introducing a library of single guide RNAs (sgRNAs) targeting thousands of genes into cells containing a Wnt activity reporter, followed by selection for cells with altered pathway activity and sequencing to identify enriched or depleted sgRNAs [44].
A representative screen design utilizes HEK293 cells stably transfected with a TCF/HSV-TK hygromycin resistance reporter. Cells in which Wnt inhibitors are knocked out become resistant to hygromycin selection due to constitutive activation of the reporter gene. This strategy has successfully identified novel Wnt pathway repressors including DHX29, USP7, and CSNK1A1 [44]. Validation experiments confirm that DHX29 knockout activates Wnt signaling, leading to upregulation of cyclin-D1, while its overexpression suppresses the pathway, establishing it as a bona fide Wnt signaling tumor suppressor [44].
For studying specific Wnt receptor-ligand interactions, multiplex CRISPR targeting enables simultaneous knockout of multiple Frizzled (FZD) receptor genes. Researchers have designed highly effective sgRNAs targeting conserved regions across FZD family members, such as the cysteine-rich domains and seventh transmembrane region [47]. By creating HEK293T cell lines with combinatorial FZD knockouts (e.g., FZD1/2/7 triple mutants), scientists can systematically map functional interactions between 10 Wnt ligands and their FZD receptors through genetic rescue experiments [47].
Table 1: CRISPR-Cas9 Applications in Wnt Signaling Research
| Application | Experimental System | Key Findings | Citation |
|---|---|---|---|
| Genome-wide knockout screen | HEK293-TCF-HygroR cells | Identified DHX29, USP7 as novel Wnt inhibitors | [44] |
| Multiplex FZD receptor knockout | HEK293T cells | Mapped functional interactions between Wnt ligands and FZD receptors | [47] |
| TMEM79/MATTRIN discovery | Human cells + Xenopus | Identified new FZD regulator promoting degradation via USP8 | [46] |
| NSD1 knockout in HCC | Hepatocellular carcinoma cells | Defined NSD1/H3/Wnt10b signaling axis in liver cancer | [48] |
| Wnt-1 knockout in insect | Dendrolimus punctatus moth | Demonstrated Wnt-1 role in segmentation and appendage development | [49] |
Lineage Tracing Methodologies
Lineage tracing technologies aim to infer ancestry relationships between cells within an organism or tissue. These approaches can be broadly classified into prospective and retrospective methods [45]. Prospective approaches introduce heritable markers into progenitor cells that are passed to descendants, while retrospective methods leverage naturally occurring or engineered genetic variations to reconstruct lineage relationships.
CRISPR-Cas9-based evolving lineage tracers represent a powerful hybrid approach that engineers cells with synthetic "scratchpad" regions (target sites) that accumulate Cas9-induced mutations over time [45]. These mutational profiles serve as heritable barcodes that can be read alongside transcriptomic data from single cells. Advanced versions like DNA Typewriter and peCHYRON introduce ordered, sequential edits to improve lineage reconstruction confidence [45].
The experimental workflow for evolving lineage tracing typically involves:
- Engineering a transgenic organism or cell line containing synthetic target sites with multiple Cas9 cut sites
- Inducing stochastic Cas9 editing at various developmental timepoints
- Performing single-cell RNA-sequencing to capture both mutation profiles and transcriptomic states
- Computational analysis to reconstruct lineage trees based on shared mutation patterns [45] [50]
These methods have been successfully applied to model organisms including zebrafish and Caenorhabditis elegans to map developmental lineages at single-cell resolution [50].
Figure 1: CRISPR-based Lineage Tracing Workflow. The process begins with engineered organisms containing target sites, followed by induction of stochastic mutations during development, single-cell sequencing, and computational lineage reconstruction.
Integration of CRISPR-Cas9 and Lineage Tracing for Wnt Signaling Studies
Advanced Computational Analysis Pipelines
The integration of CRISPR-Cas9 mutations with transcriptomic data requires sophisticated computational methods to accurately reconstruct lineage relationships. LinTIMaT (Lineage Tracing by Integrating Mutation and Transcriptomic data) represents a statistical framework that addresses key limitations of earlier approaches [50]. Unlike maximum parsimony methods that rely solely on mutation data, LinTIMaT employs a maximum-likelihood framework that combines mutational signatures with gene expression patterns to improve lineage reconstruction.
The LinTIMaT algorithm:
- Models the lineage as a rooted directed tree where CRISPR-Cas9 edits accumulate on branches
- Applies a Camin-Sokal parsimony criterion for each synthetic marker
- Computes expression likelihood using Bayesian hierarchical clustering
- Employs a heuristic search algorithm to explore tree topologies that optimize both mutation and expression agreement [50]
Benchmarking against C. elegans embryonic lineage data demonstrated that LinTIMaT achieves up to 41.64% improvement in reconstruction accuracy compared to maximum parsimony methods, particularly under conditions of low mutation rates (μ ≤ 0.15) [50]. This integrated approach also enables the construction of species-invariant lineage trees by combining data from multiple individuals, overcoming the randomness of induced mutations in single experiments.
The preprocessing of raw lineage tracing data produces a character matrix where rows represent cells, columns represent target sites, and values indicate the identity of indels observed at each site. This matrix serves as input for phylogenetic reconstruction using either character-based (maximum parsimony, maximum likelihood) or distance-based (neighbor-joining) approaches [45].
Applications to Wnt Signaling in Embryogenesis
The combination of CRISPR-Cas9 and lineage tracing has generated profound insights into Wnt signaling during early embryogenesis. In Xenopus, CRISPR screening identified TMEM79/MATTRIN as a specific inhibitor of Wnt/FZD signaling that promotes FZD degradation independent of ZNRF3/RNF43 ubiquitin ligases [46]. TMEM79 interacts with ubiquitin-specific protease 8 (USP8) to govern substrate specificity, and its inhibition is required for anterior neural development and gastrulation, demonstrating how precise genetic manipulation reveals new regulatory layers in Wnt-mediated patterning [46].
Zebrafish studies have leveraged these technologies to elucidate the roles of Dact1 and Dact2 adapter proteins in modulating noncanonical Wnt signaling during convergent extension and craniofacial morphogenesis [43]. Compound dact1/dact2 mutants exhibit convergent extension defects similar to wnt11f2 mutants, and single-cell RNA-seq analysis revealed novel regulation of calpain 8 (capn8) expression, connecting Wnt signaling to calcium-dependent proteolysis during embryogenesis [43].
Table 2: Key Findings from Integrated CRISPR-Lineage Tracing Studies of Wnt Signaling
| Biological System | Genetic Manipulation | Key Discovery | Developmental Process | |
|---|---|---|---|---|
| Xenopus embryos | TMEM79 knockout | TMEM79 promotes FZD degradation via USP8 interaction | Anterior neural development, gastrulation | [46] |
| Zebrafish embryos | dact1/2 compound mutants | Dact proteins regulate convergent extension via capn8 | Craniofacial morphogenesis, axis extension | [43] |
| Human Barrett organoids | APC knockout | Wnt activation recapitulates neoplastic transformation | Esophageal neoplasia | [51] |
| Human pluripotent stem cells | β-catenin knockout | Wnt/β-catenin required for mesendoderm but not neuroectoderm | Cardiomyocyte differentiation, lineage specification | [35] |
| Hepatocellular carcinoma | NSD1 knockout | NSD1/H3/Wnt10b signaling axis drives HCC progression | Cancer proliferation, migration, invasion | [48] |
Experimental Protocols and Methodologies
Protocol for Genome-wide CRISPR Knockout Screen of Wnt Signaling
This protocol outlines the steps for conducting a genome-scale CRISPR-Cas9 knockout screen to identify novel Wnt signaling regulators, based on the methodology successfully employed to discover DHX29 [44].
Materials:
- HEK293-TCF-HygroR reporter cell line
- GeCKO v2 CRISPR knockout library or similar
- Lentiviral packaging plasmids (psPAX2, pVSV-G)
- Hygromycin B selection antibiotic
- Tissue culture facilities for BSL-2 work
Procedure:
- Reporter Cell Line Validation: Confirm that the HEK293-TCF-HygroR cells show minimal background hygromycin resistance and appropriate responsiveness to Wnt pathway activation (e.g., via Wnt3A treatment or GSK3 inhibition).
Library Transduction:
- Produce lentivirus from the CRISPR knockout library at appropriate titer
- Transduce HEK293-TCF-HygroR cells at low MOI (≈0.3) to ensure single sgRNA integration
- Include non-transduced controls for comparison
Selection and Expansion:
- Apply hygromycin B selection (150μg/ml) for 10-14 days
- Maintain parallel non-selected transduced cells as reference
- Harvest genomic DNA from both selected and reference populations
Sequencing and Analysis:
- Amplify integrated sgRNA sequences by PCR
- Perform next-generation sequencing (Illumina platform)
- Use MAGeCK algorithm to identify significantly enriched sgRNAs in selected population
- Validate top hits through individual sgRNA knockout and functional assays
Critical Parameters:
- Maintain sufficient library coverage (≥500x) throughout the screen
- Optimize hygromycin concentration to achieve complete killing of non-transduced controls
- Include biological replicates to ensure reproducibility
Protocol for CRISPR-based Lineage Tracing in Model Organisms
This protocol describes the implementation of evolving CRISPR-Cas9 lineage tracing, adapted from zebrafish and C. elegans studies [45] [50].
Materials:
- Transgenic organism with Pol II-driven Cas9 and target site array
- sgRNAs targeting the scratchpad region
- Microinjection equipment for embryos
- Single-cell RNA-sequencing platform (10X Chromium)
- Computational resources for lineage reconstruction
Procedure:
- System Design:
- Engineer a target site array containing 8-12 identical Cas9 cut sites
- Select target sequences with minimal off-target potential in the genome
- Configure the system for either ubiquitous or tissue-specific Cas9 expression
Mutation Induction:
- Microinject sgRNAs targeting the scratchpad at early embryonic stages
- For temporal control, employ inducible Cas9 systems or staged sgRNA delivery
- Allow mutations to accumulate through development
Single-Cell Multiomic Profiling:
- Dissociate tissues of interest at desired timepoint into single-cell suspension
- Perform single-cell RNA-sequencing with custom primers to amplify target sites
- Sequence libraries to sufficient depth (≥50,000 reads/cell recommended)
Lineage Reconstruction:
- Preprocess raw sequencing data to generate character matrix of mutation states
- Align reads to reference target site sequence
- Identify insertion-deletion mutations at each cut site
- Apply LinTIMaT or similar integrated analysis pipeline
- Reconstruct lineage tree and correlate with cell type identities
Troubleshooting:
- Low mutation diversity: Increase sgRNA concentration or use multiple sgRNAs
- High cell dropout: Optimize dissociation protocol and cell viability
- Ambiguous lineage relationships: Increase target site number or employ sequential editing systems
Data Presentation and Analysis
Quantitative Analysis of CRISPR Screening Data
Systematic analysis of CRISPR screening data reveals novel regulators of Wnt signaling. The following table summarizes quantitative findings from published genome-scale screens.
Table 3: Quantitative Results from Wnt Pathway CRISPR Screens
| Gene Target | Fold Enrichment | Function in Wnt Signaling | Validation Method | Reference |
|---|---|---|---|---|
| DHX29 | 35x (hygro resistance) | Negative regulator, tumor suppressor | qPCR (cyclin-D1), Western blot | [44] |
| USP7 | 48x (hygro resistance) | Deubiquitinase, Wnt pathway repressor | TCF-binding site mutation control | [44] |
| CSNK1A1 | Significant (p<2.28e-7) | Known Wnt repressor, destruction complex | Included as positive control | [44] |
| FZD1/2/7 | Complete Wnt unresponsiveness | Redundant canonical Wnt receptors | Wnt ligand rescue experiments | [47] |
| TMEM79 | Specific FZD inhibition | Promotes FZD degradation via USP8 | Xenopus embryogenesis validation | [46] |
Lineage Tracing Performance Metrics
Evaluation of lineage tracing methods demonstrates the advantage of integrated approaches that combine mutation and expression data.
Table 4: Performance Comparison of Lineage Reconstruction Methods
| Method | Mutation Rate | Reconstruction Accuracy | Advantages | Limitations | |
|---|---|---|---|---|---|
| Maximum Parsimony | μ = 0.15 | 58.36% (C. elegans benchmark) | Simple principle, widely implemented | Sensitive to mutation saturation, random noise | [50] |
| Neighbor-Joining | μ = 0.15 | 70.55% (C. elegans benchmark) | Polynomial time complexity | Distance-based, ignores character states | [50] |
| LinTIMaT (integrated) | μ = 0.15 | 100% (C. elegans benchmark) | Combines mutations + expression, robust to noise | Computationally intensive | [50] |
| LinTIMaT (integrated) | μ = 0.2 | >12.9% improvement over MP | Better performance at high mutation rates | Requires single-cell RNA-seq data | [50] |
Visualization of Signaling Pathways and Experimental Workflows
Wnt Signaling Pathway and CRISPR Modulation Points
Figure 2: Wnt Signaling Pathway with CRISPR-Identified Regulatory Nodes. The canonical pathway shows Wnt ligand binding to FZD/LRP receptors, leading to β-catenin stabilization and target gene transcription. Blue nodes represent regulators identified through CRISPR screens.
Integrated Experimental- Computational Pipeline
Figure 3: Integrated Experimental-Computational Pipeline. The workflow begins with experimental design and perturbation, proceeds through sequencing and computational analysis, and yields biological insights into molecular mechanisms, developmental lineages, and regulatory networks.
The Scientist's Toolkit: Essential Research Reagents
Table 5: Essential Research Reagents for CRISPR-Wnt Studies
| Reagent/Category | Specific Examples | Function/Application | Key Features | |
|---|---|---|---|---|
| CRISPR Delivery Systems | lentiCRISPRv2, px459 | Vector systems for sgRNA + Cas9 expression | Puromycin selection, high efficiency | [47] [48] |
| Wnt Reporter Cells | HEK293-TCF-HygroR, HCT116 TCF4/Wnt-reporter | Readout for canonical Wnt pathway activity | Selectable markers, sensitive response | [44] |
| Lineage Tracing Systems | scGESTALT, ScarTrace, DNA Typewriter | Evolving barcodes for lineage reconstruction | Multiple target sites, sequential editing | [45] [50] |
| Organoid Culture Systems | Barrett esophageal, HCC organoids | 3D models of tissue development and disease | Genetically stable, recapitulate in vivo biology | [51] [48] |
| Computational Tools | LinTIMaT, MAGeCK, GAPML | Analysis of lineage tracing and screen data | Integration of mutation + expression data | [45] [44] [50] |
| Model Organisms | Xenopus, zebrafish, C. elegans | In vivo studies of embryogenesis | Transgenic capabilities, optical clarity | [46] [43] [50] |
| Pinol | Pinol, CAS:2437-97-0, MF:C10H16O, MW:152.23 g/mol | Chemical Reagent | Bench Chemicals | |
| KRN5 | KRN5, CAS:1800465-47-7, MF:C27H22FNO5, MW:459.5 g/mol | Chemical Reagent | Bench Chemicals |
This technical guide demonstrates how the integration of CRISPR-Cas9 genome editing with advanced lineage tracing technologies has revolutionized our ability to dissect Wnt signaling during embryogenesis. These approaches enable systematic functional genomics alongside temporal tracking of cell fate decisions, providing unprecedented resolution into how signaling pathways orchestrate complex developmental programs. As these technologies continue to evolve, they promise to unravel the remaining complexities of embryonic patterning and morphogenesis.
Biochemical Assays for Monitoring Pathway Activation
The Wnt signaling pathway is an ancient and evolutionarily conserved system that regulates crucial aspects of cell fate determination, cell migration, cell polarity, neural patterning, and organogenesis during embryonic development [10]. In early embryogenesis, Wnt proteins act as potent morphogens that direct fundamental processes including the establishment of the primary body axis and the formation of a dorsal organizing center [52] [53] [10]. The exquisite spatial and temporal regulation of Wnt signaling is particularly critical during the earliest stages of embryonic patterning, where it governs the complex cellular interactions that give rise to the three primary germ layers: ectoderm, endoderm, and mesoderm [53]. The pathway's name originates from a fusion of the Drosophila segment polarity gene wingless and its vertebrate homolog, integrated or int-1, reflecting its deep evolutionary conservation [10].
Misregulation of this finely tuned pathway has catastrophic consequences for embryonic development and is implicated in human birth defect disorders, including the most common human neural tube closure defect, spina bifida [10]. Furthermore, aberrant Wnt signaling has been linked to the onset and progression of various cancers, making its accurate monitoring not only fundamental to developmental biology but also crucial for therapeutic development [52] [10]. This technical guide provides an in-depth overview of the biochemical assays essential for monitoring Wnt pathway activation, with particular emphasis on their application in the context of early embryogenesis research.
Molecular Mechanisms of Wnt Pathway Activation
Key Signaling Branches
Wnt signaling occurs through several distinct branches, primarily categorized as canonical (β-catenin-dependent) and non-canonical (β-catenin-independent) pathways [10]. The canonical Wnt/β-catenin pathway serves as the primary mechanism regulating cell fate decisions through transcriptional control, while non-canonical pathways, including the Planar Cell Polarity and Wnt/Ca²⺠pathways, predominantly influence cell motility and polarity [10].
The core molecular machinery of the canonical pathway involves three key components [54] [10]:
- A heterodimeric receptor complex consisting of a Frizzled (Fz) receptor and a Lipoprotein Receptor-Related Protein 5/6 (LRP5/6) co-receptor.
- A cytoplasmic destruction complex that assembles Axin, Adenomatous Polyposis Coli (APC), Glycogen Synthase Kinase 3 (GSK3), Casein Kinase 1α (CK1α), and Dishevelled (Dvl).
- β-catenin, the key transcriptional co-activator whose stability is controlled by the pathway.
In the absence of Wnt stimulation, cytoplasmic β-catenin is continuously phosphorylated by the destruction complex, leading to its ubiquitination and proteasomal degradation, thereby maintaining low intracellular β-catenin levels [54]. Upon binding of Wnt ligands to the Fz/LRP5/6 receptor complex, a signal is transduced to Dvl, which triggers the recruitment of Axin from the inhibitory destruction complex to the stimulatory signalosome at the plasma membrane [10] [55]. This recruitment destabilizes the destruction complex, inhibits β-catenin degradation, and allows β-catenin to accumulate in the cytoplasm and translocate to the nucleus. There, it partners with T-cell factor/lymphoid enhancer factor (TCF/LEF) DNA-binding proteins to activate transcription of target genes [54] [10].
Visualizing the Canonical Wnt/β-catenin Pathway
The following diagram illustrates the core molecular events of the canonical Wnt/β-catenin signaling pathway, from ligand-receptor binding to target gene activation.
Biochemical Assays for Monitoring Wnt Activation
Monitoring the complex events of Wnt pathway activation requires a multifaceted biochemical approach. The following section details key methodologies, which are summarized and compared in the table below for rapid evaluation.
Table 1: Biochemical Assays for Monitoring Wnt Pathway Activation
| Assay Type | Target / Readout | Key Reagents | Applications in Embryogenesis Research | Key Advantages |
|---|---|---|---|---|
| Wnt Reporter Assay [54] | TCF/LEF transcriptional activity; Luciferase activity | TCF/LEF-responsive luciferase vector, Wnt agonists/antagonists (e.g., Wnt3a, DKK1) | Testing pathway activity in patient-specific iPSC-derived neural stem cells [54] | Functional readout of pathway endpoint; amenable to high-throughput screening |
| Immunoblotting of Secreted Wnts [52] | Secreted Wnt protein levels | Blue Sepharose (for purification), Wnt antibodies | Purifying and quantifying Wnts from cell supernatant; studying Wnt trafficking and release [52] | Direct measurement of secreted ligand availability |
| Axin Stability & Localization Assay [55] | Axin protein levels & subcellular localization; ADP-ribosylation status | Anti-Axin antibodies, Anti-V5 antibodies (for tagged Axin), Tankyrase inhibitors | In vivo analysis of Axin reprogramming in Drosophila embryos following Wg/Wnt stimulation [55] | Monitors the key scaffold protein that switches pathway function |
| Dual-Luciferase Screen [56] | β-catenin and Axin protein stability (dual luciferase readout) | β-catenin-firefly luciferase, Axin-Renilla luciferase fusion constructs | High-throughput screening for small-molecule regulators in Xenopus egg extracts [56] | Simultaneously monitors two key opposing components in a biochemical system |
TCF/LEF Wnt Reporter Assay
The Wnt reporter assay is a functional cellular assay that measures the transcriptional output of the canonical Wnt pathway, providing a direct readout of its activation status [54].
Experimental Protocol
- Reporter Construct Design: Clone a minimal promoter containing multiple TCF/LEF transcriptional response elements (TREs) upstream of a reporter gene, typically luciferase (luc2P) [54].
- Cell Transduction: Stably or transiently introduce the reporter construct into your cell model of choice. For embryogenesis research, this often involves human induced pluripotent stem cells (iPSCs) or iPSC-derived neural stem cells (NSCs) to maintain a patient-specific genetic background [54]. Both viral and non-viral (e.g., vector-based) delivery methods can be used.
- Pathway Modulation and Incubation: Treat the reporter cells with experimental conditions. This includes:
- Agonists: Recombinant Wnt3a protein to activate the pathway.
- Antagonists: Recombinant DKK1 (a LRP5/6 inhibitor) to suppress the pathway.
- Small molecule inhibitors (e.g., GSK3 inhibitors like CHIR99021) or activators [53].
- Luciferase Measurement: Lyse the cells and measure luciferase activity using a luminometer. The signal, reported as Relative Luminescence Units (RLU), is directly proportional to canonical Wnt pathway activity [54].
- Data Analysis: Normalize data to control conditions (e.g., unstimulated cells) and present as fold-change in activity. Generate dose-response curves to calculate ECâ‚…â‚€/ICâ‚…â‚€ values for pharmacological agents.
Immunoblotting of Secreted Wnt Proteins
Assessing the secretion of Wnt ligands themselves is critical, as this represents the initiating event of the pathway. This is particularly relevant when studying morphogen gradient formation in embryonic patterning.
Experimental Protocol
- Sample Collection and Preparation:
- Collect supernatant from Wnt-producing cells.
- Concentrate secreted proteins and separate them from extracellular vesicles (EVs) if analyzing specific Wnt pools (e.g., exosomal Wnts) [52].
- Wnt Purification: Due to the hydrophobic and lipid-modified nature of Wnt proteins, use Blue Sepharose chromatography to purify Wnts from the supernatant based on their hydrophobicity [52].
- Immunoblotting:
- Separate purified proteins by SDS-PAGE.
- Transfer to a membrane and probe with specific anti-Wnt antibodies.
- Compare the levels of secreted Wnt to the total cellular Wnt content from lysates to analyze secretion efficiency [52].
Analysis of Axin Stability and Localization
Axin serves as the concentration-limiting scaffold for both the destruction complex and the signalosome, making its regulation a key indicator of pathway status [55]. Wnt stimulation induces rapid, biphasic regulation of Axin.
Experimental Protocol
- In Vivo Modeling: To study Axin at near-physiological levels in an embryogenesis context, use a model like Drosophila. Generate transgenic flies expressing Axin tagged with an epitope (e.g., V5) under the control of a ubiquitous promoter (e.g., mat-Gal4) [55].
- Wnt Stimulation and Sampling: Induce Wg (the Drosophila Wnt homolog) expression and collect embryos at precise time points (e.g., from late stage 8 through stage 9) to capture early events [55].
- Visualization and Quantification:
- Immunofluorescence: Stain embryos with anti-V5 and anti-Wg antibodies. Immediately following Wg exposure, Axin levels increase 2- to 3-fold and form striping patterns centered over Wg stripes, indicating its recruitment and stabilization at the signalosome [55].
- Biochemical Analysis: Immunoblot lysates to monitor Axin's electrophoretic mobility shift, which can indicate post-translational modifications such as ADP-ribosylation by Tankyrase that enhance its interaction with LRP6 [55].
The experimental workflow for analyzing the key component Axin is visualized below, integrating both cellular and biochemical techniques.
The Scientist's Toolkit: Key Research Reagents
Successful investigation of Wnt signaling in embryogenesis relies on a core set of pharmacological and biological reagents.
Table 2: Essential Research Reagents for Wnt Pathway Analysis
| Reagent Category | Specific Example | Molecular Target / Function | Application in Research |
|---|---|---|---|
| Pathway Agonists | Recombinant Wnt3a Protein | Binds FZD/LRP6 receptor complex; stabilizes β-catenin [54] | Positive control for pathway activation in reporter assays [54] |
| Pathway Agonists | R-Spondin2 | Binds LRP5/6 and RNF43/ZNRF3; enhances surface levels of FZD receptors [53] | Potentiates Wnt signaling in stem cell cultures and organoids [53] |
| Small Molecule Inhibitors | CHIR99021 | Selective GSK-3 inhibitor; prevents β-catenin phosphorylation [53] | Activates Wnt signaling; maintains pluripotency in hPSCs [53] |
| Small Molecule Inhibitors | XAV939 | Tankyrase (Tnks) inhibitor; stabilizes Axin [53] [55] | Blocks Wnt signaling; used to study Axin regulation and for potential therapeutics [55] |
| Small Molecule Inhibitors | IWP/IWR compounds | Porcupine and Tankyrase inhibitors; block Wnt production and response [53] | Probing different stages of Wnt pathway regulation |
| Secreted Antagonists | Recombinant DKK1 | Binds LRP5/6; prevents Wnt co-receptor interaction [54] | Negative control for pathway inhibition in reporter assays [54] |
| Secreted Antagonists | SFRP-1 | Binds directly to Wnt ligands; sequesters them from receptors [53] | Studying extracellular modulation of Wnt gradient formation |
| Genetic Tools | TCF/LEF Reporter Constructs | Luciferase under TCF/LEF control; measures pathway activity [54] | Real-time monitoring of canonical signaling output |
| Genetic Tools | epitope-tagged Axin | Enables visualization and purification of endogenous Axin pools [55] | Live imaging and biochemical tracking of the key scaffold protein |
| G907 | G907, CAS:2244035-16-1, MF:C26H24ClNO3, MW:433.9 g/mol | Chemical Reagent | Bench Chemicals |
| Naspm | Naspm, CAS:122306-11-0, MF:C22H34N4O, MW:370.5 g/mol | Chemical Reagent | Bench Chemicals |
The biochemical dissection of Wnt signaling during early embryogenesis demands a synergistic application of multiple techniques. Functional reporter assays, direct protein analysis via immunoblotting, and meticulous in vivo examination of core components like Axin collectively provide a comprehensive picture of pathway dynamics. The reagents and methods detailed in this guide form a foundational toolkit for researchers aiming to elucidate the complex roles of Wnt signaling in development and disease. As the field advances, these biochemical assays will continue to be indispensable for validating new regulatory mechanisms and for screening potential therapeutic compounds aimed at modulating this critical pathway.
Blastoid and Organoid Systems for Modeling Early Development
The study of early human development has been transformed by the emergence of advanced in vitro models that recapitulate key embryonic events. Among these, blastoid and organoid systems have emerged as powerful tools for investigating the fundamental processes of blastocyst formation and implantation. These models are particularly valuable for elucidating the role of critical signaling pathways, especially the Wnt signaling pathway, which governs cell fate decisions, lineage specification, and morphogenetic events during early embryogenesis. Unlike traditional two-dimensional cell cultures, these three-dimensional structures preserve the cellular heterogeneity and spatial organization characteristic of in vivo development, providing unprecedented opportunities for mechanistic studies under controlled conditions. When framed within the context of Wnt signaling research, blastoid and organoid systems enable precise dissection of how this evolutionarily conserved pathway coordinates the complex sequence of events from pre-implantation development to uterine attachment [57] [58].
Biological Foundations: From Stem Cells to Embryo Models
The Human Blastocyst as a Developmental Blueprint
A human blastocyst forms approximately 5-7 days post-fertilization and consists of three founding lineages arranged in a characteristic structure: (1) the trophectoderm (TE), a thin epithelial sphere that forms the blastocyst cavity and later contributes to placental tissues; (2) the epiblast (EPI), a compact cluster of cells that gives rise to the embryo proper; and (3) the primitive endoderm (PrE), which generates extraembryonic structures supporting embryonic development [58]. The formation of these lineages follows a strict developmental sequence, with TE and EPI specification preceding PrE formation. Crucially, the blastocyst establishes the embryonic-abembryonic axis, characterized by the maturation of the polar TE region, which enables directional attachment to the uterine endometrium during implantation [59] [58].
Stem Cell States as Building Blocks
The fidelity of embryo models depends critically on the developmental state of the stem cells used to generate them. Human pluripotent stem cells (hPSCs) can be stabilized in different states that capture discrete developmental stages:
- Naïve hPSCs: Cultured in PXGL conditions (containing PD0325901, XAV939, Gö6983, and LIF), these cells transcriptionally reflect the blastocyst-stage epiblast and maintain the capacity to form TE analogs, mirroring the plasticity of early blastocyst cells [60] [58].
- Primed hPSCs: Maintained in FGF2 and Activin, these cells reflect the post-implantation epiblast stage and have restricted potential to form blastocyst-stage analogs [59] [60].
This distinction is crucial for blastoid generation, as only naïve hPSCs possess the developmental potency to efficiently generate all three blastocyst lineages according to the appropriate developmental sequence and timing [60].
Technical Framework for Blastoid Generation
Core Protocol for Human Blastoid Formation
The generation of blastoids from naïve hPSCs requires precise control over initial aggregate size and signaling environments to trigger the cascading sequence of blastocyst development [59] [60]. The following protocol achieves high efficiency (>70%) in forming structures that morphologically and transcriptionally resemble human blastocysts:
Day 0: Preparation of Naïve hPSCs
- Culture naïve hPSCs in PXGL medium to maintain pre-implantation epiblast identity [60] [58].
- Dissociate cells into single-cell suspension using TrypLE or similar enzyme preparation [61].
Day 1: Aggregate Formation
- Resuspend dissociated cells in blastoid medium containing lysophosphatidic acid (LPA), A83-01 (TGF-β inhibitor), PD0325901 (ERK inhibitor), LIF, and Y-27632 (ROCK inhibitor) [58].
- Seed 3,000-4,000 cells per well in non-adherent hydrogel microwells (96-well or 24-well plates) to ensure consistent aggregate size [59] [60].
- Centrifuge plates to enhance aggregate formation (300-400 × g, 2-3 minutes).
Days 1-4: Blastoid Differentiation and Maturation
- Maintain aggregates in the triple inhibition medium (Hippo/TGF-β/ERK pathway inhibition) for 96 hours [60] [58].
- Change medium every 48 hours, carefully replacing half of the spent medium with fresh pre-warmed medium.
- Monitor daily for cavitation initiation (typically visible by day 2-3) and blastocyst-like morphology formation (by day 4).
Day 4: Blastoid Harvest and Analysis
- Harvest blastoids for experimental analysis once they reach 150-250 μm in diameter with clearly visible inner cell mass and cavity [58].
- For implantation assays, use blastoids within 24 hours of full formation.
Table 1: Key Signaling Pathway Manipulations in Blastoid Formation
| Pathway | Inhibitor/Activator | Concentration | Function in Blastoid Development |
|---|---|---|---|
| Hippo | LPA (Lysophosphatidic acid) | 1-5 μM | Inhibits Hippo pathway, enabling nuclear localization of YAP1 and TE specification [58] |
| TGF-β | A83-01 | 0.5-1 μM | Blocks TGF-β/NODAL signaling, promotes trophoblast differentiation [60] |
| ERK | PD0325901 | 0.5-1 μM | Inhibits FGF/ERK signaling, supports naïve state and trophoblast specification [60] [58] |
| ROCK | Y-27632 | 5-10 μM | Enhances cell survival after dissociation, prevents anoikis [61] [58] |
Essential Quality Control Measures
To ensure blastoids faithfully recapitulate blastocyst development, implement these quality control assessments:
Morphological Criteria
- Diameter between 150-250 μm, matching human blastocysts [59] [58].
- Clear spherical structure with defined cavity and inner cell mass.
- Continuous epithelial layer of outer cells.
Lineage Specification Validation
- Immunostaining for TE markers: GATA2, GATA3, CDX2, TROP2 [58].
- Immunostaining for EPI markers: OCT4, NANOG [59] [58].
- Immunostaining for PrE markers: GATA4, SOX17, PDGFRa [59] [58].
- Sequential appearance of lineages: TE/EPI by 24-65 hours, PrE by 65-96 hours [59].
Transcriptomic Validation
- Single-cell RNA sequencing to confirm transcriptional similarity to blastocyst-stage cells (>96% blastocyst-like cells) [59] [60].
- Minimal presence (<4%) of post-implantation cell types.
Wnt Signaling in Early Development and Model Systems
Wnt Pathway Fundamentals in Embryogenesis
The Wnt signaling pathway comprises a family of 19 secreted glycoproteins that regulate critical developmental processes through canonical (β-catenin-dependent) and noncanonical (β-catenin-independent) branches [62] [5]. During early embryogenesis, Wnt signaling plays multiple essential roles:
- Lineage Specification: Canonical Wnt signaling promotes pluripotency in the epiblast while regulating trophoblast differentiation [60] [58].
- Axis Formation: Wnt signaling participates in establishing the embryonic-abembryonic axis in blastocysts [58].
- Morphogenesis: Wnt ligands regulate cell polarity, migration, and tissue patterning through both canonical and noncanonical pathways [62] [5].
In the canonical pathway, Wnt ligands bind to Frizzled receptors and LRP5/6 coreceptors, leading to stabilization and nuclear translocation of β-catenin, which partners with TCF/LEF transcription factors to activate target genes [62]. In the absence of Wnt ligands, a destruction complex comprising APC, AXIN, GSK-3β, and CK1α phosphorylates β-catenin, targeting it for proteasomal degradation [62] [63].
Monitoring Wnt Activity in Blastoid Systems
Investigating Wnt signaling in blastoids requires specific analytical approaches:
Gene Expression Analysis
- Quantitative PCR for Wnt target genes: AXIN2, SP5, MYC [62].
- Single-cell RNA sequencing to map Wnt pathway component expression across lineages.
Protein Localization Assessment
- Immunofluorescence for β-catenin localization (membrane, cytoplasmic, nuclear).
- Staining for phosphorylated LRP6 and DVL as markers of pathway activation [64].
Functional Pathway Manipulation
- Small molecule inhibitors: IWP-2 (PORCN inhibitor), XAV939 (Tankyrase inhibitor) [62] [5].
- Recombinant Wnt proteins: WNT3A (canonical activator), WNT5A (noncanonical activator) [61] [62].
- Synthetic agonists: FLAgs (Frizzled and LRP5/6 Agonists) for specific receptor activation [64].
Figure 1: Wnt/β-catenin Signaling Pathway in Early Development. This canonical pathway regulates gene expression programs governing cell fate decisions, lineage specification, and stemness maintenance during blastocyst development. Pathway activation inhibits the destruction complex, enabling β-catenin stabilization and nuclear translocation [62] [63].
Experimental Applications and Methodologies
Implantation Modeling Using Endometrial Organoids
A key application of blastoids is modeling human implantation, which can be achieved through co-culture with endometrial organoids:
Endometrial Organoid Generation
- Isolate human endometrial epithelial cells from biopsy tissue.
- Embed in Matrigel and culture with specific growth factors (FGF, WNT3A, R-spondin) to maintain stemness and differentiation capacity [65] [57].
- Hormonally prime with estrogen and progesterone to mimic the receptive endometrium [59] [58].
Implantation Assay Protocol
- Day 1: Transfer mature blastoids onto hormonally stimulated endometrial organoids.
- Day 1-3: Monitor attachment efficiency (typically 40-60% with optimized conditions).
- Day 3-6: Assess trophoblast invasion and differentiation using specific markers:
- Extravillous trophoblasts: HLA-G+
- Syncytiotrophoblasts: CGB+
- Day 6-13: Evaluate post-implantation development and lineage progression [59] [58].
Wnt Pathway Modulation Experiments
To investigate Wnt signaling functions, implement these experimental approaches:
Loss-of-Function Studies
- Treat blastoids with small molecule inhibitors: IWR-1 (AXIN stabilizer), IWP-2 (PORCN inhibitor) [62].
- Use siRNA/shRNA to knock down specific FZD receptors or downstream components.
- Apply secreted Wnt antagonists: DKK1, sFRPs [62] [63].
Gain-of-Function Studies
- Supplement culture with recombinant Wnt proteins (WNT3A, WNT5A) [61] [62].
- Utilize synthetic FLAgs to activate specific FZD receptors with tailored specificity [64].
- Employ GSK-3β inhibitors (CHIR99021) to stabilize β-catenin [5].
Lineage-Specific Analysis
- Isplicate specific lineages using FACS with surface markers (TROP2+ for TE, PDGFRa+ for PrE) [58].
- Perform transcriptomic analysis on isolated populations to identify lineage-specific Wnt responses.
- Use lineage tracing to track fate decisions following Wnt pathway manipulation.
Table 2: Research Reagent Solutions for Blastoid and Wnt Signaling Studies
| Reagent Category | Specific Examples | Application/Function | Key References |
|---|---|---|---|
| Stem Cell Culture | PXGL medium | Maintains naïve pluripotent state for blastoid formation | [59] [58] |
| Pathway Inhibitors | A83-01 (TGF-βi), PD0325901 (ERKi), LPA (Hippoi) | Triple inhibition for efficient blastoid generation | [60] [58] |
| Wnt Modulators | IWP-2, IWR-1, XAV939 | Inhibit Wnt production or signaling | [62] [5] |
| Wnt Activators | Recombinant WNT3A, CHIR99021, FLAgs | Activate canonical Wnt signaling | [62] [64] |
| Extracellular Matrix | Matrigel, synthetic hydrogels | Provides 3D scaffold for organoid culture | [61] [57] |
| Lineage Markers | TROP2, PDGFRa, GATA3, OCT4 | Isolation and validation of specific lineages | [58] |
| Synthetic Agonists | FLAgs (FZD/LRP agonists) | Specific activation of defined FZD receptors | [64] |
Data Analysis and Validation Frameworks
Computational Approaches for Blastoid Validation
Rigorous validation of blastoid systems requires multi-modal data analysis:
Morphometric Analysis
- Quantify diameter, circularity, and cavity formation using brightfield imaging.
- Track growth dynamics over time (65-200 μm range).
- Measure lineage proportions (TE: ~70%, EPI: ~20%, PrE: ~10%) [58].
Transcriptomic Evaluation
- Single-cell RNA sequencing to compare with reference blastocyst datasets.
- Project blastoid cells onto embryonic development maps to assess stage equivalence.
- Identify aberrant cell populations expressing post-implantation markers.
Functional Assessment
- Evaluate lineage potential through stem cell derivation efficiency.
- Test trophectoderm differentiation capacity into SCT and EVT lineages.
- Assess axis formation through polar TE marker expression (NR2F2, CCR7) [59].
Quantitative Benchmarking Against Human Blastocysts
Table 3: Quantitative Comparison Between Blastoids and Human Blastocysts
| Parameter | Human Blastocysts | High-Fidelity Blastoids | Assessment Method |
|---|---|---|---|
| Formation Efficiency | N/A | >70% | Morphological scoring [58] |
| Diameter Range | 150-250 μm | 150-250 μm | Brightfield microscopy [59] [58] |
| Cell Number | 100-150 cells | 129 ± 27 cells | Nuclear staining counts [58] |
| TE Analogs | GATA2+GATA3+CDX2+ | >97% of outer cells | Immunofluorescence [58] |
| EPI Analogs | OCT4+NANOG+ | 27 ± 13 cells | Immunofluorescence [58] |
| PrE Analogs | GATA4+SOX17+ | 7 ± 5 cells | Immunofluorescence [58] |
| Blastocyst Transcriptome | Reference standard | >96% similarity | scRNA-seq mapping [60] [58] |
| Implantation Competence | Directional attachment | Polar TE attachment to endometrium | Co-culture with endometrial organoids [59] [58] |
Concluding Perspectives
Blastoid and organoid systems represent a transformative approach for investigating early human development and the regulatory functions of Wnt signaling. The protocols and methodologies outlined in this technical guide provide a framework for generating high-fidelity models that recapitulate the sequence, timing, and lineage specification of blastocyst development. When integrated with advanced molecular tools for pathway manipulation—including synthetic agonists like FLAgs and precise inhibition strategies—these systems enable mechanistic dissection of Wnt signaling roles in lineage commitment, axis formation, and implantation processes. As these technologies continue to evolve, they will undoubtedly yield deeper insights into human embryogenesis while offering ethically scalable platforms for drug discovery and reproductive medicine innovation.
High-Throughput Screening for Wnt Pathway Modulators
The Wnt/β-catenin signaling pathway, often referred to as the canonical Wnt pathway, serves as a fundamental regulatory mechanism in the formation of the primary body axis during development, cellular differentiation, and tissue homeostasis. This animal-specific pathway has essential roles in the embryogenesis of higher eukaryotes, from diploblastic, radially symmetrical cnidarians to mice and humans [2] [66]. In mammalian systems, the pathway specifies pattern formation during early embryogenesis and functions critically in the differentiation and maintenance of stem cells both in vivo and in vitro [2]. The pathway's activity, which regulates transcription of downstream target genes, controls cell proliferation and differentiation, and governs proliferation and self-renewal of various stem cells [67]. Recent studies on very basal metazoan species have revealed high levels of conservation of components of both canonical and non-canonical Wnt signaling pathways, underscoring its primal importance in developmental processes [66].
The Biological Rationale: Targeting Wnt Signaling
Wnt Pathway in Development and Disease
The canonical Wnt pathway operates with 19 types of Wnt proteins as ligands and 10 types of Frizzled proteins as receptors in humans [67]. The downstream components include a co-receptor LRP 5/6, G-proteins, a multiprotein β-catenin destruction complex (containing Axin, APC, glycogen synthase kinase 3β [GSK3β] and casein kinase), and β-catenin itself, which serves as a cofactor of the key transcription factor TCF that induces expression of Wnt target genes [67]. This pathway is particularly crucial for early embryonic development, where it regulates processes such as cell adhesion and proliferation [23].
Dysregulation of this carefully orchestrated pathway contributes significantly to disease pathogenesis. Aberrant activation or hyperactivation leads to carcinogenesis in several adult organs, most notably colon and breast [67]. In triple-negative breast cancer (TNBC), Wnt/β-catenin signaling, along with other developmental signaling pathways, regulates cancer stem cells and therapy resistance [67]. Dysregulation promotes cell migration, colony formation, stem-like features, and chemoresistance, playing important roles throughout tumor progression [67]. Detailed studies on gene expression profiles have subdivided TNBC into six subtypes, all demonstrating aberrant overexpression of Wnt-associated genes [67].
The Case for High-Throughput Screening
The urgent need for targeted therapies against Wnt-dependent cancers like TNBC, coupled with the pathway's complexity offering multiple potential intervention points, makes it an ideal candidate for high-throughput screening (HTS) approaches. HTS is a method for scientific discovery especially used in drug discovery that leverages robotics, data processing/control software, liquid handling devices, and sensitive detectors to quickly conduct millions of chemical, genetic, or pharmacological tests [68]. This approach allows researchers to rapidly recognize active compounds, antibodies, or genes that modulate a particular biomolecular pathway, providing starting points for drug design and for understanding the role of a particular location within the pathway [68].
Table 1: Key Characteristics of High-Throughput Screening
| Feature | Description | Significance for Wnt Screening |
|---|---|---|
| Throughput | Capability to test up to 100,000 compounds per day in modern systems [68] | Enables rapid screening of diverse compound libraries against multiple Wnt pathway targets |
| Automation | Integrated robot systems transport assay-microplates between stations for sample and reagent addition, mixing, incubation, and detection [68] | Reduces human error and enables consistent handling of delicate cellular models like stem cells |
| Miniaturization | Microplates with 96, 192, 384, 1536, 3456, or 6144 wells; recent advances enable screening using picoliter volumes in drop-based microfluidics [68] | Conserves precious reagents, including specialized Wnt ligands and stem cell-derived materials |
| Data Generation | Specialized analysis machines can measure dozens of plates in minutes, generating thousands of data points quickly [68] | Provides statistical power for identifying subtle modulators in complex biological systems |
HTS Assay Development for Wnt Pathway
Reporter System Design
The cornerstone of effective HTS for Wnt pathway modulators is a robust reporter system that accurately reflects pathway activity. The most established approach utilizes TCF/LEF promoter elements driving easily quantifiable reporters. In one validated system, researchers generated a sensitive, HTS-compatible Wnt/β-catenin signaling reporter system in homogeneous, expandable neural progenitor cells (NPCs) derived from human induced pluripotent stem cells (iPSCs) [69]. This system demonstrated dose-responsive stimulation by several known Wnt/β-catenin signaling pathway modulators, including Wnt3a, GSK3 inhibitors, and lithium, with responses that were robust and reproducible over time across many repeated assays [69].
Similarly, in triple-negative breast cancer research, investigators have employed TNBC cell lines (e.g., BT-20) stably transfected with the TopFlash reporter construct, which expresses firefly luciferase under the Wnt-dependent TCF-promoter, so that luminescence activity directly reflects Wnt signaling intensity [67]. These cells can be stimulated by exogenous Wnt3a, typically provided as Wnt3a-conditioned medium, which serves as a good substitute for purified Wnt3a for cell culturing purposes and effectively activates the cascade in luciferase-based assays [67].
Cellular Models for Wnt Screening
Choosing appropriate cellular models is critical for physiologically relevant screening outcomes. Several advanced cellular systems have been successfully implemented:
- Human iPSC-Derived Neural Progenitors: These provide ideal disease-relevant contexts for neuropsychiatric disorders and can be adapted for HTS-compatible formats [69].
- Airway Basal Stem Cells (ABSCs): Both mouse and human ABSCs have been used to model Wnt pathway dysregulation observed in squamous lung cancer progression, forming heaping morphology resembling premalignant lesions when treated with GSK3β inhibitors [70].
- Triple-Negative Breast Cancer Cells: Native TNBC cell lines harbor intrinsic Wnt pathway activation, making them ideal for identifying compounds that suppress pathological signaling [67].
- Embryonic Development Models: Porcine embryos have been utilized to study IGF-1 interaction with Wnt/β-catenin signaling during trophectoderm development, revealing that IGF-1 promotes β-catenin levels and TE cell differentiation through Wnt pathway activation [23].
Implementation of Wnt-Targeted HTS
Screening Protocols and Workflows
Successful implementation of HTS for Wnt pathway modulators requires meticulously optimized protocols. The following workflow exemplifies a robust approach used in identifying Wnt inhibitors in airway stem cells:
Primary Screening Protocol [70]:
- Cell Preparation: BEAS2B cells (normal human bronchial epithelial cell line) stably transduced with TCF/LEF luciferase reporter.
- Compound Treatment: Cells treated with DMSO (control), 5 μM CHIR99021 (GSK3β inhibitor to activate Wnt signaling), or 5 μM CHIR99021 + 10 μM screen compound for 24 hours.
- Viability Assessment: Parallel Hoechst staining to assess toxicity; compounds yielding fewer than 80% of nuclei compared to DMSO controls considered toxic and removed from analysis.
- Activity Measurement: TCF/LEF activity measured via luciferase assay.
- Hit Identification: Compounds decreasing TCF/LEF activity without toxicity considered initial hits.
Advanced Model System Protocol [70]:
- Airway Basal Stem Cell Culture: Mouse or human ABSCs seeded in collagen-coated transwells.
- Wnt Activation: Treatment with GSK3β inhibitor CHIR99021 under submerged culture conditions for 3-4 days to activate canonical Wnt signaling and induce hyperproliferation.
- Differentiation Phase: Transition to air-liquid interface (ALI) conditions with continued CHIR treatment for 14 days to assess differentiation capacity.
- Phenotypic Assessment: Evaluation of proliferation (EdU incorporation), differentiation (ciliated cell markers via acetylated β-tubulin), and pathway activation (nuclear p-β-cateninY489 localization).
Quantitative HTS (qHTS) Approaches
Recent advances have led to quantitative HTS (qHTS), a paradigm that pharmacologically profiles large chemical libraries through generation of full concentration-response relationships for each compound [68]. With accompanying curve fitting and cheminformatics software, qHTS data yields half maximal effective concentration (EC50), maximal response, and Hill coefficient (nH) for entire libraries, enabling assessment of nascent structure activity relationships (SAR) from primary screening data [68].
Table 2: Key Experimental Parameters in Wnt Pathway HTS
| Parameter | Typical Implementation | Variations and Considerations |
|---|---|---|
| Activation Method | GSK3β inhibitors (CHIR99021, GSK3XV) [70] | Recombinant Wnt3a [70], Wnt3a-conditioned medium [67], endogenous pathway activation in cancer models [67] |
| Readout System | TCF/LEF-driven luciferase reporter [70] [67] | GFP/fluorescent reporters, endogenous target measurement (AXIN2, MYCBP) [66], phospho-β-catenin localization [70] |
| Cell Models | Engineered cell lines (BEAS2B, BT-20) [70] [67] | Primary stem cells (ABSCs) [70], iPSC-derived neural progenitors [69], embryonic models [23] |
| Screening Scale | 20,000 compounds for initial screen [70] | ~1,500 FDA-approved compounds for focused libraries [69], natural product extracts from marine sources [67] |
| Secondary Validation | Proliferation assays (EdU), differentiation capacity, nuclear β-catenin localization [70] | Gene expression analysis, apoptosis assays, additional pathway specificity testing [23] |
Data Analysis and Hit Selection
Quality Control Metrics
High-quality HTS assays are critical for successful screening campaigns. Three important means of quality control include: (i) good plate design, (ii) selection of effective positive and negative controls, and (iii) development of effective QC metrics to identify assays with inferior data quality [68]. For Wnt pathway screens, effective controls typically consist of:
- Positive Controls: GSK3β inhibitors (CHIR99021) or Wnt3a-conditioned medium to activate signaling [70] [67]
- Negative Controls: DMSO vehicle alone to establish baseline signaling
- Inhibition Controls: Known Wnt inhibitors (where available) for inhibitor screens
Several quality-assessment measures have been proposed to evaluate data quality, including signal-to-background ratio, signal-to-noise ratio, signal window, assay variability ratio, and Z-factor [68]. The Z-factor is particularly important, with values above 0.5 indicating excellent assays; one Wnt-focused screen achieved a Z' factor of 0.82, indicating a highly reliable platform [70]. Strictly standardized mean difference (SSMD) has recently been proposed for assessing data quality in HTS assays and may provide advantages over traditional metrics [68].
Hit Selection Methods
The process of selecting compounds with desired effects (hits) employs different statistical approaches depending on screening design:
- Primary Screens Without Replicates: z-score method or SSMD, which capture data variability based on the assumption that every compound has the same variability as a negative reference [68]. Robust methods like z-score, SSMD, B-score, and quantile-based methods address outlier sensitivity [68].
- Screens with Replicates: t-statistic or SSMD that directly estimate variability for each compound without relying on the strong assumptions of z-score methods [68].
For Wnt pathway screens, additional confirmation steps are crucial. In the ABSC screen, initial hits that decreased TCF/LEF activity were further tested for their ability to reduce ABSC proliferation, induce ciliated cell differentiation, and decrease nuclear p-β-cateninY489 [70]. This multiparameter validation ensures identified compounds produce physiologically relevant pathway modulation.
Research Reagent Solutions
Table 3: Essential Research Reagents for Wnt Pathway HTS
| Reagent Category | Specific Examples | Function in Wnt Screening |
|---|---|---|
| Pathway Activators | CHIR99021 [70], GSK3XV [70], Wnt3a-conditioned medium [67], Recombinant Wnt3a [70] | Induce canonical Wnt signaling activation for inhibitor screens; establish positive controls and assay windows |
| Reporter Systems | TCF/LEF-firefly luciferase constructs (TopFlash) [67], TCF/LEF-GFP reporters | Quantify pathway activity through luminescent or fluorescent readouts; enable high-throughput measurement |
| Cell Models | BEAS2B bronchial epithelial cells [70], BT-20 TNBC cells [67], Human iPSC-derived neural progenitors [69], Primary airway basal stem cells [70] | Provide cellular context for screening; disease-relevant models for specific applications |
| Detection Reagents | Luciferase assay substrates, Hoechst stain [70], EdU proliferation assay [70], Antibodies for p-β-cateninY489 [70] | Enable measurement of pathway activity, viability, proliferation, and mechanistic follow-up |
| Specialized Media | Porcine zygote medium-3 (PZM-3) for embryonic cultures [23], Air-liquid interface (ALI) differentiation media [70] | Support specialized culture conditions for specific screening models |
Case Studies and Applications
Successful Identification of Wnt Inhibitors
Several screening campaigns have successfully identified novel Wnt pathway modulators:
Wnt Inhibitor Compound 1 (WIC1) [70]:
- Source: High-throughput drug screen of 20,000 small molecules
- Mechanism: Suppresses TCF/LEF activity, reduces ABSC proliferation, induces ciliated cell differentiation, decreases nuclear p-β-cateninY489
- Validation: Demonstrated efficacy in restoring airway homeostasis in models of dysregulated Wnt signaling
Marine-Derived Wnt Inhibitors [67]:
- Source: Ethanol extracts from 81 marine invertebrate specimens collected in the Kuril Basin
- Screening Model: TNBC cell line BT-20 with TopFlash reporter stimulated with Wnt3a-conditioned medium
- Outcome: Identification of highly specific anti-Wnt activities from Ophiura irrorata and other Pacific brittle stars targeting multiple levels within the Wnt pathway
iPSC-Derived Neural Progenitor Screen [69]:
- Scale: ~1,500 compounds from FDA-approved drugs and known bioactives
- Validation: Dose-responsive stimulation by known Wnt modulators (Wnt3a, GSK3 inhibitors, lithium) with robust reproducibility
- Outcome: Identification of multiple chemical and biological classes of novel small-molecule probes of Wnt/β-catenin signaling
Integration with Embryogenesis Research
The connection between HTS for Wnt modulators and embryogenesis research is particularly strong, as the Wnt pathway plays essential roles in early mammalian embryogenesis and stem cell maintenance/differentiation [2]. Studies in porcine embryos have revealed that IGF-1 promotes trophectoderm cell proliferation through activation of the Wnt/β-catenin pathway [23]. This interaction was demonstrated through rescue experiments where the suppressive effects of an IGF-1 receptor inhibitor (picropodophyllin) on developmental parameters and β-catenin levels were reversed by both IGF-1 and the Wnt/β-catenin signaling activator CHIR99021 [23]. Such embryogenesis models provide critical physiological context for evaluating Wnt modulators identified through HTS campaigns.
High-throughput screening for Wnt pathway modulators represents a powerful approach for identifying potential therapeutic compounds targeting this fundamental developmental pathway. The integration of physiologically relevant cellular models, robust reporter systems, and sophisticated data analysis methods has enabled successful identification of novel modulators with potential applications in regenerative medicine, cancer therapy, and basic research. As screening technologies continue advancing—with innovations like quantitative HTS, ultra-miniaturized platforms, and improved stem cell models—the pace of discovery is likely to accelerate. Furthermore, the intersection of Wnt-focused HTS with embryogenesis research continues to provide fundamental insights into both normal development and disease processes, creating a virtuous cycle of discovery and therapeutic development.
Overcoming Technical Challenges in Wnt Pathway Research
Addressing Pathway Crosstalk and Compensatory Mechanisms
Within the context of early embryogenesis, the precise regulation of cell fate, proliferation, and differentiation is paramount. The Wnt/β-catenin signaling pathway is a cornerstone of these processes, and its activity is therefore subject to stringent, multi-layered control. A critical aspect of this regulation involves intricate crosstalk with other signaling pathways and compensatory mechanisms that ensure robust developmental outcomes. This whitepaper provides an in-depth technical guide to the core principles, experimental methodologies, and analytical frameworks for investigating these complex interactions, with a specific focus on research in early embryogenesis.
The Wnt signaling pathway is an evolutionarily conserved system that plays a fundamental role in early mammalian embryogenesis and stem cell maintenance [2]. The pathway is broadly categorized into the canonical (Wnt/β-catenin-dependent) and noncanonical (β-catenin-independent) branches. The canonical pathway, the primary focus of this guide, culminates in the stabilization and nuclear translocation of β-catenin, where it partners with T-cell factor/lymphoid enhancer factor (TCF/LEF) transcription factors to activate target genes governing cell proliferation and differentiation [19] [9].
Given its pivotal role, aberrant Wnt pathway activity—whether due to mutation, epigenetic modification, or dysregulated crosstalk with other pathways—is a major contributor to tumor initiation and progression [19]. Consequently, understanding the mechanisms of crosstalk is not only essential for developmental biology but also for identifying novel therapeutic targets in oncology.
Key Mechanisms of Crosstalk and Compensation
Crosstalk between the Wnt pathway and other cellular systems adds a critical layer of regulatory complexity. The following mechanisms are among the most significant.
Crosstalk with Long Non-Coding RNAs (lncRNAs)
Long non-coding RNAs (lncRNAs), defined as RNA transcripts longer than 200 nucleotides that do not code for proteins, have emerged as crucial transcriptional and post-transcriptional regulators. They can fine-tune the outcome of the Wnt pathway through diversified mechanisms [71]. For instance:
- Positive Regulation: Some lncRNAs, such as those involved in a positive feedback loop with ASCL2, can reinforce Wnt pathway activity to control intestinal stem cell fate [71].
- Complex Formation: LncRNAs like ASBEL can interact with transcription factors such as TCF3, forming complexes essential for the tumorigenicity of colorectal cancer cells, thereby creating an alternative node for pathway modulation [71].
This regulatory layer allows for precise, context-specific control of Wnt pathway output, which is crucial during the finely-tuned stages of embryonic development.
Interaction with the IGF-1 Signaling Pathway
Recent research on porcine embryos has revealed a critical crosstalk mechanism between Insulin-like Growth Factor-1 (IGF-1) and the canonical Wnt pathway, particularly in the development of the trophectoderm (TE) [23]. The TE is the outer cell layer of the blastocyst, essential for implantation and placental formation. The study demonstrated that IGF-1 treatment significantly enhanced blastocyst formation rates and TE cell proliferation. These effects were mechanistically linked to the activation of the Wnt/β-catenin pathway, as evidenced by increased β-catenin levels and the expression of related genes. Crucially, the inhibition of the IGF-1 receptor suppressed developmental parameters and β-catenin levels, an effect that was rescued by a Wnt/β-catenin pathway activator [23]. This provides a compelling model of compensatory activation where one pathway can directly influence the core machinery of another.
Table 1: Quantitative Effects of IGF-1 on Porcine Embryo Development and Wnt Pathway Markers
| Parameter | Control Group | IGF-1 Treated Group | Measurement Method |
|---|---|---|---|
| Blastocyst Formation Rate | Baseline | Significantly Enhanced | Embryo morphology assessment [23] |
| Trophectoderm Cell Proliferation | Baseline | Increased | Cell counting/Immunochemistry [23] |
| β-catenin Protein Levels | Baseline | Increased | Immunofluorescence, Protein quantification [23] |
| Rescue of IGF-1R Inhibition | Not Applicable | Successful rescue by Wnt activator CHIR99021 | Blastocyst development assessment [23] |
Experimental Protocols for Investigating Crosstalk
To rigorously study these interactions, robust and reproducible experimental protocols are required. Below is a detailed methodology for investigating IGF-1/Wnt crosstalk in a porcine embryo model, which can be adapted to other systems.
Detailed Protocol: Investigating IGF-1/Wnt Crosstalk in Embryos
Objective: To assess the functional interaction between IGF-1 signaling and the Wnt/β-catenin pathway during early embryonic development, specifically focusing on trophectoderm formation.
Materials and Reagents:
- Porcine Oocytes: Collected from ovaries obtained from a local slaughterhouse.
- Culture Media: In vitro maturation (IVM) medium, Porcine Zygote Medium-3 (PZM-3).
- Chemical Reagents:
- Recombinant IGF-1 (dissolved in distilled water).
- IGF-1 Receptor Inhibitor: Picropodophyllin (PPP, dissolved in DMSO).
- Wnt/β-catenin Pathway Activator: CHIR99021 (ChiR, dissolved in DMSO).
- Parthenogenetic Activation Reagents: Ionomycin, Cytochalasin B, 6-Dimethylaminopurine.
- Assay Kits: In Situ Cell Death Detection Kit (TUNEL assay).
- Antibodies: Primary antibodies for CDX2 (TE marker), YAP1, and β-catenin.
Methodology:
- In Vitro Maturation (IVM): Collect cumulus-oocyte complexes (COCs) from porcine ovaries. Culture selected COCs in IVM medium for 44 hours under controlled conditions (38.5°C, 5% CO₂) [23].
- Parthenogenetic Activation & In Vitro Culture (IVC):
- Activate matured oocytes with 15 µM ionomycin for 5 minutes.
- Subsequently, culture oocytes in PZM-3 medium containing cytochalasin B and 6-dimethylaminopurine for 4 hours.
- Transfer activated oocytes to fresh IVC medium with experimental treatments [23].
- Chemical Treatment:
- Treatment Groups: Include control, IGF-1 (e.g., 50 µM), PPP (IGF-1R inhibitor, e.g., 0.1 µM), and PPP + ChiR (1 µM) groups.
- Treatment Timing: To study late-stage effects, apply treatments on day 3 of culture and continue until day 6 [23].
- Outcome Assessment:
- Blastocyst Analysis: On day 6, image blastocysts and measure parameters like formation rate and blastocyst area.
- Immunofluorescence (IF): Fix blastocysts and stain with antibodies against CDX2 (to mark TE cells) and β-catenin. Use fluorescence microscopy to quantify protein levels and localization [23].
- TUNEL Assay: Fix and permeabilize blastocysts, then incubate with TUNEL enzyme to label apoptotic cells. Counterstain with DAPI to count total cell numbers and calculate apoptosis rates [23].
- Gene Expression Analysis: Islect RNA and perform RT-qPCR for Wnt target genes (e.g., AXIN2) and TE-specific markers.
Advanced Techniques: Quantitative Live-Cell Imaging and Computational Modeling
For a dynamic and quantitative understanding of pathway dynamics, cutting-edge techniques are required.
- Quantitative Live-Cell Imaging: Using CRISPR/Cas9-mediated genome editing, an endogenous CTNNB1 (β-catenin) pool can be tagged with a fluorescent protein (e.g., SGFP2) in haploid HAP1 cells. This allows for the quantification of CTNNB1 stabilization, translocation, and diffusion characteristics in real-time upon pathway activation using confocal microscopy and Fluorescence Correlation Spectroscopy (FCS) [72].
- Computational Modeling: Data from live-cell imaging (e.g., protein concentrations, diffusion coefficients) can be used to build computational models of WNT/CTNNB1 signaling. These models can predict the distribution of CTNNB1 across cellular compartments and identify key regulatory nodes, such as cytoplasmic destruction, nucleocytoplasmic shuttling, and nuclear retention [72].
The Scientist's Toolkit: Essential Research Reagents
A curated list of essential reagents for studying Wnt pathway crosstalk is provided below.
Table 2: Key Research Reagent Solutions for Wnt Crosstalk Studies
| Reagent / Tool | Function / Application | Example Use in Investigation |
|---|---|---|
| Recombinant IGF-1 | Activates IGF-1 receptor signaling; used to stimulate the pathway. | Investigating crosstalk with Wnt/β-catenin in trophectoderm proliferation [23]. |
| Picropodophyllin (PPP) | Selective inhibitor of the IGF-1 Receptor (IGF-1R). | Used to inhibit IGF-1 signaling and test dependence of Wnt activation on IGF-1R [23]. |
| CHIR99021 (ChiR) | Potent and selective inhibitor of GSK-3, activating Wnt/β-catenin signaling. | Used to directly activate the Wnt pathway and rescue effects of IGF-1R inhibition [23]. |
| CRISPR/Cas9 with HDR | Genome editing tool for endogenous protein tagging. | Used to create cell lines expressing fluorescently tagged endogenous β-catenin for live-cell imaging [72]. |
| WNT3A Protein | Canonical Wnt ligand; used to physiologically activate the pathway. | Stimulating Wnt signaling in cell culture models to study downstream effects [72]. |
| Signaling Hypergraphs | Computational representation modeling multi-molecule reactions and complexes. | Used for advanced pathway analysis to identify essential proteins and interactions for a specific response [73]. |
| (+)-trans-C75 | (+)-trans-C75, CAS:218137-86-1, MF:C14H22O4, MW:254.32 g/mol | Chemical Reagent |
| LY134046 | LY134046, CAS:849662-80-2, MF:C28H28N2O3S, MW:472.6 g/mol | Chemical Reagent |
Visualization and Computational Analysis of Pathway Networks
Traditional graph representations of pathways are limited in modeling complex interactions involving more than two molecules. Signaling hypergraphs provide a superior framework.
This hypergraph approach allows researchers to formulate and solve problems related to identifying the minimal set of proteins and interactions required to stimulate a specific downstream response, providing deeper insight into pathway logic and compensatory routes than traditional graphs [73].
Optimizing Temporal Control of Wnt Signaling in Experimental Models
The canonical Wnt signaling pathway is a fundamental regulator of cell fate decisions, embryonic development, and tissue homeostasis [2]. In early embryogenesis, precise temporal control of Wnt signaling is not merely beneficial but essential, as the duration of Wnt activation can determine specific cell lineage commitments [74] [23]. The pathway's function in mammalian embryogenesis and stem cell maintenance underscores its importance in developmental biology [2]. Recent advances in optogenetics and quantitative imaging have revealed that Wnt signal duration serves as a critical encoding mechanism for cellular information transmission, with cells interpreting the length of Wnt exposure to activate distinct genetic programs [74]. This technical guide synthesizes current methodologies for achieving precise temporal control of Wnt signaling in experimental models, with particular emphasis on applications within early embryogenesis research and drug development. We provide a comprehensive framework for optimizing temporal parameters, complete with quantitative benchmarks, experimental protocols, and visualization tools to enable researchers to precisely manipulate this crucial signaling pathway.
Molecular Mechanisms of Wnt/β-catenin Signaling
Core Pathway Components and Dynamics
The canonical Wnt/β-catenin pathway comprises an intricate network of molecular interactions that ultimately regulate the stability and nuclear localization of β-catenin, the pathway's key mediator [75]. In the absence of Wnt ligands, cytoplasmic β-catenin forms a complex with scaffold proteins Axin and Adenomatous Polyposis Coli (APC), leading to its phosphorylation by glycogen synthase kinase-3β (GSK3β) and subsequent proteasomal degradation [76] [75]. This degradation complex maintains low cytoplasmic β-catenin levels in unstimulated cells.
Upon Wnt activation, the pathway undergoes precise spatial and temporal dynamics. Wnt ligands bind to Frizzled receptors and LRP co-receptors, disrupting the degradation complex and allowing β-catenin to accumulate [76] [75]. The stabilized β-catenin then translocates to the nucleus, where it forms a complex with T-cell factor (TCF) to activate transcription of target genes [75]. Research in HEK293T cells has demonstrated that with Wnt3A stimulation, the total amount of β-catenin rises throughout the cell, with the increase occurring initially faster in the nuclear compartment [76]. This compartment-specific dynamics underscores the importance of spatial considerations in temporal control strategies.
Pathway Visualization
Figure 1: Canonical Wnt/β-catenin Signaling Pathway. The pathway exists in two states: OFF (without Wnt ligand, red) where β-catenin is degraded, and ON (with Wnt ligand, green) where β-catenin stabilizes and activates gene transcription.
Quantitative Analysis of Wnt Signaling Dynamics
Temporal Parameters and Gene Expression Relationships
Understanding the quantitative relationships between Wnt signal duration and downstream responses is fundamental to optimizing temporal control. Research using optogenetic Wnt activation in HEK293T cells has demonstrated that output gene expression (measured by TopFlash reporter) exhibits predictable scaling with signal duration [74].
Table 1: Quantitative Relationship Between Wnt Signal Duration and Gene Expression Output
| Signal Duration (hours) | Mean Expression (μg(t)) | Expression Variance (σg²(t)) | Optimal Information Capacity |
|---|---|---|---|
| 0 | Baseline | Low | N/A |
| 4 | ∠t | ∠t² | Limited |
| 12 | ∠t | ∠t² | Intermediate |
| 20 | ∠t | ∠t² | High |
The data reveal that mean output expression μg(t) scales linearly with signal duration (t), while variance σg²(t) scales approximately quadratically [74]. This relationship can be described by a gamma distribution, providing a mathematical framework for predicting cellular responses to timed Wnt exposures.
Information Transmission Capacity
The information capacity of the Wnt pathway is directly influenced by temporal encoding strategies. Studies have demonstrated that it is possible to reach an information capacity beyond 1 bit only through appropriate, discrete encoding of signals [74]. As effective noise decreases through cellular averaging mechanisms, the optimal encoding comprises more discrete input signals, transitioning into a continuous code in the small-noise limit.
Table 2: Information Transmission Optimization Strategies
| Noise Condition | Optimal Signal Encoding | Maximum Information Capacity | Experimental Approach |
|---|---|---|---|
| High noise | Binary (2 levels) | ~1 bit | Single-cell measurement |
| Moderate noise | Discrete (3+ levels) | 1-2 bits | Population averaging |
| Low noise | Continuous | >2 bits | Tissue-level synchronization |
These quantitative frameworks enable researchers to design temporal stimulation protocols that maximize information transmission for specific experimental contexts, whether studying binary cell fate decisions or more nuanced differentiation outcomes.
Experimental Approaches for Temporal Control
Optogenetic Control Systems
Optogenetic control represents the most precise method for temporal regulation of Wnt signaling. The opto-Wnt system utilizes light-sensitive domains fused to Wnt signaling components, enabling exquisite temporal precision down to minute-scale resolution [74]. The experimental workflow typically involves:
- Cell Preparation: Engineering HEK293T cells or other cell types to express optogenetic Wnt components [74]
- Stimulation Protocol: Applying light pulses of defined duration using high-throughput devices like LITOS plates [74]
- Signal Termination: Implementing precise cool-down periods (e.g., 4 hours) to allow pathway effectors to return to baseline [74]
- Output Measurement: Quantifying downstream responses using reporters like TopFlash or endogenous target genes [74]
This approach allows researchers to systematically explore how signal duration patterns influence cellular decision-making, particularly in embryonic stem cell models where Wnt dynamics direct lineage specification [41].
Small Molecule and Protein-Based Approaches
Chemical modulation provides an accessible alternative for temporal control, particularly in systems where genetic engineering is challenging. Key approaches include:
- GSK3β Inhibitors (e.g., CHIR99021): Sustain pathway activation by preventing β-catenin phosphorylation [41]. Temporal control is achieved through timed wash-out or concentration gradients.
- Wnt Ligand Treatment: Recombinant Wnt proteins (e.g., Wnt3a) applied for defined durations, followed by thorough wash-out [41].
- Secreted Inhibitors (e.g., IWP2): Block Wnt production or secretion to terminate signaling at precise timepoints [41].
In porcine embryo studies, IGF-1 treatment during specific developmental windows (days 3-6 vs. days 0-3) demonstrated stage-specific effects on trophectoderm proliferation through Wnt/β-catenin activation [23]. This highlights the importance of developmental timing in intervention strategies.
Experimental Workflow Visualization
Figure 2: Experimental Workflow for Temporal Control of Wnt Signaling. The process involves preparation, stimulation with precise temporal parameters, and quantitative analysis with appropriate cool-down periods.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Temporal Control of Wnt Signaling
| Reagent Category | Specific Examples | Function in Temporal Control | Applications |
|---|---|---|---|
| Optogenetic Systems | Opto-Wnt constructs [74] | Light-activated pathway components for minute-scale precision | Information capacity studies, fate mapping |
| Chemical Activators | CHIR99021 [41], Wnt3a protein [41] | Pharmacological pathway activation with wash-out control | Embryonic stem cell differentiation, tissue patterning |
| Pathway Inhibitors | IWP2 [41], DKK1 [75] | Timed pathway inhibition to define critical windows | Developmental timing studies, cancer models |
| Reporters | TCF-GFP [41], TopFlash [74] | Real-time monitoring of pathway activity dynamics | Live-cell imaging, FACS analysis |
| Biosensors | FRAP, FCS, FDAP [77] | Quantitative measurement of ligand dynamics and binding | Extracellular trafficking studies |
| Experimental Models | HEK293T [74] [76], hESCs [41], Porcine embryos [23] | Physiologically relevant systems with defined responses | Developmental biology, translational research |
| 17-PA | 17-PA, CAS:694438-95-4, MF:C25H34O, MW:350.5 g/mol | Chemical Reagent | Bench Chemicals |
| 7BIO | 7BIO, MF:C16H10BrN3O2, MW:356.17 g/mol | Chemical Reagent | Bench Chemicals |
Applications in Embryogenesis and Disease Modeling
Lineage Specification in Embryonic Stem Cells
Temporal control of Wnt signaling has revealed profound insights into lineage specification during early embryogenesis. In human embryonic stem cells (hESCs), heterogeneous endogenous Wnt signaling creates an equilibrium of distinct lineage-specified progenitors [41]. Wnthigh hESCs exhibit enhanced clonogenic potential and preferentially differentiate into endodermal and cardiac lineages, while Wntlow hESCs generate primarily neuroectodermal cells [41]. By manipulating Wnt signaling duration using small molecules or recombinant proteins, researchers can direct differentiation toward specific lineages with enhanced efficiency.
Notably, the effects of temporal Wnt activation are context-dependent, influenced by culture conditions and concurrent signaling pathways. For example, in defined media, Wnt activation promotes hESC differentiation, while in serum-containing conditions, it supports self-renewal [41]. This underscores the importance of optimizing both temporal parameters and microenvironmental conditions.
Trophectoderm Development in Embryos
In porcine embryos, IGF-1 promotes trophectoderm cell proliferation by activating the Wnt/β-catenin pathway, with effects more pronounced when treatment occurs during specific developmental windows (days 3-6 vs. days 0-3) [23]. This demonstrates how developmental timing influences responsiveness to Wnt pathway modulation. Inhibition experiments using picropodophyllin (PPP) confirmed the specific involvement of Wnt signaling in trophectoderm formation, with rescued development upon Wnt activation [23].
Technical Considerations and Optimization Strategies
Addressing Experimental Noise and Variability
Cell-to-cell variability presents a significant challenge in temporal control experiments. Several strategies can enhance signal-to-noise ratio:
- Population Averaging: Increasing sample sizes to account for intrinsic noise [74]
- Tissue-level Synchronization: Coordinating signaling responses across cell populations [74]
- Multiple Output Measurements: Monitoring several pathway targets to improve inference accuracy [74]
- Mathematical Modeling: Using gamma distributions or similar frameworks to describe and predict noise characteristics [74]
Spatial Considerations in Temporal Control
The spatial dynamics of Wnt signaling introduce additional complexity to temporal control strategies. Research in HEK293T cells has revealed compartment-specific β-catenin dynamics, with nuclear accumulation occurring at different rates than cytoplasmic changes [76]. Computational models that incorporate spatial dimensions provide more accurate representations of pathway behavior and can inform temporal optimization approaches [76] [78].
Protocol Standardization and Reproducibility
The development of standardized protocols and provenance tracking, as implemented in tools like WebProv, enhances reproducibility across studies [78]. Documenting model relationships, experimental parameters, and validation approaches allows researchers to build upon existing temporal control strategies and compare results across different experimental systems.
Future Directions and Concluding Remarks
The optimization of temporal control in Wnt signaling represents a rapidly advancing frontier with significant implications for developmental biology, regenerative medicine, and disease modeling. Emerging approaches include:
- Multiplexed Temporal Control: Simultaneous regulation of Wnt signaling with complementary pathways
- In Vivo Optogenetics: Applying temporal precision to intact model organisms
- Machine Learning Optimization: Using computational approaches to identify optimal timing patterns for specific outcomes
- Clinical Translation: Applying temporal control principles to enhance therapeutic Wnt modulation
As these methodologies mature, the ability to precisely manipulate Wnt signaling timing will continue to provide fundamental insights into embryonic development and create new opportunities for therapeutic intervention in cancer, regenerative medicine, and congenital disorders. The frameworks and protocols outlined in this technical guide provide a foundation for advancing these efforts through rigorous, reproducible, and quantitatively sophisticated experimental design.
Navigating Species-Specific Differences in Pathway Regulation
The Wnt signaling pathway is an evolutionarily conserved mechanism that plays an essential role in the embryogenesis of higher eukaryotes, from diploblastic, radially symmetrical cnidarians to mice and humans [2]. This pathway functions as a critical regulator of cell proliferation, differentiation, and embryonic development, with particular importance in neural development, stem cell maintenance, and tissue morphogenesis [79] [2] [23]. While the core components of the pathway remain consistent across species, significant differences in protein concentrations, regulatory mechanisms, and functional outcomes present substantial challenges for researchers extrapolating findings across model organisms.
Understanding species-specific variations in Wnt pathway regulation is particularly crucial for drug development professionals aiming to translate preclinical findings into clinical applications. The pathway's involvement in both developmental processes and disease states, including various cancers and congenital disorders, underscores the importance of accurately modeling its behavior in relevant systems [79] [76]. This technical guide provides a comprehensive framework for identifying, quantifying, and addressing species-specific differences in Wnt pathway regulation, with specific emphasis on applications in early embryogenesis research.
Core Pathway Mechanisms and Comparative Biology
Canonical Wnt/β-catenin Signaling Architecture
The canonical Wnt/β-catenin pathway operates through a carefully regulated degradation complex that controls the stability of β-catenin, a dual-function protein involved in both cell adhesion and transcriptional regulation [76]. In the absence of Wnt signaling, β-catenin forms a complex with scaffold proteins Axin and Adenomatous Polyposis Coli (APC), facilitating phosphorylation by glycogen synthase kinase-3β (GSK3β) [76]. This phosphorylation targets β-catenin for proteasomal degradation, maintaining low cytoplasmic concentrations [76].
Upon activation by Wnt ligands, the degradation complex is disrupted, leading to stabilized β-catenin that accumulates in the cytoplasm and translocates to the nucleus [76]. Nuclear β-catenin forms a complex with T-Cell Factor (TCF) transcription factors, activating gene expression programs responsible for cell survival, proliferation, and differentiation [76]. The precise regulation of this pathway ensures proper embryonic development, with dysregulation leading to significant malformations and disease states [79].
Key Species-Specific Variations in Pathway Components
Table 1: Comparative Analysis of Wnt Pathway Components Across Species
| Component | Xenopus laevis | Mus musculus | Homo sapiens | Functional Implications |
|---|---|---|---|---|
| Axin | Low concentration relative to other pathway proteins [76] | Higher concentration [76] | Higher concentration [76] | Alters degradation complex kinetics and β-catenin turnover rates |
| APC | Not specified | Low concentration relative to other pathway proteins [76] | Low concentration [76] | Impacts complex formation and β-catenin phosphorylation efficiency |
| β-catenin degradation rate | Rapid [76] | Slower [76] | Slower [76] | Influences temporal dynamics of pathway activation and resolution |
| Response to HCMV infection | Not applicable | Neural progenitor disruption [79] | Wnt pathway dysregulation, neural damage [79] | Species-specific viral pathogenesis affecting embryonic development |
Recent studies have revealed that these compositional differences significantly impact pathway behavior. For instance, the degradation rate of mammalian β-catenin is much slower than in Xenopus oocyte cell-free extracts, suggesting important functional distinctions between these systems [76]. Additionally, research on Human Cytomegalovirus (HCMV) infection has demonstrated species-specific effects on Wnt signaling, with infections disrupting neural progenitor development in human systems and contributing to fetal malformations through Wnt pathway dysregulation [79].
Quantitative Spatial Dynamics: A Case Study in Cross-Species Analysis
Compartmental Modeling of β-catenin Dynamics
Advanced imaging and computational approaches have revealed significant spatial aspects of Wnt signaling that exhibit species-specific characteristics. Research in Human Epithelial Kidney cells (HEK293T) has demonstrated that β-catenin dynamics follow distinct patterns in cellular compartments [76]. Following Wnt3A stimulation, the total amount of β-catenin increases throughout the cell, but this increase occurs more rapidly in the nuclear compartment during the initial phase (approximately the first hour) [76]. When protein synthesis is inhibited, β-catenin decreases at similar rates in both nucleus and cytosol-membrane compartments, suggesting that diffusional transport occurs rapidly compared to β-catenin degradation in the cytosol [76].
These spatial dynamics have been formalized through computational models that account for compartment-specific behaviors. While earlier models based on Xenopus extracts utilized single-compartment approaches, more recent models incorporating mammalian data require two-compartment structures (nucleus and cytosol-membrane) to accurately reproduce experimental observations [76]. The inclusion of active transport mechanisms alongside passive diffusion has proven essential for modeling mammalian Wnt pathway dynamics.
Table 2: Quantitative Parameters of β-catenin Dynamics in HEK293T Cells
| Parameter | Cytosol-Membrane Compartment | Nuclear Compartment | Experimental Conditions |
|---|---|---|---|
| Initial increase rate | Slow | Fast (~first hour) [76] | Wnt3A stimulation |
| Decrease rate | Same as nuclear | Same as cytosol [76] | Cycloheximide treatment |
| Steady-state distribution | Variable | Variable | Cell-type specific |
| Transport mechanism | Passive diffusion and active transport [76] | Passive diffusion and active transport [76] | Model-dependent |
Experimental Protocol: 3D Confocal Quantification of Spatial Signaling Dynamics
Purpose: To acquire spatial and temporal quantitative data on target protein (e.g., β-catenin) concentrations in specific cellular compartments during pathway perturbation [76].
Materials:
- HEK293T cells (or other relevant cell line)
- Wnt pathway modulators (e.g., Wnt3A for activation, cycloheximide for protein synthesis inhibition)
- Fluorescent markers for cellular compartments (e.g., DAPI for nuclei, N-cadherin antibodies for cell boundary)
- Intensity calibration standards (e.g., InSpeck microspheres)
- Confocal microscopy system with 3D imaging capability
- Image analysis software (e.g., ImageJ, proprietary solutions)
Procedure:
- Cell Culture and Treatment: Culture HEK293T cells under standard conditions. Apply perturbations according to experimental design (e.g., Wnt3A stimulation or cycloheximide treatment) for designated time points (0, 1, 2, 4 hours) [76].
- Sample Preparation: Fix cells and stain with fluorescent markers to identify specific cellular compartments. Use DAPI for nuclear identification and appropriate antibodies (e.g., N-cadherin) for cell boundary demarcation [76].
- Intensity Calibration: Include 0.3% rated InSpeck microspheres as intensity calibration standards to standardize intensity levels between samples and time points [76].
- Image Acquisition: Acquire 3D confocal image stacks using standardized settings across all samples. Ensure adequate z-resolution to capture complete cellular architecture [76].
- Image Analysis: Process 3D image stacks to quantify protein concentrations in specific compartments (nucleus vs. cytosol-membrane). Combine intensities from all 2D images comprising the 3D signals rather than relying on single 2D sections [76].
- Data Normalization: Normalize fluorescence intensities using calibration standards to account for technical variations between imaging sessions [76].
- Computational Modeling: Construct compartment-specific models of signaling pathways based on quantitative spatial and temporal data [76].
Applications: This protocol enables researchers to generate cell-specific compartment models of signaling pathways, revealing transport dynamics and activation kinetics that may vary between species [76].
Pathway Cross-Talk and Context-Specific Regulation
IGF-1 and Wnt Signaling Interdependence
Research in porcine embryos has revealed significant cross-talk between IGF-1 signaling and the Wnt/β-catenin pathway, with important implications for early embryonic development [23]. IGF-1 treatment during early embryonic stages significantly enhances developmental parameters, particularly blastocyst formation rates, and specifically increases trophectoderm (TE) cell proliferation [23]. The TE is an essential component of the blastocyst, maintaining its structure and facilitating implantation [23].
Notably, these effects demonstrate temporal specificity, with IGF-1 treatment during the last 3 days of embryonic development (days 3-6) producing more pronounced effects compared to treatment during the first 3 days (days 0-3) [23]. This temporal variation underscores the importance of developmental context in pathway regulation. Mechanistically, IGF-1 promotes activation of the Wnt/β-catenin signaling pathway, increasing β-catenin levels and related gene expression [23]. When IGF-1 signaling is inhibited using picropodophyllin (PPP), developmental parameters are suppressed along with reduced β-catenin levels, impaired TE cell differentiation, and disrupted tight junction formation [23]. These effects can be rescued by both IGF-1 and the Wnt/β-catenin signaling activator CHIR99021 [23].
Experimental Protocol: Analyzing Ligand-Receptor Mechanisms in Paracrine Noncanonical Wnt Signaling
Purpose: To evaluate paracrine noncanonical Wnt signaling interactions between signal-sending and signal-receiving cells of different types [80].
Materials:
- Signal-sending cells (e.g., neural crest cells)
- Signal-receiving cells (e.g., C2C12 myoblasts)
- Cell culture reagents (DMEM, FBS, penicillin/streptomycin)
- Molecular biology tools for genetic manipulation (e.g., siRNA, expression vectors)
- Pharmacologic inhibitors/activators
- Immunostaining equipment (fixatives, permeabilization agents, antibodies)
- Wound-healing assay materials
- Imaging systems for phenotypic analysis
Procedure:
- Cell Culture Establishment: Culture signal-sending and signal-receiving cells separately using appropriate media and conditions. For C2C12 myoblasts, use DMEM with 10% FBS and 1% penicillin/streptomycin [80].
- Non-contact Coculture System: Establish a coculture system that allows paracrine interactions without direct cell contact between signal-sending and signal-receiving cells [80].
- Perturbation Experiments: Implement specific genetic, molecular, or pharmacologic perturbations to dissect the role of signal-sending versus signal-receiving molecules. For example, target the Wnt5a-ROR2 axis as a crucial noncanonical Wnt signaling pathway [80].
- Functional Assessment: Conduct wound-healing assays to evaluate directional migration of signal-receiving cells in response to paracrine signaling [80].
- Morphological Analysis: Perform immunostaining to assess changes in cellular architecture, including phalloidin staining to visualize cytoplasmic filopodia and lamellipodia formation [80].
- Rescue Experiments: Apply specific molecular or pharmacologic approaches to rescue phenotypic effects observed following perturbation [80].
Applications: This protocol enables researchers to model complex paracrine signaling interactions observed in vivo, identify specific ligand-receptor mechanisms, and evaluate cell-specific functional responses that may vary across species [80].
Pathophysiological Implications: Viral Infection and Neural Development
HCMV Infection and Wnt Pathway Dysregulation
Human Cytomegalovirus (HCMV) infection provides a clinically relevant example of Wnt pathway dysregulation with significant species-specific implications. As the most prevalent intrauterine infectious agent in low- and middle-income countries, HCMV disrupts the development of neural stem cells, leading to fetal malformations and abnormal structural and physiological functions in the fetal brain [79]. Research has demonstrated that HCMV infection dysregulates the canonical Wnt/β-catenin signaling pathway, disrupting normal embryonic development [79].
The mechanisms underlying HCMV-induced pathology include impaired neural rosette development in brain organoids, reduced nidogen-1 expression, and disruption of calcium signaling in neural progenitor cells and organoids [79]. These effects highlight the vulnerability of developing neural systems to pathway disruption and underscore the importance of appropriate model systems for studying these processes. Mouse models of CNS disease following congenital HCMV infections have provided valuable insights, but significant differences in viral pathogenesis and pathway regulation necessitate careful interpretation when extrapolating to human systems [79].
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Research Reagents for Studying Species-Specific Wnt Pathway Regulation
| Reagent Category | Specific Examples | Function/Application | Species Considerations |
|---|---|---|---|
| Pathway Activators | Wnt3A ligand [76], CHIR99021 [23] | Activate canonical Wnt/β-catenin signaling | Concentration effects may vary between species |
| Pathway Inhibitors | Picropodophyllin (PPP) [23], IWP-2 | Inhibit specific pathway components (e.g., IGF-1 receptor) | Inhibitor sensitivity may differ across species |
| Cell Lines | HEK293T [76], C2C12 myoblasts [80], Neural crest cells [80] | Model specific cellular contexts and responses | Endogenous pathway component expression varies |
| Detection Reagents | β-catenin antibodies [76], TUNEL assay kits [23] | Quantify protein localization and cellular responses | Antibody cross-reactivity must be verified across species |
| Spatial Analysis Tools | InSpeck microspheres [76], Compartment-specific markers [76] | Enable quantitative spatial analysis of pathway components | May require optimization for different model systems |
| Gene Expression Tools | TRAP-Seq, Bulk RNA sequencing [81] | Profile pathway activity and downstream responses | Transcriptional programs show species-specific differences |
| iMDK | iMDK, MF:C21H13FN2O2S, MW:376.4 g/mol | Chemical Reagent | Bench Chemicals |
Visualizing Pathway Architecture and Experimental Approaches
Canonical Wnt/β-catenin Pathway and Cross-Species Variation
Experimental Workflow for Cross-Species Pathway Analysis
The investigation of Wnt signaling pathway regulation across species reveals both conserved mechanisms and critical differences that significantly impact embryonic development and disease pathogenesis. Researchers and drug development professionals must account for species-specific variations in protein concentrations, pathway kinetics, and functional responses when designing experiments and interpreting results. The integrated application of spatial quantification techniques, computational modeling, and carefully designed comparative studies provides a robust framework for navigating these complexities. As research in this field advances, continued attention to species-specific pathway regulation will enhance the translational potential of preclinical findings and support the development of targeted therapeutic interventions for developmental disorders and diseases involving Wnt signaling dysregulation.
Challenges in Targeting Wnt Signaling for Therapeutic Development
The Wnt signaling pathway is a highly conserved system critically involved in orchestrating fundamental cellular functions during early embryogenesis, including proliferation, migration, survival, and cell fate determination [82]. This pathway is categorized into canonical (β-catenin-dependent) and non-canonical (β-catenin-independent) branches, both playing pivotal roles in developmental processes [82] [83]. During embryonic development, Wnt signaling provides both quantitative and directional information that coordinates growth and patterning, essential for the formation of complex multi-cellular organisms [83]. The pathway's regulation is particularly crucial in pluripotent stem cells (PSCs), where it influences self-renewal, potency, and somatic cell reprogramming [84]. Given its fundamental role in development and tissue homeostasis, dysregulated Wnt signaling has been extensively linked to the pathogenesis of various diseases, particularly cancer, making it a promising therapeutic target [82] [63]. However, developing effective therapies faces substantial challenges due to the pathway's complexity, pleiotropic effects, and essential physiological functions.
Molecular Mechanisms of Wnt Signaling
Canonical Wnt/β-Catenin Pathway
The canonical Wnt/β-catenin pathway serves as a critical regulatory mechanism for target gene expression within the nucleus [82]. In the OFF state (absence of Wnt ligands), β-catenin undergoes phosphorylation by a multiprotein destruction complex comprising Axin, adenomatous polyposis coli (APC), glycogen synthase kinase 3β (GSK3β), casein kinase 1α (CK1α), and other components [82] [63]. This phosphorylation marks β-catenin for ubiquitination by β-TrCP and subsequent proteasomal degradation, preventing its accumulation [82].
In the ON state (presence of Wnt ligands), Wnt proteins bind to Frizzled (Fzd) family receptors and lipoprotein receptor-related protein (LRP)-5/6 co-receptors [82] [85]. This binding recruits cytosolic disheveled (Dvl) proteins, disrupting the destruction complex and preventing β-catenin degradation [82]. The stabilized β-catenin then accumulates in the cytoplasm and translocates to the nucleus, where it associates with transcriptional coactivators and T-cell factor/lymphoid enhancer factor (TCF/LEF) transcription factors to initiate the expression of Wnt target genes that regulate cell cycle progression and survival [82] [63].
Non-Canonical Wnt Pathways
The non-canonical Wnt pathways function independently of β-catenin and are essential for regulating cell polarity, migration, and calcium signaling [82]. The Wnt/planar cell polarity (PCP) pathway involves Wnt ligands (e.g., Wnt5a, Wnt7, Wnt11) binding to Fzd receptors, initiating signaling through Dvl that triggers downstream effectors including Rho/Rac small GTPases and Jun N-terminal kinase (JNK) to regulate tissue polarity and cell motility [82]. The Wnt/calcium (Ca²âº) pathway also begins with Wnt binding to Fzd, activating phospholipase C (PLC) through G-protein signaling, resulting in the release of intracellular Ca²âº, which influences processes ranging from early embryonic development to inflammatory responses [82].
Diagram 1: Wnt Signaling Pathways Overview. This diagram illustrates the key components and flow of both canonical and non-canonical Wnt signaling pathways, highlighting the critical decision points that determine pathway activation and cellular outcomes.
Key Challenges in Therapeutic Targeting
Pathway Pleiotropy and On-Target Toxicity
The pleiotropic nature of Wnt signaling represents a fundamental challenge in therapeutic development. The pathway regulates diverse cellular functions including self-renewal, stemness, lineage commitment, and cell cycle regulation in pluripotent stem cells [84]. This functional diversity means that systemic inhibition of Wnt signaling inevitably disrupts essential physiological processes in healthy tissues. Clinical trials with Wnt inhibitors have reported significant adverse effects including bone fractures and gastrointestinal toxicity, consequences of disrupting Wnt's normal roles in bone homeostasis and intestinal epithelium maintenance [85]. The pathway's critical involvement in embryonic development and tissue homeostasis creates a narrow therapeutic window where effective anti-tumor concentrations often overlap with toxic thresholds [82] [85].
Intricate Pathway Crosstalk and Compensation
Wnt signaling engages in extensive crosstalk with other key signaling pathways, creating a complex regulatory network that complicates therapeutic targeting [82]. The pathway interacts with Hedgehog (Hh), Notch, Hippo, TGF-β/Smad, NF-κB, and PI3K/AKT signaling cascades, forming interdependent relationships that ensure precise cellular regulation under both physiological and pathological conditions [82]. For example, Wnt and Hh pathways collaboratively regulate growth factor expression during embryonic limb development [82]. This intricate network means that inhibiting a single component may trigger compensatory mechanisms through alternative pathways, potentially diminishing therapeutic efficacy or even promoting resistance [82] [85]. The dynamic interplay between canonical and non-canonical Wnt pathways adds another layer of complexity, as inhibition of one branch may enhance activity in the other [82].
Technical Challenges in Drug Development
Targeting protein-protein interactions involving β-catenin presents significant technical hurdles. The interaction between β-catenin and TCF/LEF transcription factors occurs through large, relatively flat surfaces that are difficult to disrupt with small molecules [63] [85]. Developing inhibitors with sufficient binding affinity and specificity for these interfaces has proven challenging. Additionally, the structural similarity between different Fzd receptors makes achieving subtype specificity difficult, potentially leading to off-target effects [85]. The absence of reliable biomarkers for patient stratification and response monitoring further impedes clinical development, as it complicates the identification of patients most likely to benefit from Wnt-targeted therapies [63].
Table 1: Major Challenges in Wnt-Targeted Therapeutic Development
| Challenge Category | Specific Challenges | Potential Consequences |
|---|---|---|
| Pathway Biology | Pleiotropic effects across tissues and developmental stages [84] | On-target toxicity in bone, gastrointestinal tract, and other healthy tissues [85] |
| Signaling Complexity | Extensive crosstalk with Hedgehog, Notch, Hippo, and other pathways [82] | Compensatory activation of alternative pathways and development of resistance [85] |
| Molecular Targeting | Difficulties in disrupting β-catenin/TCF protein-protein interactions [63] | Limited efficacy of small molecule inhibitors and lack of drug binding pockets [85] |
| Clinical Development | Lack of reliable biomarkers for patient stratification [63] | Inability to identify responsive patients and monitor treatment efficacy [63] [85] |
| Therapeutic Window | Essential physiological functions in stem cell maintenance and tissue homeostasis [84] [82] | Narrow therapeutic index limiting tolerable dosing [85] |
Current Therapeutic Strategies and Limitations
Targeted Inhibition Approaches
Several strategic approaches have emerged to target different components of the Wnt signaling pathway, each with distinct mechanisms and limitations:
Table 2: Current Therapeutic Strategies Targeting Wnt Signaling
| Therapeutic Approach | Molecular Targets | Mechanism of Action | Development Challenges |
|---|---|---|---|
| Porcupine (PORCN) Inhibitors | Wnt ligand secretion [85] | Blocks palmitoylation of Wnt ligands, preventing their secretion and activation of receptors [85] | Gastrointestinal toxicity due to disruption of gut stem cell maintenance [85] |
| Tankyrase (TNKS) Inhibitors | Axin stabilization [85] | Inhibits tankyrase-mediated Axin degradation, promoting β-catenin destruction [85] | Limited efficacy as monotherapy and potential bone toxicity [85] |
| FZD-Targeted Antibodies | Frizzled receptors [63] [85] | Blocks Wnt binding to FZD receptors; some specificity for FZD subtypes (e.g., FZD7) [63] | Specificity challenges due to receptor similarity; vantictumab showed bone-related adverse events [63] [85] |
| β-catenin/TCF Complex Inhibitors | Nuclear β-catenin transcription [63] [85] | Disrupts formation of β-catenin/TCF transcriptional complex [63] | Difficulty in achieving effective intracellular concentrations and target disruption [63] |
| Antibody-Drug Conjugates (ADCs) | FZD7 and other surface targets [63] | Targeted delivery of cytotoxic payloads to Wnt-activated cells [63] | Limited to tumors with specific receptor expression patterns [63] |
Emerging Technologies and Innovative Approaches
Recent advances have introduced novel targeting modalities that address some traditional challenges. Photoswitchable agonists represent an innovative approach enabling spatiotemporal regulation of Wnt signaling [86]. These compounds, such as the azo derivative based on BML-284, undergo reversible trans-cis isomerization upon visible light irradiation, with only the cis isomer activating Wnt signaling [86]. This technology allows precise activation of the pathway at specific regions of interest in model systems, achieving ~88% agonist activity compared to non-photoswitchable controls [86]. Such approaches could potentially enable more localized modulation of Wnt signaling, reducing systemic toxicity.
Combination therapy strategies represent another promising direction. Integrating Wnt inhibitors with other therapeutic modalities, such as immune checkpoint blockers, conventional chemotherapy, or targeted agents against complementary pathways, may enhance efficacy while allowing dose reduction of individual components [85]. The development of FZD7-targeted antibody-drug conjugates like septuximab vedotin (F7-ADC) demonstrates the potential of leveraging Wnt pathway components for targeted delivery of cytotoxic payloads, showing strong anti-tumor activity and favorable safety profiles in preclinical models [63].
Experimental Models and Methodologies
Research Reagent Solutions
Table 3: Essential Research Reagents for Wnt Signaling Studies
| Research Reagent | Category/Type | Experimental Function | Application Context |
|---|---|---|---|
| BML-284 | Small molecule agonist [86] | Canonical Wnt pathway activation; reference compound for photoswitchable derivatives [86] | Baseline Wnt activation in control experiments; comparator for novel agonist development [86] |
| Photoswitchable Azo Derivatives | Optochemical agonists [86] | Spatiotemporal control of Wnt activation via light-induced isomerization [86] | Precision manipulation of Wnt signaling in specific cell populations or regions [86] |
| Reporter Plasmid Constructs | Molecular biology reagent | Luminescence-based quantification of Wnt pathway activity via TCF/LEF-responsive elements [86] | High-throughput screening of Wnt modulators; mechanistic studies of pathway regulation [86] |
| FZD7-Targeting Antibodies | Biological inhibitors [63] | Specific blockade of FZD7 receptor function; component of ADC development [63] | Functional studies of FZD7 role in cancer; validation of FZD7 as therapeutic target [63] |
| PORCN Inhibitors | Small molecule antagonists [85] | Inhibition of Wnt ligand palmitoylation and secretion [85] | Evaluation of Wnt ligand-dependent signaling; assessment of secretory pathway targeting [85] |
Key Experimental Protocols
Photoswitchable Agonist Development and Testing
The development of photoswitchable Wnt agonists follows a structured methodology that enables precise spatiotemporal control of pathway activation [86]:
Design and Synthesis: Based on the known Wnt agonist BML-284, researchers designed and synthesized photoswitchable azo derivative compounds containing diazene units that undergo reversible trans-cis isomerization [86].
Photochemical Characterization: The compounds are subjected to irradiation with visible light of specific wavelengths to confirm reversible isomerization and determine optimal conditions for cis-state formation and stability [86].
Biological Activity Assessment:
- Utilize luminescence-based reporter assays in cultured cells with TCF/LEF-responsive elements driving luciferase expression
- Compare agonist activity between irradiated (cis-dominant) and non-irradiated (trans-dominant) conditions
- Quantify efficacy relative to non-photoswitchable BML-284 control (e.g., ~88% activity for compound 2) [86]
Spatiotemporal Validation: Demonstrate selective activation of Wnt signaling in specific regions of interest in model cell culture systems using patterned light irradiation, confirming the ability for precise spatial control [86].
Biomarker Identification and Validation
The identification of reliable biomarkers for predicting Wnt signaling sensitivity involves multi-platform approaches [63]:
Genetic Alteration Analysis: Screen for mutations in key Wnt pathway components (APC, CTNNB1, AXIN1/2) using sequencing technologies and correlate with pathway activation status [63].
Expression Profiling:
- Quantify mRNA levels of Wnt target genes (e.g., c-Myc, cyclin D1, Survivin) via qPCR or RNA-seq
- Assess protein expression and localization (e.g., nuclear β-catenin accumulation) through immunohistochemistry and Western blotting [63]
Functional Assays:
- Implement reporter assays to directly measure pathway activity
- Correlate pathway activity with therapeutic response in preclinical models [63]
Proteomic Approaches: Utilize advanced proteomics to identify protein signatures associated with Wnt pathway activation, including post-translational modifications that influence pathway activity [63].
Diagram 2: Experimental Workflow for Wnt-Targeted Therapeutic Development. This diagram outlines the key stages in the research and development pipeline for Wnt-targeted therapies, from initial target identification through translational research approaches aimed at clinical application.
Future Perspectives and Concluding Remarks
The future of Wnt-targeted therapeutic development lies in overcoming the fundamental challenge of achieving pathway modulation in diseased tissues while preserving essential physiological functions. Several promising directions are emerging. First, tissue-specific targeting approaches utilizing antibody-drug conjugates, nanoparticles, or tissue-restricted delivery systems may enhance therapeutic windows by concentrating drug effects at disease sites [63] [85]. Second, the development of context-dependent inhibitors that leverage differential pathway activation states between normal and diseased cells could improve specificity [85]. Third, advanced biomarker strategies incorporating multi-omic profiling and functional readouts are essential for identifying patient subsets most likely to benefit from Wnt-targeted therapies [63].
The integration of emerging technologies such as optochemical tools [86], proteolysis-targeting chimeras (PROTACs), and RNA-based therapeutics may provide novel avenues for targeting previously "undruggable" components of the pathway. Furthermore, sophisticated combination therapy regimens that simultaneously modulate Wnt signaling and complementary pathways offer potential for enhanced efficacy and reduced toxicity [85]. As our understanding of the pleiotropic functions of Wnt signaling in development and disease continues to advance [84] [82] [83], so too will our ability to develop safer, more effective therapeutic interventions for cancer and other diseases driven by aberrant Wnt pathway activation.
Strategies for Precise Modulation of Pathway Activity Levels
The Wnt signaling pathway is an evolutionarily conserved cascade that orchestrates fundamental cellular processes including proliferation, differentiation, polarity, migration, metabolism, and survival [87]. Its precise regulation is particularly critical during early mammalian embryogenesis, where it specifies pattern formation and functions in the differentiation and maintenance of stem cells both in vivo and in vitro [2]. The pathway is categorized into the canonical (β-catenin-dependent) and non-canonical (β-catenin-independent) branches, with the canonical Wnt/β-catenin pathway playing an indispensable role in establishing dorso-ventral and anterior-posterior axes and being essential for normal gastrulation [82] [88]. Research has demonstrated that β-catenin knockout embryos are lethal as they fail to develop mesodermal and endodermal germ layers [88], highlighting the absolute requirement for precise pathway activity regulation during these critical developmental windows. Beyond development, aberrant Wnt activation is causally linked to cancer, degenerative disorders, metabolic syndromes, and developmental abnormalities [87], making the strategic modulation of this pathway a paramount focus for basic research and therapeutic development.
Molecular Mechanisms of Wnt Pathway Activity
Canonical Wnt/β-Catenin Signal Transduction
The canonical Wnt/β-catenin signaling pathway centers on the regulation of β-catenin stability and nuclear translocation [82]. In the absence of Wnt ligands, cytoplasmic β-catenin is constantly phosphorylated by a multiprotein destruction complex comprising Axin, Adenomatous Polyposis Coli (APC), Glycogen Synthase Kinase 3β (GSK3β), Casein Kinase 1α (CK1α), and other proteins [82] [88]. This phosphorylation marks β-catenin for ubiquitination by β-TrCP and subsequent proteasomal degradation, maintaining low cytoplasmic levels [75]. When Wnt ligands bind to Frizzled (Fzd) family receptors and LRP5/6 co-receptors, they initiate a signaling cascade that recruits cytosolic Dishevelled (Dvl) and disrupts the destruction complex [82]. This disruption stabilizes β-catenin, allowing it to accumulate in the cytoplasm and translocate to the nucleus, where it associates with T-cell factor/Lymphoid enhancer factor (TCF/LEF) transcription factors and co-activators to initiate transcription of Wnt target genes [75] [82].
Non-Canonical Wnt Pathways
The non-canonical Wnt pathway functions independently of β-catenin and comprises two major intracellular signaling cascades: the Wnt/Planar Cell Polarity (PCP) pathway and the Wnt/Calcium (Ca²âº) pathway [82]. In the PCP pathway, Wnt ligands such as Wnt5a, Wnt7, and Wnt11 bind to Fzd receptors, initiating signaling through Dvl that triggers downstream Rho/Rac small GTPases and Jun N-terminal kinase (JNK) to modulate cell polarity and migration [82]. In the Wnt/Ca²⺠pathway, Wnt binding activates phospholipase C (PLC) through G-protein signaling, resulting in the release of intracellular Ca²âº, which can influence cell adhesion, motility, and immune responses [82].
Figure 1: Wnt Signaling Pathway Mechanisms. The diagram illustrates the core components and flow of both canonical (β-catenin-dependent) and non-canonical (β-catenin-independent) Wnt signaling pathways.
Quantitative Profiling of Wnt Modulation Strategies
Small Molecule Inhibitors
Small molecules represent the most diverse category of Wnt pathway modulators, with compounds targeting various pathway components through distinct mechanisms of action.
Table 1: Small Molecule Inhibitors of Wnt Signaling
| Target | Compound Examples | Mechanism of Action | Effective Concentration Range | Developmental Stage Applicability |
|---|---|---|---|---|
| Porcupine | LGK974, IWP compounds | Inhibits Wnt ligand secretion by blocking Porcupine-mediated palmitoylation | 0.1-1 µM | Early embryogenesis, stem cell maintenance |
| Tankyrase | XAV939, IWR compounds | Stabilizes Axin by inhibiting Tankyrase, enhancing β-catenin degradation | 1-10 µM | Mid-late embryogenesis, cancer models |
| β-catenin/TCF | PRI-724, FOG-001 (clinical) | Disrupts β-catenin-CBP or β-catenin-TCF interactions | 0.5-5 µM | Late embryogenesis, differentiation studies |
| CK1α | Pyrvinium | Activates CK1α, promoting β-catenin phosphorylation and degradation | 10-100 nM | Early patterning, axis specification |
| GSK3β | CHIR99021, BIO | Inhibits GSK3β activity, stabilizing β-catenin | 1-10 µM | Pluripotency maintenance, neural differentiation |
Biologics and Genetic Modulators
Biological and genetic approaches offer high specificity for targeting extracellular pathway components or achieving permanent transcriptional control.
Table 2: Biologics and Genetic Modulators of Wnt Signaling
| Modulator Type | Target | Examples | Mechanism | Application Context |
|---|---|---|---|---|
| Monoclonal Antibodies | Wnt Ligands | OMP-54F28 (Ipafricept) | Binds and sequesters multiple Wnt ligands | Cancer, tissue regeneration |
| Monoclonal Antibodies | Frizzled Receptors | OMP-18R5 (Vantictumab) | Blocks Wnt binding to Fzd receptors | Organoid development, cancer |
| Secreted Inhibitors | Extracellular regulators | DKK1, sFRPs, WIF1 | Natural antagonists that prevent receptor activation | Embryonic patterning, stem cell niches |
| Gene Therapy | Intracellular pathway | CRISPR/Cas9, RNAi | Permanent genetic modification of pathway components | Functional screening, disease modeling |
| Nanoparticles | Drug delivery | LNPs, polymeric nanocarriers | Targeted co-delivery of multiple modulators | In vivo studies, therapeutic applications |
Experimental Framework for Wnt Activity Modulation
Protocol for Small Molecule Inhibition in Embryonic Stem Cells
Objective: To precisely modulate Wnt/β-catenin signaling activity in mouse embryonic stem cells (mESCs) to study its role in pluripotency and differentiation.
Materials:
- mESC line (e.g., E14 or GS1)
- Serum + LIF medium or defined pluripotency medium
- Small molecule inhibitors (select based on target):
- Porcupine inhibitor: LGK974 (1 µM stock in DMSO)
- Tankyrase inhibitor: XAV939 (5 mM stock in DMSO)
- GSK3β inhibitor: CHIR99021 (10 mM stock in DMSO)
- Control: DMSO vehicle (0.1% final concentration)
- Cell culture reagents (PBS, trypsin, etc.)
Methodology:
- Culture mESCs in appropriate medium and passage to maintain pluripotency.
- Seed cells at 5×10ⴠcells/cm² in tissue culture-treated plates.
- After 24 hours, replace medium with fresh medium containing:
- Experimental condition: Target inhibitor at optimized concentration
- Control condition: DMSO vehicle only
- Incubate cells for desired duration (typically 24-72 hours).
- Assess pathway modulation through:
- Viability assay: MTT or CellTiter-Glo at 24h and 48h
- Gene expression: qRT-PCR for Axin2, Lef1, Sp5 (direct targets)
- Protein analysis: Western blot for β-catenin, phospho-β-catenin
- Functional readouts: Pluripotency marker staining, differentiation potential
Optimization Notes:
- Perform dose-response curves (typically 0.1-10 µM) to establish EC₅₀ for each inhibitor
- Time-course experiments (6h-72h) to determine optimal treatment duration
- Combine with pathway activation (e.g., CHIR99021) for rescue experiments
- For developmental studies, apply inhibitors during specific differentiation windows
Protocol for Genetic Modulation Using CRISPR/Cas9
Objective: To create stable Wnt pathway component knockout or knockin mESC lines for developmental studies.
Materials:
- CRISPR/Cas9 plasmid system (e.g., px330 with gRNA expression)
- gRNAs targeting Wnt pathway genes (APC, β-catenin, Axin, etc.)
- mESC line with high transfection efficiency
- Selection markers (puromycin, blasticidin, etc.)
- SURVEYOR or T7E1 mutation detection kit
Methodology:
- Design and validate 3-5 gRNAs per target using online tools (e.g., CRISPOR)
- Clone gRNAs into CRISPR/Cas9 vector with selection marker
- Transfect mESCs using appropriate method (electroporation, lipofection)
- Select transfected cells with appropriate antibiotic (48-72 hours)
- Isolate single clones by limiting dilution or FACS
- Screen clones by:
- Genomic DNA extraction and sequencing of target locus
- SURVEYOR assay to detect editing efficiency
- Western blot to confirm protein level changes
- Characterize validated clones for:
- Wnt pathway activity (TOPFlash reporter, target gene expression)
- Pluripotency maintenance (Nanog, Oct4 expression)
- Differentiation potential (embryoid body formation)
Validation Criteria:
- Sanger sequencing confirmation of indels or precise edits
- >70% reduction in target protein expression by Western blot
- Altered Wnt pathway activity in reporter assays
- Maintained pluripotency in baseline conditions
- Changed differentiation capacity in directed differentiation protocols
Figure 2: Experimental Workflow for Wnt Pathway Modulation. The diagram outlines key decision points and methodological steps for implementing different Wnt modulation strategies in research settings.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents for Wnt Pathway Studies
| Reagent Category | Specific Examples | Function/Application | Key Considerations |
|---|---|---|---|
| Pathway Reporters | TOPFlash/FOPFlash, AXIN2-d2EGFP, TCF/LEF-GFP | Readout of β-catenin/TCF transcriptional activity | Normalize to Renilla luciferase; account for signal stability in live-cell imaging |
| Cell Lines | HEK293T (high transfection efficiency), L-Wnt3A (Wnt ligand source), RKO (APC mutant), mESCs (developmental studies) | Model systems for pathway manipulation | Verify baseline Wnt activity; check for endogenous mutations in pathway components |
| Detection Antibodies | Anti-β-catenin (total), anti-phospho-β-catenin (Ser33/37, Thr41), anti-Axin2, anti-active-β-catenin (ABC) | Assessment of pathway component expression and activation | Use phospho-specific antibodies for destruction complex activity; validate specificity |
| Recombinant Proteins | Wnt3a, Wnt5a, Dkk1, sFRP1 | Pathway activation or inhibition | Verify bioactivity; consider carrier protein effects; optimize concentration for specific cell type |
| Critical Assays | Co-IP for β-catenin/TCF interactions, RNA-seq for target genes, chromatin immunoprecipitation (ChIP) | Mechanistic studies of pathway function | Include appropriate controls (e.g., IgG for IP, input samples for ChIP) |
Advanced Applications and Technical Considerations
Nanomedicine Approaches for Targeted Modulation
Advanced drug delivery systems represent the cutting edge of Wnt pathway modulation strategies, particularly for in vivo applications and potential therapeutic translation. Lipid nanoparticles (LNPs), polymeric nanocarriers, and exosome-based platforms enhance the targeted accumulation of metabolic inhibitors and immunomodulatory agents while minimizing systemic toxicity [87]. These systems can be engineered to exploit tumor-specific metabolic markers (e.g., GLUT1, macropinocytic vesicles) or tissue-specific surface proteins, improving drug accumulation in desired locations while minimizing off-target effects [87]. For embryonic studies, such targeted approaches could theoretically enable tissue-specific pathway modulation, though this application remains largely exploratory. The development of nanocarriers for co-delivery of multiple Wnt pathway inhibitors (e.g., Porcupine inhibitor with tankyrase inhibitor) presents a promising strategy for overcoming the metabolic plasticity often observed in Wnt-driven systems [87].
Integration with Other Signaling Pathways
Effective Wnt pathway modulation requires consideration of its extensive crosstalk with other developmental signaling pathways. Wnt engages in complex interactions with Hedgehog (Hh), Notch, Hippo, TGF-β/Smad, NF-κB, and PI3K/AKT pathways [82]. For instance, during embryonic limb development, Wnt and Hh pathways collaboratively regulate growth factor expression, influencing cell differentiation and tissue morphology [82]. Research indicates that Hh signaling can potentiate Wnt pathway activity, while Wnt signaling modulates Hh effectors—a dynamic interplay essential in tissue regeneration and cancer progression [82]. Additionally, Wnt signaling intersects with the Hippo pathway through β-catenin and YAP/TAZ interactions, forming a complex feedback regulatory network vital for tissue size control and stem cell maintenance [82]. These interactions necessitate comprehensive pathway activity monitoring beyond just Wnt readouts when implementing modulation strategies.
Validation and Troubleshooting Framework
Robust validation of Wnt pathway modulation requires a multi-parameter approach that assesses both on-target efficacy and potential compensatory mechanisms. Essential validation steps include:
- Direct Target Engagement: Use cellular thermal shift assays (CETSA) or drug affinity responsive target stability (DARTS) to confirm compound binding to intended targets
- Pathway Activity Mapping: Employ multiple complementary readouts (transcriptional reporters, protein localization, target gene expression) to capture comprehensive pathway status
- Specificity Profiling: Evaluate effects on related pathways (e.g., Hedgehog, Notch) to identify off-target consequences
- Phenotypic Anchoring: Correlate molecular changes with functional outcomes (differentiation status, proliferation rates, metabolic profiles)
Common technical challenges include cell line-specific responses, temporal dynamics of pathway inhibition/activation, and compensatory pathway engagement. These can be addressed through time-course experiments, dose-response analyses, and combination approaches targeting multiple pathway nodes simultaneously. For developmental studies specifically, the timing of intervention is particularly critical, as Wnt pathway requirements shift dramatically during different embryological stages.
Cross-Species Analysis and Clinical Translation of Wnt Research
Pluripotent stem cells (PSCs) possess the remarkable capacity to self-renew indefinitely while maintaining the potential to differentiate into all cell types of the three germ layers. Research over the past decade has revealed that PSCs exist in multiple distinct but interrelated metabolic states—naive, formative, and primed—that correspond to different stages of early embryonic development [89]. The Wnt signaling pathway, an evolutionarily conserved system that regulates complex biological processes across all metazoan species, plays fundamentally different roles in maintaining these pluripotent states in mouse and human models [90] [91]. This divergence presents significant challenges for translational research, particularly in drug development and regenerative medicine, where human-relevant models are paramount. Understanding these species-specific differences is not merely an academic exercise but a critical prerequisite for developing effective therapeutic strategies. This review synthesizes current evidence on the divergent Wnt signaling requirements in human and rodent pluripotency paradigms, providing technical guidance for researchers navigating this complex landscape.
Fundamental Dichotomies in Pluripotency Regulation
Distinct Pluripotent States and Their Characteristics
Mouse and human pluripotent stem cells exhibit fundamental differences in their signaling requirements, epigenetic regulation, and transcriptional networks. While mouse embryonic stem cells (mESCs) represent a naive pluripotency state derived from the pre-implantation inner cell mass, conventional human embryonic stem cells (hESCs) typically reside in a primed pluripotency state more analogous to the post-implantation epiblast [89]. These states demonstrate different morphological characteristics, with naive mESCs forming dome-shaped colonies and primed hESCs exhibiting flatter colony morphology [89]. Female naive ESCs maintain two activated X chromosomes (XaXa), while primed cells display X chromosome inactivation (XaXi) [89]. Most significantly for drug development applications, these states show markedly different responses to core signaling pathways including Wnt, FGF, and TGFβ/Activin [90].
Table 1: Core Characteristics of Pluripotent States in Mouse and Human Systems
| Characteristic | Mouse Naive (mESCs) | Human Primed (hESCs) |
|---|---|---|
| Developmental Equivalence | Pre-implantation inner cell mass | Post-implantation epiblast |
| Colony Morphology | Dome-shaped, three-dimensional | Flat, two-dimensional |
| X-Chromosome Status | XaXa (both active) | XaXi (one inactive) |
| LIF/STAT3 Signaling | Required for self-renewal | Not required |
| FGF/Erk Signaling | Differentiation signal | Required for self-renewal |
| Wnt/β-catenin Pathway | Controversial role in self-renewal | Supports pluripotency in primed state |
| Chimera Formation | Efficient contribution | Limited or no contribution |
Divergent Wnt Signaling Requirements
The Wnt/β-catenin pathway exhibits opposing net effects on mouse versus human naive pluripotency [90]. For human naive PSC induction, synergistic inhibition of WNT/β-CATENIN, protein kinase C (PKC), and SRC signaling consolidates the induction of teratoma-competent naive human PSCs [90]. This requirement for Wnt inhibition stands in stark contrast to mouse naive PSCs, where Wnt/β-catenin signaling activity safeguards epigenetic stability and homeostasis [88].
In mouse ESCs, Wnt/β-catenin signaling plays a crucial role in maintaining epigenetic stability by ensuring normal DNA methylation at imprinting control regions (ICRs) [88]. When Wnt signaling activity is progressively silenced during extended in vitro culture, mESCs show loss of DNA methylation at ICRs, loss of chromatin repressor recruitment, and activation of retrotransposons, resulting in impaired differentiation capacity [88]. Sustained Wnt/β-catenin signaling in mESCs maintains normal ICR methylation and cellular homeostasis, positioning this pathway as a key regulator of genomic stability [88].
Table 2: Comparative Wnt Pathway Requirements in Pluripotency
| Aspect | Mouse System | Human System |
|---|---|---|
| Naive State Induction | Requires Wnt activation via GSK3 inhibition | Requires Wnt inhibition via TNKSi/IWR1 |
| Epigenetic Stability | Maintained by Wnt/β-catenin signaling | Mechanisms less dependent on Wnt |
| Role in Differentiation | Essential for mesoderm and endoderm formation | Context-dependent roles |
| Response to TNKSi | Compromises naive pluripotency | Boosts naive pluripotency markers |
| Response to GSK3i | Supports self-renewal in naive state | Compromises naive marker expression |
Experimental Paradigms and Methodologies
Human Naive Pluripotency Induction Protocol
The establishment of robust human naive pluripotency requires specific inhibition of the Wnt pathway, contrary to rodent paradigms. The following methodology has been demonstrated to effectively induce and maintain human naive PSCs:
Baseline Culture Conditions: Begin with conventional human PSCs maintained in primed conditions using mTeSR1 or equivalent medium on Matrigel or Geltrex-coated plates [90].
Transition Media Formulation: Prepare human enhanced naive stem cell medium (HENSM) containing:
- Tankyrase inhibitor (XAV939 or IWR-1): Stabilizes AXIN in the cytoplasm by inhibiting Tankyrase enzyme, effectively inhibiting Wnt/β-catenin signaling [90]
- SRC inhibitor (CGP77675): Works synergistically with Wnt inhibition to stabilize dome-like OCT4+ cells [90]
- Protein kinase C inhibitor (PKCi): Cooperates with Wnt and SRC inhibition to consolidate naive pluripotency [90]
- Optional boosters: JNK/P38 inhibition enhances naive pluripotency marker expression and cell viability [90]
Induction and Maintenance:
- Passage primed hPSCs as single cells into HENSM medium
- Culture at 37°C with 5% O₂ and 7% CO₂ for 7-14 days
- Monitor for emergence of dome-shaped colonies with distinct morphology
- Confirm naive status via ΔPE-OCT4-GFP reporter activation and surface marker expression [90]
Validation Assays:
- Assess teratoma formation capacity in immunocompromised mice
- Evaluate differentiation potential into trophoblast stem cells and extraembryonic naive endodermal cells [90]
- Analyze DNA methylation status at key imprinting control regions
Mouse Naive Pluripotency Preservation Protocol
Maintaining mouse ESCs in a naive state requires distinct conditions that often include Wnt pathway activation:
Standard mESC Culture:
Wnt Activity Monitoring:
Epigenetic Stability Assessment:
Wnt Signaling Pathway Visualization
Wnt Signaling Pathways and Species-Specific Pluripotency Regulation
The diagram illustrates the core Wnt signaling pathways and their divergent effects on mouse versus human naive pluripotency. The canonical Wnt/β-catenin pathway controls cell fate decisions through regulation of β-catenin stability and nuclear translocation, where it associates with TCF/LEF transcription factors to activate target genes [91] [4]. Non-canonical pathways, including planar cell polarity and Wnt/Ca²⺠pathways, regulate cytoskeletal organization and cell polarity through different intracellular messengers [91] [4]. Crucially, activation of canonical signaling promotes mouse naive pluripotency and epigenetic stability [88] while inhibiting human naive pluripotency induction [90], highlighting fundamental species-specific differences.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Investigating Wnt Signaling in Pluripotency
| Reagent Category | Specific Examples | Function/Application | Species Specificity |
|---|---|---|---|
| Wnt Pathway Inhibitors | IWR-1, XAV939 (TNKSi) | Stabilizes AXIN, inhibits Wnt/β-catenin signaling | Human naive induction [90] |
| Wnt Pathway Activators | CHIR99021 (GSK3i) | Inhibits GSK3, stabilizes β-catenin | Mouse naive maintenance [88] |
| Signaling Modulators | CGP77675 (SRCi), PKCi | Synergize with Wnt inhibition for naive state | Human specific [90] |
| Cytokines/Growth Factors | LIF, FGF2, ACTIVIN A | Support pluripotency in state-specific manner | Context-dependent [90] [89] |
| Reporter Systems | ΔPE-OCT4-GFP, Top-Gal | Monitor naive status and Wnt activity | Both species [90] [91] |
| Epigenetic Tools | DNA methylation assays, chromatin immunoprecipitation | Assess epigenetic stability and imprinting | Mouse emphasis [88] |
Implications for Drug Development and Disease Modeling
The divergent Wnt signaling requirements between human and rodent pluripotency paradigms have profound implications for pharmaceutical development and disease modeling. Understanding these differences is critical for:
Toxicity Screening: Compounds that modulate Wnt signaling may show species-specific toxicities due to differential effects on stem cell populations and tissue homeostasis [91].
Disease Modeling: Neurological disorders, congenital heart defects, and skeletal malformations associated with Wnt signaling defects [79] [92] [91] require human-specific models for accurate pathological recapitulation.
Regenerative Medicine: Cellular therapies derived from PSCs must account for species-specific signaling requirements to ensure safety and efficacy [90].
Cancer Research: Given Wnt pathway dysregulation in various cancers [91] [93], human-specific pluripotency models provide more relevant platforms for drug discovery.
Researchers must carefully consider these fundamental differences when designing preclinical studies and selecting appropriate model systems for specific applications.
Validating Therapeutic Targets Through Preclinical Models
Abstract The Wnt signaling pathway, a cornerstone of early embryogenesis governing cell fate, polarity, and migration, represents a prime target for therapeutic intervention in diseases like cancer. This technical guide delineates a structured preclinical framework for validating novel Wnt pathway targets. We detail the deployment of advanced in vitro and in vivo models, with a focus on human induced pluripotent stem cell (iPSC)-derived systems that recapitulate ontogenic processes. The document provides explicit protocols for key functional assays, standardized data presentation tables, and visual workflows to equip researchers with the methodologies necessary for robust, reproducible target validation in drug discovery pipelines.
The Wnt signaling pathway is an evolutionarily conserved system fundamental to early embryogenesis, including anterior-posterior axis patterning, neural tube morphogenesis, and stem cell differentiation [54] [14]. This pathway's pivotal role in regulating cell proliferation, fate determination, and tissue homeostasis makes it a compelling therapeutic target in oncology, given that its dysregulation is a hallmark of numerous cancers [94] [19] [95]. The pathway operates through two primary branches: the canonical (β-catenin-dependent) pathway, which regulates gene transcription, and the non-canonical (β-catenin-independent) pathways, which encompass the planar cell polarity (PCP) and Wnt/Ca²⺠pathways, controlling cell movement and polarity [96] [94] [14]. Validating modulators of this complex pathway requires preclinical models that can capture its intricate feedback loops and context-dependent activity. This guide outlines a strategic approach to this validation process, leveraging modern tools such as iPSC-derived models and luciferase reporter assays to bridge embryogenesis research and therapeutic development.
Core Principles of Preclinical Wnt Target Validation
A successful validation strategy rests on three pillars:
- Target Identification: Leveraging multi-omics data (e.g., from RNA-Seq and miRNA microarrays) to identify dysregulated Wnt pathway components (e.g., ligands, receptors, destruction complex members) in diseased versus normal tissues [97].
- Model Selection: Choosing model systems with high physiological relevance. iPSC-derived somatic cells preserve a patient-specific genetic background, making them superior for investigating Wnt pathway dynamics in a tissue-specific context [54].
- Functional Assessment: Implementing a suite of assays to measure changes in pathway activity, downstream transcriptional outputs, and resultant phenotypic consequences like cell proliferation, stemness, and migration [54] [95].
Preclinical Model Systems for Wnt Signaling
3.1 In Vitro Models
- Human iPSC-Derived Neural Stem Cells (NSCs): iPSCs from patients or healthy donors can be differentiated into NSCs, providing a limitless, genetically defined, and tissue-relevant source of human cells. These models are particularly apt for studying the role of Wnt in neurodevelopment and CNS disorders while maintaining the donor's genetic profile [54].
- Cancer Cell Lines: Established cell lines (e.g., from colorectal cancers with known APC mutations) are valuable for initial, high-throughput screening of therapeutic compounds. However, they may not fully represent the genetic heterogeneity of primary tumors [97] [95].
3.2 In Vivo Models
- Genetically Engineered Mouse Models (GEMMs): These models, engineered with conditionally activated or inactivated Wnt pathway genes (e.g., APC, CTNNB1), are indispensable for studying tumorigenesis, metastasis, and therapy response within a complete tumor microenvironment [95].
- Patient-Derived Xenografts (PDXs): PDX models, established by implanting patient tumor tissue into immunodeficient mice, better preserve the original tumor's histopathological and genetic characteristics, providing a robust platform for evaluating drug efficacy [95].
Key Experimental Protocols and Workflows
4.1 Protocol: TCF/LEF Luciferase Reporter Assay in iPSC-Derived Neural Stem Cells This assay quantitatively measures the activity of the canonical Wnt/β-catenin pathway [54].
- Objective: To functionally assess the activity of the canonical Wnt signaling pathway in response to agonists (e.g., Wnt3a), antagonists (e.g., DKK1), or genetic manipulation.
- Materials:
- Neural Stem Cells (NSCs) derived from human iPSCs.
- TCF/LEF Reporter Vector: A plasmid containing a firefly luciferase gene (luc2P) under the control of a minimal promoter and tandem TCF/LEF transcriptional response elements (TRE) [54].
- Control Renilla luciferase vector (e.g., pRL-TK for normalization).
- Lipofectamine or another non-viral transfection reagent.
- Wnt pathway agonist (e.g., recombinant Wnt3a) and antagonist (e.g., recombinant DKK1).
- Dual-Luciferase Reporter Assay System and a luminometer.
- Methodology:
- Cell Culture: Maintain iPSC-derived NSCs in appropriate neural expansion media.
- Transfection: Co-transfect cells with the TCF/LEF-firefly luciferase reporter vector and the control Renilla luciferase vector using a non-viral transfection method. A virus-free approach is recommended to preserve the native genetic background of the cells [54].
- Treatment: After transfection, treat cells with the experimental compounds (agonists/antagonists) or corresponding vehicle controls for a defined period (e.g., 24-48 hours).
- Luciferase Measurement: Lyse cells and measure firefly and Renilla luciferase activities using the Dual-Luciferase Assay System.
- Data Analysis: Normalize firefly luciferase activity to Renilla luciferase activity to control for transfection efficiency and cell viability. Express results as Relative Luminescence Units (RLU) or fold-change over control.
- Expected Outcome: A dose-dependent increase in normalized luciferase activity with Wnt agonist treatment and a decrease with antagonist treatment, confirming functional canonical pathway activity [54].
4.2 Protocol: Gene Expression Profiling of Wnt Pathway Components
- Objective: To identify and quantify dysregulation of Wnt signaling pathway genes and their associated miRNAs.
- Materials: Total RNA from paired diseased and normal tissue or treated/untreated cells, RNA-Seq or qRT-PCR platforms, miRNA microarray (e.g., Agilent Human miRNA Microarray) [97].
- Methodology: Isolate high-quality RNA, perform reverse transcription, and analyze using RNA-Seq or targeted qRT-PCR for mRNAs and microarrays for miRNAs. Use the KEGG pathway database for gene classification [97].
- Expected Outcome: Identification of significantly up- or down-regulated Wnt pathway genes and miRNAs, suggesting potential therapeutic targets. For example, in colorectal cancer, 59 of 138 Wnt pathway genes were found to be significantly dysregulated [97].
Data Presentation and Analysis
Table 1: Experimentally Validated miRNA-mRNA Interactions in the Wnt Pathway of Colorectal Cancer This table summarizes data from a study of 217 CRC cases, linking dysregulated Wnt pathway genes with associated miRNAs, highlighting potential post-transcriptional regulatory nodes for therapeutic intervention [97].
| Gene Symbol | Gene Name | Fold Change in CRC | Regulation in CRC | Associated miRNA(s) | miRNA Fold Change | Potential Interaction |
|---|---|---|---|---|---|---|
| MYC | MYC proto-oncogene | >1.5 | Up | hsa-miR-145-5p, hsa-miR-17-5p, hsa-miR-20a-5p | Varied | Indirect/Direct [97] |
| CCND1 | Cyclin D1 | >1.5 | Up | hsa-miR-193-3p, hsa-miR-19b-3p, hsa-miR-203a | Varied | Indirect/Direct [97] |
| SFRP4 | Secreted Frizzled Related Protein 4 | <0.67 | Down | hsa-miR-150-5p, hsa-miR-20b-5p | Varied | Indirect/Direct [97] |
| ROCK2 | Rho Associated Coiled-Coil Containing Protein Kinase 2 | >1.5 | Up | hsa-miR-221-3p, hsa-miR-27a-3p | Varied | Indirect/Direct [97] |
Table 2: The Scientist's Toolkit: Essential Reagents for Wnt Pathway Validation This table catalogs key reagents, their functions, and example applications in preclinical validation studies, as derived from the cited literature.
| Reagent / Tool | Category | Function in Validation | Example Application |
|---|---|---|---|
| TCF/LEF Reporter Plasmid | Molecular Tool | Measures canonical Wnt/β-catenin pathway transcriptional activity. | Quantifying pathway activation/inhibition in response to drug candidates [54]. |
| Recombinant Wnt3a | Pathway Agonist | Activates the canonical Wnt pathway by binding to FZD and LRP5/6 receptors. | Used as a positive control in reporter assays and functional studies [54]. |
| Recombinant DKK1 | Pathway Antagonist | Inhibits canonical signaling by binding to LRP5/6 co-receptors. | Used to establish baseline inhibition and study pathway repression [54] [14]. |
| iPSC-Derived Neural Stem Cells | Cellular Model | Provides a human, tissue-relevant, and genetically defined system. | Modeling Wnt signaling in neurodevelopment and CNS cancers [54]. |
| Anti-β-catenin Antibody | Detection Reagent | Visualizes and quantifies β-catenin protein levels and localization (cytoplasmic/nuclear). | Immunofluorescence, Western blot to confirm pathway activation [98]. |
| miRNA Microarray | Profiling Tool | Identifies miRNAs associated with dysregulated Wnt pathway genes. | Discovering post-transcriptional regulators of the pathway in specific cancers [97]. |
Visualizing Workflows and Pathways
Diagram 1: Preclinical Validation Workflow for Wnt Targets
This workflow outlines the sequential stages of target validation, from initial discovery to candidate selection.
Diagram 2: Core Canonical Wnt/β-catenin Signaling Pathway
This logic diagram summarizes the key mechanism of the canonical Wnt pathway, highlighting the critical switch controlled by β-catenin stability [94] [19] [14].
Wnt Pathway Dysregulation in Developmental Disorders
The Wnt signaling pathway is a highly conserved, animal-specific signal transduction cascade that functions as a central regulator of morphogenesis during early vertebrate development [99] [66]. It governs crucial processes including anterior-posterior axis formation, neural patterning, cell fate determination, proliferation, and migration [99] [100]. The pathway derives its name from the fusion of "Wingless" (Wg), the Drosophila segment polarity gene, and "Int-1," a vertebrate proto-oncogene first identified as an integration site for mouse mammary tumor virus (MMTV) [11] [100]. Wnt proteins are secreted glycoproteins that are lipid-modified, making them hydrophobic despite their secretion, which presents a unique challenge for their travel through the aqueous extracellular space [100]. The evolutionary conservation of Wnt pathway components from diploblastic cnidarians to mammals underscores its fundamental role in establishing the primary body axis and coordinating tissue self-organization [2] [66]. Within the context of early embryogenesis, precise spatial and temporal regulation of Wnt signaling is critical for normal development, and its dysregulation underlies a spectrum of human genetic disorders and developmental anomalies [99] [11].
Molecular Mechanisms of Wnt Signal Transduction
Wnt signaling proceeds through several distinct branches, primarily classified based on their dependence on the key effector β-catenin.
The Canonical Wnt/β-Catenin Pathway
The canonical pathway, centered on the regulation of β-catenin stability, is the most extensively characterized Wnt signaling branch [20] [30].
- Off-State (No Wnt Ligand): In the absence of a Wnt ligand, cytoplasmic β-catenin is targeted for proteasomal degradation by a multi-protein "destruction complex." This complex includes the scaffold proteins Axin and Adenomatous Polyposis Coli (APC), and the kinases Glycogen Synthase Kinase 3β (GSK-3β) and Casein Kinase Iα (CK1α). These kinases sequentially phosphorylate β-catenin, leading to its recognition by the E3 ubiquitin ligase β-TrCP and subsequent degradation. In the nucleus, transcription factors of the T-cell factor/Lymphoid enhancer factor (TCF/LEF) family are bound by transcriptional co-repressors, keeping Wnt target genes silent [20] [30] [100].
- On-State (Wnt Ligand Present): Signaling is initiated when a Wnt ligand binds to a seven-transmembrane Frizzled (Fz) receptor and a single-pass co-receptor from the Low-Density Lipoprotein Receptor-Related Proteins 5 or 6 (LRP5/6, also known as Arrow in Drosophila) [99] [20]. This ligand-receptor interaction promotes the formation of a signalosome at the plasma membrane, recruiting the cytoplasmic phosphoprotein Dishevelled (Dvl). Dvl, in turn, inhibits the destruction complex, preventing β-catenin phosphorylation and degradation. Consequently, β-catenin accumulates in the cytoplasm and translocates into the nucleus. There, it displaces co-repressors from TCF/LEF, recruits co-activators, and initiates the transcription of target genes such as c-Myc, cyclin D1, and Axin2, which regulate cell proliferation, survival, and differentiation [20] [11] [30].
Non-Canonical Wnt Pathways
The β-catenin-independent, non-canonical pathways regulate cell polarity, migration, and calcium signaling [99] [11]. The two primary non-canonical branches are:
- The Planar Cell Polarity (PCP) Pathway: This pathway regulates the polarization of cells within the plane of an epithelial sheet. It is activated when specific Wnt ligands (e.g., Wnt5a) bind to Frizzled receptors along with co-receptors such as Ryk (Related to tyrosine kinase) or Ror (Receptor tyrosine kinase-like orphan receptor). This interaction recruits and activates Dishevelled, which then signals through small GTPases like RhoA and Rac. Downstream effectors including Rho-associated kinase (ROCK) and c-Jun N-terminal kinase (JNK) are activated, leading to cytoskeletal reorganization and coordinated cell orientation, which is critical for processes like convergent extension during gastrulation [99] [11] [100].
- The Wnt/Ca²⺠Pathway: Activation of this pathway also involves Frizzled and specific co-receptors, but it leads to the activation of heterotrimeric G-proteins. This triggers phospholipase C (PLC), which generates inositol 1,4,5-trisphosphate (IP₃) and diacylglycerol (DAG). IP₃ mediates the release of calcium ions from intracellular stores into the cytoplasm. The increased intracellular calcium then activates calcium-sensitive enzymes such as Protein Kinase C (PKC), Calmodulin-dependent Kinase II (CamKII), and the phosphatase Calcineurin. This cascade influences cell adhesion, motility, and can have anti-inflammatory effects [99] [11].
The specific signaling outcome is often determined by the combinatorial interaction of different Wnt ligands with various Frizzled receptors and co-receptors in a specific cellular context [99] [100].
Table: Wnt-Related Developmental Disorders and Genetic Lesions
Dysregulation of Wnt signaling, through either gain-of-function or loss-of-function mutations, is a well-established cause of various congenital disorders. The table below summarizes key developmental disorders linked to aberrant Wnt pathway activity.
Table 1: Developmental Disorders Associated with Wnt Signaling Dysregulation
| Disorder | Affected Gene(s) | Molecular Lesion | Key Clinical/Phenotypic Features | Reference |
|---|---|---|---|---|
| Tetra-amelia Syndrome | WNT3 | Loss-of-function mutation | Complete absence of all four limbs, severe craniofacial and urogenital malformations | [11] |
| Osteoporosis-Pseudoglioma Syndrome | LRP5 | Loss-of-function mutation | Decreased bone mineral density, predisposition to fractures, and eye vascular abnormalities | [11] |
| Robinow Syndrome | ROR2, WNT5A | Mutations in non-canonical pathway components | Short-limbed dwarfism, distinctive facial features, vertebral segmentation defects | [66] |
| Caudal Regression Syndrome | WNT3A, other Wnt genes | Likely loss-of-function mutation | Sacral agenesis, lower limb defects, and urogenital anomalies | [99] (Inferred) |
| Hippocampal Defects (e.g., in mouse models) | WNT3A, LEF1, LRP6 | Loss-of-function mutation | Failure of hippocampal formation, specifically affecting the dentate gyrus | [99] |
| Skeletal Dysplasias | PCP pathway components (e.g., VANGL) | Mutations affecting planar cell polarity | Defects in bone growth and patterning, skeletal asymmetry | [11] |
Experimental Analysis of Wnt Signaling in Development
Research into the role of Wnt signaling in development and its dysregulation relies on a suite of well-established molecular, cellular, and biochemical techniques.
Key Methodologies and Workflows
A common experimental approach involves modulating the pathway in model systems and analyzing the phenotypic and molecular consequences. The workflow below outlines a typical experiment using a small molecule activator.
A recent study on porcine embryos provides a concrete example of such a methodology [23]. The researchers investigated the crosstalk between Insulin-like Growth Factor-1 (IGF-1) and the canonical Wnt pathway in trophectoderm development.
Detailed Experimental Protocol:
- In Vitro Embryo Culture: Porcine embryos were generated via in vitro maturation and parthenogenetic activation. They were cultured in Porcine Zygote Medium-3 (PZM-3) for 6 days [23].
- Chemical Treatment: Embryos were treated with:
- IGF-1: To activate its receptor signaling.
- Picropodophyllin (PPP): A specific IGF-1 receptor inhibitor, to suppress IGF-1 signaling.
- CHIR99021: A potent and selective GSK-3β inhibitor that acts as a canonical Wnt pathway activator by stabilizing β-catenin [23].
- Phenotypic Analysis:
- Blastocyst Development: Rates of blastocyst formation and total blastocyst area were quantified using image analysis software (e.g., ImageJ) [23].
- Trophectoderm (TE) Proliferation: The total cell number in blastocysts was assessed, and TE-specific proliferation was evaluated using immunofluorescence for TE-specific markers like CDX2 [23].
- Apoptosis Assay: A Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay was performed on fixed blastocysts to quantify the rate of apoptotic cell death [23].
- Molecular Analysis:
- Immunofluorescence: Blastocysts were stained with antibodies against β-catenin and YAP1 to analyze protein levels and subcellular localization (cytoplasmic vs. nuclear) [23].
- Gene Expression Analysis: Quantitative real-time PCR (RT-qPCR) was used to measure the mRNA levels of Wnt target genes and TE-specific functional genes [23].
The Scientist's Toolkit: Key Research Reagents
The following table lists essential reagents and their applications for studying Wnt signaling in developmental contexts.
Table 2: Essential Research Reagents for Wnt Pathway Analysis
| Reagent / Tool | Category | Function / Mechanism of Action | Example Use Case |
|---|---|---|---|
| CHIR99021 | Small Molecule Agonist | GSK-3β inhibitor; stabilizes β-catenin to activate canonical signaling | Rescue of β-catenin levels in inhibition models; study of pathway activation effects [23]. |
| IWP-2 / IWR-1 | Small Molecule Inhibitor | Porcupine (PORCN) inhibitor; blocks Wnt ligand secretion and activity | Determining if a phenotype is Wnt ligand-dependent [11]. |
| XAV939 | Small Molecule Inhibitor | Tankyrase inhibitor; stabilizes the Axin destruction complex to promote β-catenin degradation | Investigating consequences of pathway inhibition; modeling loss-of-function [11]. |
| Recombinant Wnt Proteins | Protein | Purified, active Wnt ligands (e.g., Wnt3a, Wnt5a) | Directly activating Wnt signaling in cell culture or ex vivo systems. |
| siRNA/shRNA vs. Ryk, Ror2 | Molecular Biology Tool | Gene knockdown to deplete specific Wnt co-receptors | Elucidating the role of non-canonical pathways in cell migration and polarity [99]. |
| Anti-β-catenin Antibody | Antibody | Detects total and active (non-phospho) β-catenin via Western Blot/IF | Assessing pathway activity through protein level and nuclear localization [23]. |
| TCF/LEF Reporter Plasmid | Reporter Assay | Luciferase construct with TCF/LEF binding sites; measures transcriptional activity | Quantifying canonical Wnt pathway output in live cells [99]. |
Therapeutic Targeting of Wnt Signaling
Given its pivotal role in development and disease, the Wnt pathway is an active area for therapeutic intervention. Strategies are primarily focused on inhibiting aberrantly active signaling, particularly in cancer, but also on modulating the pathway for regenerative purposes [11] [30].
Inhibitory Strategies:
- Porcupine (PORCN) Inhibitors: These small molecules (e.g., IWP-2) block the palmitoylation of Wnt ligands by the PORCN enzyme in the endoplasmic reticulum, which is essential for their secretion and activity. This provides a broad, upstream inhibition of Wnt signaling [11].
- Secreted Antagonists: Naturally occurring extracellular inhibitors like Dickkopf-1 (DKK1) and Secreted Frizzled-related Proteins (SFRPs) can be exploited therapeutically. DKK1 binds to LRP5/6 co-receptors, preventing formation of the active receptor complex, while SFRPs act as decoy receptors by sequestering Wnt ligands [11] [30].
- Tankyrase Inhibitors: Drugs like XAV939 inhibit tankyrase, a protein that promotes the degradation of Axin. By stabilizing Axin, these inhibitors enhance the formation of the β-catenin destruction complex, leading to increased β-catenin degradation [11].
- β-catenin/TCF Complex Disruptors: Compounds such as PRI-724 and ICG-001 interfere with the interaction between β-catenin and its transcriptional co-activator CBP in the nucleus, specifically blocking the transcription of pro-proliferative target genes without affecting β-catenin's adhesion function at the membrane [11] [30].
Emerging Research and Challenges: A significant challenge in targeting the canonical Wnt pathway is its fundamental role in adult tissue homeostasis, particularly in stem cell maintenance of the intestinal crypts. Systemic inhibition can therefore lead to severe on-target toxicity [30]. Emerging approaches aim to achieve greater specificity, such as using miRNA-based therapies (e.g., miR-34a mimics) to fine-tune oncogenic Wnt signaling, or developing biologics like the anti-sclerostin antibody romosozumab, which indirectly enhances Wnt signaling in bone to treat osteoporosis [11] [30]. Understanding the complex crosstalk between Wnt and other developmental pathways like Notch, Hedgehog, and TGF-β remains a critical focus for developing safe and effective treatments for both developmental disorders and cancer [11] [30].
The Wnt signaling pathway is a highly conserved system that orchestrates critical physiological processes, including cell proliferation, differentiation, migration, and apoptosis [101] [19]. Its name originates from a fusion of the Drosophila segment polarity gene wingless and the vertebrate homolog, integrated or int-1 [10]. This pathway plays a paramount role in early mammalian embryogenesis and stem cell maintenance, dictating cell fate decisions and pattern formation during development [2]. Given its fundamental role in regulating tissue homeostasis, dysregulation of Wnt signaling is a key driver in the pathogenesis of diverse human diseases, most notably cancer [101] [19]. This whitepaper delves into the molecular mechanics of the Wnt pathway, explores its dual role in human in vitro fertilization (IVF) and oncogenesis, summarizes key experimental methodologies for its study, and highlights emerging therapeutic strategies that target this pathway.
Molecular Mechanisms of Wnt Signaling
The Wnt pathway transduces signals from the extracellular environment to the nucleus, primarily through two branches: the canonical (β-catenin-dependent) pathway and the non-canonical (β-catenin-independent) pathways, which include the Planar Cell Polarity (PCP) and Wnt/Ca²⺠pathways [10] [14]. The following diagram illustrates the core components and flow of information in the canonical Wnt/β-catenin pathway.
The Canonical Wnt/β-catenin Pathway
In the absence of a Wnt signal, cytoplasmic β-catenin levels are kept low by a multiprotein destruction complex. This complex, composed of the scaffolding proteins Axin and Adenomatous Polyposis Coli (APC), along with the kinases Glycogen Synthase Kinase 3β (GSK3β) and Casein Kinase 1α (CK1α), facilitates the phosphorylation of β-catenin. Phosphorylated β-catenin is recognized by the E3 ubiquitin ligase β-TrCP, leading to its ubiquitination and subsequent degradation by the proteasome. In the nucleus, T-cell Factor/Lymphoid Enhancer-binding Factor (TCF/LEF) transcription factors associate with transcriptional repressors to keep Wnt target genes silent [75] [10] [19].
Upon binding of a Wnt ligand to a Frizzled (FZD) receptor and its co-receptor, Low-Density Lipoprotein Receptor-Related Protein 5/6 (LRP5/6), the signal is transduced to the protein Dishevelled (DVL). DVL becomes activated and recruits the Axin/GSK3β/CK1α complex to the plasma membrane, effectively disrupting the destruction complex. This disruption prevents β-catenin phosphorylation, allowing it to accumulate in the cytoplasm and subsequently translocate into the nucleus. Inside the nucleus, β-catenin binds to TCF/LEF, displacing corepressors and recruiting co-activators such as B-cell lymphoma 9 (BCL9), Pygopus, and CREB-binding protein (CBP/p300) to initiate the transcription of target genes like MYC and CCND1 (cyclin D1), which promote cell proliferation and survival [101] [19].
Non-canonical Wnt Pathways
The non-canonical pathways operate independently of β-catenin and LRP5/6. The Wnt/PCP pathway regulates cytoskeletal organization and cell polarity by activating small GTPases like Rho and Rac, which in turn influence actin polymerization and cell motility. The Wnt/Ca²⺠pathway triggers the release of intracellular calcium, activating kinases such as Protein Kinase C (PKC) and Calcium/calmodulin-dependent kinase II (CaMKII), which can influence cell adhesion and movements [101] [14]. These pathways are often activated by specific Wnt ligands, such as Wnt5a and Wnt11.
Extracellular Regulation and Ligand Dynamics
Wnt signaling is tightly regulated in the extracellular space. Wnt proteins themselves are secreted glycoproteins that undergo lipid modification (palmitoylation) by the enzyme Porcupine (PORCN) in the endoplasmic reticulum, a process critical for their secretion and activity [75] [19]. Their diffusion and distribution are influenced by interactions with Heparan Sulfate Proteoglycans (HSPGs) on cell surfaces [77]. Furthermore, a suite of secreted antagonists fine-tunes the pathway: the sFRP class (e.g., sFRPs, WIF-1) binds directly to Wnt ligands, while the Dickkopf (Dkk) class binds to the LRP5/6 co-receptor, preventing signal transduction [75] [14].
The Wnt Pathway in Early Embryogenesis and In Vitro Fertilization (IVF)
The Wnt signaling pathway is a master regulator of early mammalian embryogenesis. It is indispensable for pattern formation, axis specification, and cell fate determination during the initial stages of development [2] [10]. The pathway's role in maintaining pluripotency and self-renewal in stem cells is of particular relevance to the field of assisted reproduction, as these processes are fundamental to the viability of early embryos [2].
Clinical Implications for IVF
In the context of In Vitro Fertilization (IVF), a profound understanding of Wnt signaling could translate into significant improvements in clinical outcomes.
- Enhancing Embryo Quality and Viability: Precise modulation of Wnt signaling could be leveraged to improve the developmental competence of embryos in culture. By creating conditions that mimic the in vivo embryonic microenvironment, it may be possible to enhance blastocyst formation rates and embryo quality.
- Regulation of Trophoblast Development: The Wnt pathway is critically involved in the formation and function of the trophectoderm, the precursor to the placenta. Optimizing Wnt signaling could promote better trophoblast invasion and uterine implantation, thereby increasing the success rates of embryo transfers.
- Stem Cell-Based Applications: The use of embryonic stem cells (ESCs) derived from IVF embryos in research and regenerative medicine is heavily reliant on controlling their pluripotent state. Wnt signaling is a key pathway in maintaining ESC pluripotency and directing their differentiation into specific lineages [2].
Table 1: Key Wnt Pathway Components and Their Roles in Early Development and Cancer
| Component | Role in Early Embryogenesis | Dysregulation in Cancer | Associated Cancers |
|---|---|---|---|
| β-catenin | Key mediator of cell fate; nuclear accumulation drives target gene expression. | Mutations (e.g., in CTNNB1) or aberrant stabilization lead to constitutive proliferation. | Colorectal, Hepatocellular, Breast, Medulloblastoma [101] [19] |
| APC | Part of the destruction complex; regulates β-catenin turnover. | Loss-of-function mutations are a primary driver of colorectal carcinogenesis. | Colorectal Cancer (∼80% of cases) [102] [19] |
| AXIN | Scaffold protein in the destruction complex; crucial for β-catenin degradation. | Inactivating mutations prevent β-catenin degradation, leading to pathway activation. | Colorectal, Hepatocellular [19] |
| DVL | Downstream effector of Frizzled; transduces signal to inhibit destruction complex. | Overexpression is common and can hyperactivate both canonical and non-canonical pathways. | Breast, Lung, Prostate [101] |
| TCF/LEF | Nuclear transcription factors that bind β-catenin to activate target genes. | Overactive TCF/LEF-driven transcription promotes cell cycle progression and stemness. | Various solid and hematological malignancies [19] |
Dysregulation of Wnt Signaling in Carcinogenesis
The very pathways that orchestrate careful development in the embryo, when dysregulated, become powerful engines of tumorigenesis. Aberrant activation of the Wnt signaling pathway, particularly the canonical branch, is a hallmark of numerous cancers [19]. This can occur through mutations in key pathway components (e.g., APC, CTNNB1, AXIN), epigenetic modifications, or crosstalk with other signaling cascades within the tumor microenvironment (TME) [101] [102].
Mechanisms of Oncogenic Activation
In cancer, the finely tuned regulation of β-catenin is lost. Mutations in the APC tumor suppressor gene are the most common cause of colorectal cancer, leading to a truncated protein that cannot effectively scaffold the destruction complex [102] [19]. Similarly, oncogenic mutations in CTNNB1 (β-catenin) render it resistant to phosphorylation and degradation. The result is the constitutive stabilization and nuclear translocation of β-catenin, where it perpetually drives the transcription of genes that fuel uncontrolled proliferation, evasion of apoptosis, and maintenance of cancer stem cells (CSCs) [101] [19]. CSCs, which share properties with embryonic stem cells, are often responsible for tumor recurrence, metastasis, and therapy resistance.
The Wnt Pathway in the Tumor Microenvironment (TME)
The role of Wnt signaling extends beyond the cancer cell itself. There is a complex, bidirectional crosstalk between tumor cells and the surrounding TME. For instance, in breast cancer, Wnt signaling within the TME promotes immune evasion by driving the polarization of macrophages towards an immunosuppressive M2 phenotype (Tumor-Associated Macrophages, or TAMs) and hindering T-cell infiltration [101]. Furthermore, physical factors in the TME, such as hypoxia and mechanical stress, can further modulate Wnt signaling, creating a feed-forward loop that accelerates tumor progression and metastasis [101].
Table 2: Quantitative Dynamics of Wnt Pathway Components in Experimental Systems
| Parameter / Component | Experimental System / Context | Quantitative Finding | Implication |
|---|---|---|---|
| β-catenin Dynamics | HEK293T cells (Human Kidney Epithelial) [76] | Upon Wnt3a stimulation, total β-catenin rises; initial increase is faster in the nucleus (~first hour). | Suggests rapid nuclear signaling and potential for active transport mechanisms. |
| β-catenin Half-life | HEK293T cells with protein synthesis inhibition (Cycloheximide) [76] | β-catenin decreases at the same rate in nucleus and cytosol-membrane compartments. | Indicates diffusional transport between compartments is fast compared to degradation rate. |
| Wnt8 Ligand Transport | Xenopus embryos [77] | Wnt8 disperses over distances of ~15 cell diameters. A small proportion diffuses freely, while most are bound to cell surfaces. | Supports a "restricted diffusion" or "bucket brigade" model for Wnt gradient formation. |
| Wnt8 Ligand Dynamics | Xenopus embryos (Fluorescence Correlation Spectroscopy) [77] | Surface-bound Wnt8 ligands decrease exponentially, indicating dynamic exchange between bound and free populations. | Ligand distribution is controlled by exchange kinetics, not just simple diffusion. |
Experimental Protocols for Investigating Wnt Signaling
To dissect the complex roles of the Wnt pathway, researchers employ a range of sophisticated techniques. The following section outlines a detailed protocol for quantifying the spatial dynamics of β-catenin, a key readout for canonical Wnt signaling.
Detailed Protocol: Quantifying β-catenin Spatial Dynamics via 3D Confocal Imaging
This protocol is adapted from a study investigating Wnt signaling in HEK293T cells and provides a methodology for acquiring temporal and spatial quantitative data on β-catenin concentrations in different cellular compartments [76].
Objective: To quantitatively measure changes in endogenous β-catenin protein levels in the nucleus and cytosol-membrane compartments of intact adherent cells in response to pathway activation (Wnt3A) or inhibition of protein synthesis (Cycloheximide).
Materials and Reagents:
- Cell Line: HEK293T cells (human embryonic kidney cells expressing SV40 large T antigen), known to respond to Wnt3A stimulation.
- Culture Medium: Standard DMEM or RPMI supplemented with fetal bovine serum (FBS).
- Stimuli/Inhibitors:
- Wnt3A Conditioned Medium: To activate the canonical Wnt pathway.
- Cycloheximide (CHX) Stock Solution: A protein synthesis inhibitor (e.g., 100 mg/mL in DMSO) to assess β-catenin degradation kinetics.
- Fixative: 4% Paraformaldehyde (PFA) in Phosphate Buffered Saline (PBS).
- Permeabilization/Blocking Buffer: PBS containing a detergent (e.g., 0.1% Triton X-100) and a blocking agent (e.g., 1% Bovine Serum Albumin - BSA).
- Primary Antibody: Monoclonal or polyclonal anti-β-catenin antibody.
- Secondary Antibody: Fluorescently conjugated antibody (e.g., Alexa Fluor 488) specific to the host species of the primary antibody.
- Nuclear Stain: DAPI (4',6-diamidino-2-phenylindole).
- Cell Boundary Stain: Antibody against a membrane protein (e.g., N-cadherin), conjugated to a different fluorophore (e.g., Alexa Fluor 555 or 647).
- Intensity Calibration Standard: Fluorescent microspheres (e.g., 0.3% rated InSpeck microspheres) with stable and specific fluorescent intensity.
Equipment:
- Confocal Laser Scanning Microscope with capabilities for 3D Z-stack acquisition and multiple fluorescence channels.
- Cell culture incubator (37°C, 5% CO₂).
- Laminar flow hood.
- Centrifuge.
- Image analysis software (e.g., ImageJ/FIJI, Volocity, Imaris).
Procedure:
- Cell Seeding and Culture: Seed HEK293T cells onto glass-bottom culture dishes or chambered coverslips at an appropriate density. Culture the cells until they reach ~70% confluence.
- Perturbation/Treatment:
- Wnt3A Stimulation: Replace the culture medium with Wnt3A conditioned medium. Incubate for defined time points (e.g., 0, 1, 2, 4 hours).
- CHX Inhibition: Treat cells with a working concentration of CHX (e.g., 50-100 µg/mL) for the same time points to inhibit new protein synthesis.
- Include an untreated control (time 0) for both conditions.
- Cell Fixation and Staining:
- At each time point, carefully aspirate the medium and wash the cells gently with pre-warmed PBS.
- Fix the cells with 4% PFA for 15-20 minutes at room temperature.
- Permeabilize and block the cells using the Permeabilization/Blocking Buffer for 30-60 minutes.
- Incubate with the primary anti-β-catenin antibody (diluted in blocking buffer) overnight at 4°C.
- Wash thoroughly with PBS to remove unbound primary antibody.
- Incubate with the fluorescent secondary antibody, DAPI, and the membrane stain (e.g., anti-N-cadherin) for 1-2 hours at room temperature in the dark.
- Perform final washes with PBS.
- Microscopy and Intensity Calibration:
- Add a solution containing the InSpeck intensity calibration microspheres to the sample.
- Using the confocal microscope, acquire 3D Z-stack images for each time point and condition. Ensure images capture the entire cell volume, the DAPI-stained nuclei, the membrane marker, the β-catenin signal, and the calibration microspheres. Use identical laser power, gain, and offset settings for all acquisitions to allow for quantitative comparison.
- Image and Data Analysis:
- Use the image analysis software to define two distinct cellular compartments based on the fluorescence signals:
- Nucleus: Defined by the DAPI signal.
- Cytosol-Membrane: Defined by the area between the nuclear boundary (DAPI) and the cell boundary (N-cadherin signal).
- Measure the total fluorescence intensity of β-catenin (Alexa Fluor 488 channel) within each of these two compartments for every cell analyzed across all Z-slices.
- Use the intensity of the calibration microspheres to standardize the β-catenin fluorescence intensities across different samples and time points, converting arbitrary fluorescence units into calibrated intensity values.
- Calculate the mean concentration of β-catenin (in calibrated units/µm³) for the nuclear and cytosol-membrane compartments at each time point for both Wnt3A and CHX treatments.
- Use the image analysis software to define two distinct cellular compartments based on the fluorescence signals:
Expected Outcomes:
- Wnt3A Stimulation: A time-dependent increase in total cellular β-catenin, with a potentially faster initial accumulation in the nuclear compartment [76].
- CHX Inhibition: A time-dependent decrease in β-catenin levels in both compartments at a similar rate, reflecting the degradation of the existing protein pool without new synthesis [76].
The Scientist's Toolkit: Key Research Reagents
Table 3: Essential Research Reagents for Investigating Wnt Signaling
| Reagent / Tool | Category | Primary Function in Wnt Research | Example Application |
|---|---|---|---|
| Recombinant Wnt3a | Pathway Agonist | Activates the canonical Wnt/β-catenin pathway by binding to FZD and LRP5/6 receptors. | Stimulating pathway in cell culture to study downstream gene activation [76]. |
| Recombinant Dkk1 | Pathway Antagonist | Binds to LRP5/6 co-receptor, preventing Wnt ligand binding and pathway initiation. | Validating the specificity of a Wnt-driven cellular response [75] [14]. |
| IWP-2 / IWR-1 | Small Molecule Inhibitor | IWP-2 inhibits PORCN, blocking Wnt ligand secretion; IWR-1 stabilizes Axin, promoting β-catenin degradation. | Determining dependence on autocrine/paracrine Wnt signaling; probing destruction complex function. |
| Anti-β-catenin Antibody | Immunoassay Reagent | Detects and quantifies total and active (non-phospho) β-catenin protein levels. | Western blotting, immunofluorescence (as in protocol above) [76]. |
| TCF/LEF Reporter Plasmid | Gene Reporter | Contains TCF/LEF binding sites driving luciferase or GFP; reports on canonical pathway activity. | High-throughput screening for Wnt pathway modulators. |
| Cycloheximide (CHX) | Protein Synthesis Inhibitor | Blocks new protein synthesis, allowing study of protein half-life and degradation kinetics. | Measuring the degradation rate of β-catenin [76]. |
| GSK3β Inhibitors (e.g., CHIR99021) | Small Molecule Agonist | Inhibits GSK3β kinase activity, leading to β-catenin stabilization and pathway activation. | Maintaining pluripotency in stem cells; mimicking oncogenic Wnt activation [2]. |
Therapeutic Targeting and Future Directions
The centrality of the Wnt pathway in cancer and development makes it an attractive therapeutic target. However, its ubiquitous role in normal tissue homeostasis poses a significant challenge, as global inhibition could lead to severe on-target toxicities. Current strategies are focused on developing agents that target specific nodes of the pathway.
Current Therapeutic Strategies:
- Targeting Wnt Ligand Secretion: Inhibitors of PORCN (e.g., LGK974) prevent the palmitoylation and subsequent secretion of all Wnt ligands, effectively reducing signaling in the TME [19].
- Targeting Receptor Complexes: Monoclonal antibodies against FZD receptors or RSPO3 (a pathway agonist) are being explored to block signal reception at the membrane [19].
- Disrupting the β-catenin/TCF Complex: Small molecules that interfere with the critical nuclear interaction between β-catenin and its transcription factor partners, such as TCF, are under investigation to selectively inhibit oncogenic transcription [19].
- Natural Compounds: Dietary flavonoids (e.g., quercetin, apigenin) and other natural products exhibit anticancer activity partly through modulating Wnt signaling and the expression of related microRNAs [102].
Future Outlook and Personalized Medicine: The future of Wnt-targeted therapy lies in patient stratification and combination treatments. Identifying tumors with specific genetic alterations in the pathway (e.g., APC or RSPO mutations) will help select patients most likely to benefit. Furthermore, combining Wnt inhibitors with immune checkpoint blockers, chemotherapy, or radiation may overcome resistance mechanisms and improve therapeutic efficacy. As our understanding of the pathway's complexity deepens, so too will our ability to therapeutically harness it for diseases ranging from infertility to cancer.
Visualizing the Experimental Workflow
The following diagram summarizes the key experimental workflow for quantifying β-catenin dynamics, as described in the detailed protocol above.
Comparative Analysis of TCF/LEF Expression Across Species
The T-cell factor/lymphoid enhancer factor (TCF/LEF) family of transcription factors represents the major endpoint mediators of Wnt/β-catenin signaling throughout metazoans, playing indispensable roles in early embryogenesis, stem cell maintenance, and tissue differentiation [26] [2]. This technical analysis reveals a fundamental evolutionary divergence: while invertebrate genomes typically possess a single TCF/LEF ortholog capable of both activation and repression functions, vertebrate lineages have expanded to four family members (five in zebrafish) that have undergone functional specialization [26] [103]. This gene duplication event has enabled complex tissue-specific and temporal expression patterns during mammalian embryonic development, with profound implications for organogenesis and stem cell niche maintenance. Understanding these cross-species conservation and specialization mechanisms provides critical insights for developmental biology research and therapeutic targeting of Wnt-related pathologies.
TCF/LEF proteins are multifunctional transcription factors that utilize sequence-specific DNA-binding and context-dependent interactions to specify which genes are regulated by Wnt signals [26]. All TCF/LEF family members share conserved structural domains that have been maintained across evolutionary lineages, though with distinct modifications that underlie their functional specialization.
Conserved Protein Domains
β-catenin binding domain: Located at the N-terminus, this domain mediates interaction with β-catenin through formation of an alpha helix and salt bridges with charged residues in the central Armadillo repeat domain [103]. Deletion of this domain abrogates TCF-mediated transcriptional activation and acts as a dominant negative regulator of Wnt signaling [103].
HMG DNA-binding domain: Contains a high-mobility group (HMG) box that recognizes specific DNA sequences (5′-SCTTTGATS-3′) with nanomolar affinity and enforces a 90°-130° bend in the DNA helix [26] [103]. This domain also includes a nuclear localization signal recognized by importin alpha subunits [103].
C-clamp domain: A second DNA-binding domain located carboxy-terminal to the HMG domain that provides specificity for GC-rich "Helper sites" with variable spacing and orientation relative to the primary Wnt response element [26]. This domain is not present in all vertebrate TCF/LEF family members.
Context-dependent regulatory domain: Exhibits only 15-20% identity between family members and recruits repressor proteins like Groucho/transducin-like enhancer of split (Gro/TLE) in the absence of Wnt signaling [26] [103].
Table 1: TCF/LEF Family Members Across Species
| Species | TCF/LEF Members | Key Characteristics | Developmental Functions |
|---|---|---|---|
| Invertebrates | Single ortholog | Dual activator/repressor functions | Pattern formation, cell fate specification |
| Zebrafish | 5 members | Expanded family | Early embryogenesis, axis patterning |
| Mice | 4 members (TCF7, LEF1, TCF7L1, TCF7L2) | Functional specialization | Limb development, osteogenesis, intestinal homeostasis |
| Humans | 4 members (TCF7, LEF1, TCF7L1, TCF7L2) | Tissue-specific expression | Stem cell maintenance, cancer progression |
Species-Specific Expression Patterns and Functional Diversification
Murine Models (Mus musculus)
Murine models demonstrate precise spatiotemporal expression of TCF/LEF family members during embryogenesis, with specialized roles in skeletal development and organ formation:
LEF1: Detected at embryonic day 14.5 in caudal, hip osteoprogenitor, and surrounding cochlear mesenchymal cells [103]. LEF1 knockout mice are smaller than normal littermates, display defects in tissues formed by epithelial-mesenchymal interactions (lack of teeth, body hair, and beard), and die within two weeks after birth [103].
TCF7: Expressed in prechondrocytes of the mandible, palate, nasal bone, occipital bone, vertebrae, and ribs at embryonic stages [103]. TCF7 knockout mice show slight decreases in bone mineral density postnatally [103].
TCF7L2: Detected in mesenchymal cell regions around embryonal cartilage at day 10.5 and in embryonal osteoblasts at day 16.5 [103]. TCF7L2-deficient mice display developmental defects in intestinal epithelial cells and crypt cells, leading to postnatal death [103].
Compound mutants: Animals lacking both LEF1 and TCF7 resemble wnt3a-/- mutants, failing to develop limbs and dying at approximately embryonic day 10.5 [103].
Porcine Models (Sus scrofa)
Porcine embryogenesis provides valuable insights for mammalian translational research, particularly in trophectoderm development:
- IGF-1 promotes trophectoderm cell proliferation through activation of the Wnt/β-catenin pathway, with treatment during days 3-6 of embryonic development proving more effective than days 0-3 [23].
- Inhibition of IGF-1 receptor with picropodophyllin suppresses developmental parameters, β-catenin levels, TE cell differentiation, and tight junction formation - effects rescued by CHIR99021-mediated Wnt activation [23].
Human Systems (Homo sapiens)
Human TCF/LEF expression demonstrates clinical relevance in disease contexts, particularly cancer:
- Colorectal cancer: LEF1 expression found in 26% and TCF7L2 in 46% of tumors, with heterogeneous distribution throughout tumor tissue [104].
- Prognostic significance: TCF7L2 expression correlates with shorter overall survival (p = 0.020), while LEF1 expression associates with longer survival (p = 0.015) in colorectal cancer patients [104].
- Therapeutic implications: Recent evidence demonstrates that transcriptional activity mediated by β-CATENIN and TCF/LEF family members is completely dispensable for survival and propagation of multiple human colorectal cancer cell lines, challenging their broad utility as drug targets [105].
Table 2: TCF/LEF Expression During Murine Embryonic Development
| Developmental Stage | TCF/LEF Member | Expression Location | Functional Consequences of Knockout |
|---|---|---|---|
| E10.5 | TCF7L2 | Mesenchymal cell region around embryonal cartilage | Postnatal death due to intestinal defects |
| E14.5 | LEF1 | Caudal, hip osteoprogenitor, cochlear mesenchymal cells | Neonatal lethality, missing teeth, hair, beard |
| E16.5 | TCF7L2 | Embryonal osteoblasts | Abnormal bone development |
| Various stages | TCF7 | Prechondrocytes in multiple skeletal structures | Reduced bone mineral density |
Experimental Approaches and Methodologies
Genetic Manipulation Techniques
CRISPR/Cas9-Mediated Gene Inactivation
- Application: Generation of TCF/LEF-deficient human colorectal cancer cell lines [105]
- Protocol: Frame-shift inducing exon deletion strategy targeting TCF7, TCF7L1, and TCF7L2 genes
- Validation: Western blot analysis confirming absence of TCF/LEF protein expression
- Outcome: Demonstration that CRC cells survive and can be propagated in complete absence of TCF/LEF expression [105]
Nephron Progenitor Cell (NPC) Culture System
- Culture Conditions: Chemically defined nephron progenitor expansion medium (NPEM) with varying CHIR99021 concentrations [106]
- CHIR Titration: Low CHIR (1.25 µM) for NPC maintenance vs. high CHIR (5 µM) for induction of nephrogenic program
- Time Course: Rapid inductive response within 3 days of elevated CHIR exposure
- Analysis Methods: mRNA-seq, chromatin confirmation studies, TCF/LEF engagement profiling [106]
Porcine Embryo Culture and Treatment
- Parthenogenetic Activation: Ionomycin (15 µM, 5 min) followed by culture in PZM-3 medium with cytochalasin B and 6-dimethylaminopurine [23]
- IGF-1 Treatment: Recombinant IGF-1 at 0, 10, 50, and 100 µM concentrations during specific developmental windows
- Inhibition Studies: IGF-1 receptor inhibitor picropodophyllin (0-0.5 µM) with rescue experiments using CHIR99021 (1 µM)
- Assessment Methods: TUNEL assay, immunofluorescence for CDX2 and YAP1, protein quantification [23]
Signaling Mechanisms and Transcriptional Regulation
The canonical model of Wnt signaling invokes two transcriptional states governed by TCF/LEF factors. In the absence of Wnt ligand, TCF/LEFs recruit co-repressors (Tle, Ctbp) to silence target gene expression. Upon Wnt pathway activation, β-catenin accumulates and translocates to the nucleus where it partners with TCF/LEF proteins to activate transcription [106]. Recent research reveals additional complexity in this paradigm:
Figure 1: TCF/LEF-Mediated Transcriptional Switch in Wnt Signaling
β-Catenin-Driven TCF/LEF Switching Mechanism
Research in nephron progenitor cells reveals a sophisticated mechanism where β-catenin levels act as a key regulatory switch modifying TCF/LEF complex engagement:
- Low β-catenin conditions: TCF7L1/TCF7L2 complexes maintain repression at target gene enhancers
- Elevated β-catenin conditions: TCF7/LEF1/β-catenin complexes replace TCF7L1/TCF7L2 binding on enhancers of differentiation-promoting genes [106]
- Chromatin architecture: Pre-established promoter-enhancer connections poise target genes for rapid initiation of differentiation programs
- Functional specialization: TCF7L1 predominantly acts as a repressor, TCF7L2 as context-dependent activator/repressor, and TCF7/LEF1 as transcriptional activators [106]
Research Reagent Solutions Toolkit
Table 3: Essential Research Reagents for TCF/LEF Studies
| Reagent/Chemical | Function | Application Examples | Species Utility |
|---|---|---|---|
| CHIR99021 | GSK3β inhibitor stabilizing β-catenin | NPC culture (1.25-5 µM) [106]; Porcine embryo culture [23] | Multispecies |
| Recombinant IGF-1 | Activator of Wnt/β-catenin pathway | Trophectoderm proliferation studies (10-100 µM) [23] | Porcine, mammalian models |
| Picropodophyllin (PPP) | IGF-1 receptor inhibitor | Validation of IGF-1/Wnt pathway crosstalk (0.05-0.5 µM) [23] | Porcine, mammalian models |
| CRISPR/Cas9 System | Gene editing | Generation of TCF/LEF-deficient cell lines [105] | Human, murine cell lines |
| Anti-LEF1 Antibody | Immunodetection | IHC on tissue microarrays (1:150) [104] | Human, murine tissues |
| Anti-TCF7L2 Antibody | Immunodetection | IHC on tissue microarrays (1:50) [104] | Human, murine tissues |
Discussion and Research Implications
The comparative analysis of TCF/LEF expression across species reveals both remarkable conservation of core structural domains and significant functional diversification in vertebrate lineages. The evolutionary expansion from a single TCF/LEF ortholog in invertebrates to multiple family members in vertebrates has enabled sophisticated regulatory mechanisms for spatial and temporal control of Wnt-responsive gene expression during embryogenesis.
The context-dependent functions of TCF/LEF members, particularly their cell-type-specific expression and capacity to form repressive or activating complexes based on β-catenin availability, highlights their importance as developmental regulators. Furthermore, the dispensability of TCF/LEF transcriptional activity in certain cancer contexts [105] challenges conventional understanding of Wnt pathway essentiality and necessitates reevaluation of therapeutic strategies targeting this pathway.
Future research directions should focus on elucidating the precise mechanisms governing TCF/LEF switching in different developmental contexts, understanding how chromatin architecture influences TCF/LEF binding specificity, and exploiting species-specific differences to identify conserved regulatory modules that could be targeted for therapeutic intervention in developmental disorders and cancers.
Conclusion
The Wnt signaling pathway emerges as a master regulator of early embryogenesis, with distinct functions in maintaining pluripotency, guiding cell lineage specification, and coordinating tissue patterning. Critical differences between human and rodent Wnt signaling mechanisms underscore the necessity of human-specific models for translational research. While technical challenges in pathway modulation and species-specific validation remain, recent advances in stem cell biology, targeted protein degradation, and high-precision inhibitors offer promising avenues for therapeutic development. Future research should focus on resolving the precise spatiotemporal control of Wnt signaling, developing more specific pharmacological tools, and translating these findings into clinical applications for regenerative medicine, cancer therapy, and improved assisted reproductive technologies. The integration of basic mechanistic studies with advanced disease modeling will be essential for unlocking the full therapeutic potential of targeting this evolutionarily conserved pathway.
References
- 1. Interrelationship of canonical and non-canonical Wnt signalling pathways in chronic metabolic diseases - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC5752873/]
- 2. The canonical Wnt in pathway mammalian early and... embryogenesis [https://pubmed.ncbi.nlm.nih.gov/15380245/]
- 3. Signaling - GeeksforGeeks Wnt [https://www.geeksforgeeks.org/biology/wnt-signaling/]
- 4. Signalling Wnt Pathway [https://www.news-medical.net/life-sciences/Wnt-Signalling-Pathway.aspx]
- 5. Developmental Signals - Wnt - Embryology [https://embryology.med.unsw.edu.au/embryology/index.php/Developmental_Signals_-_Wnt]
- 6. Canonical and Non-Canonical Wnt Signaling in Immune Cells - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7367500/]
- 7. Components of the Canonical and Non-Canonical Wnt Pathways Are Not Mis-Expressed in Pituitary Tumors - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3637156/]
- 8. Analysis of Wnt signaling β-catenin spatial dynamics in HEK293T cells - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4108056/]
- 9. Wnt signaling pathway - Wikipedia [https://en.wikipedia.org/wiki/Wnt_signaling_pathway]
- 10. Wnt signal transduction pathways - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2634250/]
- 11. : Key Mechanisms & Roles | Danaher Life... Wnt Signaling Pathway [https://lifesciences.danaher.com/us/en/library/wnt-signaling-pathway.html]
- 12. Frontiers | Wnt Signaling and Its Significance Within the Tumor ... [https://www.frontiersin.org/articles/10.3389/fimmu.2019.02872]
- 13. in biology and disease: mechanisms and... Wnt signaling pathways [https://pmc.ncbi.nlm.nih.gov/articles/PMC11968978/]
- 14. | Abcam Wnt signaling pathway [https://www.abcam.com/en-us/technical-resources/pathways/wnt-signaling-pathway]
- 15. Non-canonical WNT in cardiovascular disease: mechanisms... signalling [https://pmc.ncbi.nlm.nih.gov/articles/PMC9191761/]
- 16. Wnt Signaling Pathway - an overview | ScienceDirect Topics [https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/wnt-signaling-pathway]
- 17. / Wnt - β signalling: function, biological mechanisms, and... catenin [https://www.nature.com/articles/s41392-021-00762-6?error=cookies_not_supported&code=0742e167-57b5-4524-a09c-c19858c39c74]
- 18. / Wnt - β -Mediated PPARδ Expression during... catenin Pathway [https://pmc.ncbi.nlm.nih.gov/articles/PMC7918746/]
- 19. / Wnt - β signaling catenin in carcinogenesis and cancer therapy pathway [https://jhoonline.biomedcentral.com/articles/10.1186/s13045-024-01563-4]
- 20. Wnt/β-Catenin Signaling | Cell Signaling Technology [https://www.cellsignal.com/pathways/wnt-beta-catenin-signaling-pathway]
- 21. Understanding Wnt Through Signaling - β Localization in Live... Catenin [https://www.promegaconnections.com/understanding-wnt-signaling-in-live-cells/]
- 22. Wg/ Wnt -signaling-induced nuclear of translocation - β is... catenin [https://pmc.ncbi.nlm.nih.gov/articles/PMC11311196/]
- 23. IGF-1 promotes trophectoderm cell proliferation of porcine embryos by... [https://biosignaling.biomedcentral.com/articles/10.1186/s12964-025-02191-2]
- 24. WNT-pathway components as predictive markers useful for diagnosis, prevention and therapy in inflammatory bowel disease and sporadic colorectal cancer - PubMed [https://pubmed.ncbi.nlm.nih.gov/24657851/]
- 25. /beta-catenin WNT in nephron progenitors and their epithelial... signaling [https://pubmed.ncbi.nlm.nih.gov/18633347/]
- 26. / TCF and LEFs Signaling in the Nucleus - PMC Wnt [https://pmc.ncbi.nlm.nih.gov/articles/PMC3536346/]
- 27. TCF/LEF Transcription Factors Identified as Promising Drug Targets in Wnt [https://bioengineer.org/tcf-lef-transcription-factors-identified-as-promising-drug-targets-in-wnt-signaling-for-fibrosis-and-cancer-treatment/]
- 28. / TCF family - Wikipedia LEF [https://en.wikipedia.org/wiki/TCF/LEF_family]
- 29. Complete sequence-based pathway analysis by differential on-chip DNA and RNA extraction from a single cell | Scientific Reports [https://www.nature.com/articles/s41598-017-10704-4]
- 30. in colorectal cancer: pathogenic Wnt and therapeutic... signaling role [https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-022-01616-7]
- 31. Impact of WNT on tissue lineage differentiation in the early... signaling [https://pubmed.ncbi.nlm.nih.gov/21762130/]
- 32. Frontiers | The Canonical Wnt Pathway in Osteoporosis: A Scoping Review of Key Compounds and Proteins Modulating Wnt-Induced Osteogenesis [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2025.1669222/abstract]
- 33. Mutations and mechanisms of WNT pathway tumour suppressors in cancer | Nature Reviews Cancer [https://www.nature.com/articles/s41568-020-00307-z]
- 34. Regulation of Wnt Stem Cell Self-Renewal - PMC Pathway Embryonic [https://pmc.ncbi.nlm.nih.gov/articles/PMC3428775/]
- 35. Interrogating Canonical Wnt in Human Pluripotent... Signaling Pathway [https://link.springer.com/article/10.1007/s12195-016-0453-8]
- 36. : A comprehensive Wnt signaling pathway review [https://pubmed.ncbi.nlm.nih.gov/35297539/]
- 37. mediates self-organization and axis formation in... Wnt signaling [https://pmc.ncbi.nlm.nih.gov/articles/PMC2683270/]
- 38. Embryoid bodies get organized | Nature Reports Stem Cells [https://www.nature.com/articles/stemcells.2008.146]
- 39. Bulk cell density and Wnt/TGFbeta signalling regulate mesendodermal patterning of human pluripotent stem cells | Nature Communications [https://www.nature.com/articles/ncomms13602]
- 40. The Role of Wnt in an Signaling Generation Protocol... Embryoid Body [https://odr.chalmers.se/items/4ea454b7-22a8-44b8-b215-6cbc505caa43]
- 41. Endogenous Wnt signalling in human embryonic stem cells generates an equilibrium of distinct lineage-specified progenitors | Nature Communications [https://www.nature.com/articles/ncomms2064]
- 42. Endogenous suppression of WNT in signalling embryonic... human [https://www.nature.com/articles/s41598-021-85447-4?error=cookies_not_supported&code=2fe38694-f5bc-4590-ac00-53885f16873a]
- 43. Genetic requirement of dact1/2 to regulate noncanonical Wnt ... [https://elifesciences.org/articles/91648]
- 44. A CRISPR knockout screen reveals new regulators of canonical Wnt signaling | Oncogenesis [https://www.nature.com/articles/s41389-021-00354-7]
- 45. 15 Lineage — Single-cell best practices tracing [https://www.sc-best-practices.org/trajectories/lineage_tracing.html]
- 46. TMEM79/MATTRIN defines a pathway for Frizzled regulation and is... [https://pubmed.ncbi.nlm.nih.gov/32924931/]
- 47. Mapping of Wnt-Frizzled interactions by multiplex CRISPR targeting of receptor gene families - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC5636703/]
- 48. / CRISPR -mediated knockout of NSD1 suppresses the... Cas 9 [https://jeccr.biomedcentral.com/articles/10.1186/s13046-019-1462-y]
- 49. Frontiers | Genome Editing of Wnt -1, a Gene Associated with... [https://www.frontiersin.org/journals/physiology/articles/10.3389/fphys.2016.00666/full]
- 50. Single-cell lineage by integrating tracing - CRISPR mutations with... Cas 9 [https://www.nature.com/articles/s41467-020-16821-5?error=cookies_not_supported]
- 51. Modeling Wnt signaling by CRISPR - Cas genome editing recapitulates... 9 [https://pubmed.ncbi.nlm.nih.gov/30144514/]
- 52. to Analyze Biochemical Protein Secretion Methods Wnt [https://pubmed.ncbi.nlm.nih.gov/27590148/]
- 53. Methods to Manipulate and Monitor Signaling in... | SpringerLink Wnt [https://link.springer.com/protocol/10.1007/978-1-4939-6393-5_16]
- 54. for a Protocol reporter assay to measure its activity in human... Wnt [https://pmc.ncbi.nlm.nih.gov/articles/PMC10329100/]
- 55. by ADP-ribosylation | Nature Communications Wnt pathway activation [https://www.nature.com/articles/ncomms11430?error=cookies_not_supported&code=39976770-2cca-4217-8c46-3e2e0a19aaf0]
- 56. (PDF) A Biochemical Screen for Identification of Small-Molecule... [https://www.academia.edu/128343608/A_Biochemical_Screen_for_Identification_of_Small_Molecule_Regulators_of_the_Wnt_Pathway_Using_Xenopus_Egg_Extracts]
- 57. Organoids as Novel Models for Embryo Implantation Study - PubMed [https://pubmed.ncbi.nlm.nih.gov/33650092/]
- 58. Human blastoids model blastocyst development and... | Nature [https://www.nature.com/articles/s41586-021-04267-8?error=cookies_not_supported]
- 59. Generating human blastoids modeling blastocyst-stage embryos and implantation - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7617227/]
- 60. for Human Protocol Modeling Blastocyst Development and... Blastoids [https://app.jove.com/t/63388/protocol-for-human-blastoids-modeling-blastocyst-development]
- 61. 5a suppresses colorectal cancer progression via... Wnt [https://biosignaling.biomedcentral.com/articles/10.1186/s12964-025-02364-z]
- 62. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities | Signal Transduction and Targeted Therapy [https://www.nature.com/articles/s41392-021-00762-6]
- 63. Wnt signaling in cancer: from biomarkers to targeted therapies and clinical translation | Molecular Cancer | Full Text [https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-025-02306-w]
- 64. Tailored tetravalent antibodies potently and specifically activate... [https://elifesciences.org/articles/46134]
- 65. The Notch and Wnt regulate stemness and differentiation in... pathways [https://pubmed.ncbi.nlm.nih.gov/26643275/]
- 66. The origin and evolution of Wnt | Nature Reviews Genetics signalling [https://www.nature.com/articles/s41576-024-00699-w?error=cookies_not_supported&code=7e015cf5-d93a-4e9c-82f3-8b6bd360c6da]
- 67. - High targeted throughput in triple-negative breast cancer... screening [https://www.nature.com/articles/s41598-017-12232-7?error=cookies_not_supported&code=4328ceb1-cc9f-45b6-a4de-9d69f310d0d7]
- 68. High-throughput screening - Wikipedia [https://en.wikipedia.org/wiki/High-throughput_screening]
- 69. A high - throughput for screen /β-catenin signaling Wnt ... pathway [https://pubmed.ncbi.nlm.nih.gov/22923789/]
- 70. - High Drug Throughput Identifies a Potent Screening Inhibitor... Wnt [https://pmc.ncbi.nlm.nih.gov/articles/PMC7050206/]
- 71. between the Crosstalk and... mechanisms WNT signaling pathway [https://pubmed.ncbi.nlm.nih.gov/30159439/]
- 72. live-cell imaging and computational modeling shed new... Quantitative [https://elifesciences.org/articles/66440]
- 73. Pathway Analysis with Signaling Hypergraphs - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5810418/]
- 74. information transmission in the canonical Optimizing pathway Wnt [https://arxiv.org/html/2506.22633v1]
- 75. Wnt signaling pathway diagram | The WNT Homepage [https://wnt.stanford.edu/wnt-signaling-pathway-diagram]
- 76. Analysis of Wnt β-catenin spatial dynamics in HEK293T cells signaling [https://bmcsystbiol.biomedcentral.com/articles/10.1186/1752-0509-8-44]
- 77. analyses reveal extracellular dynamics of Quantitative ligands in... Wnt [https://elifesciences.org/articles/55108]
- 78. Relating simulation studies by provenance—Developing a family of... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8407594/]
- 79. Human Cytomegalovirus Infection and Embryonic Malformations: The... [https://www.besjournal.com/article/doi/10.3967/bes2025.103]
- 80. Modeling Paracrine Noncanonical Wnt In Vitro Signaling [https://www.jove.com/t/63247/modeling-paracrine-noncanonical-wnt-signaling-in-vitro]
- 81. Microglia-astrocyte crosstalk regulates synapse remodeling via Wnt ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10871360/]
- 82. Wnt signaling pathways in biology and disease: mechanisms and therapeutic advances | Signal Transduction and Targeted Therapy [https://www.nature.com/articles/s41392-025-02142-w]
- 83. Wnt signaling in development and disease - PubMed [https://pubmed.ncbi.nlm.nih.gov/22520685/]
- 84. The Pleiotropic Effects of the Canonical Wnt in Pathway ... Early [https://pubmed.ncbi.nlm.nih.gov/29443926/]
- 85. in cancer: molecular mechanisms and potential... WNT signaling [https://link.springer.com/article/10.1186/s43556-025-00327-x]
- 86. Photoswitchable agonists for visible-light activation of the Wnt ... [https://pubs.rsc.org/en/content/articlelanding/2025/ob/d4ob01827c]
- 87. Frontiers | Targeting Wnt -driven metabolic adaptations in cancer... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1622218/full]
- 88. Wnt/β-catenin signaling pathway safeguards epigenetic stability and homeostasis of mouse embryonic stem cells | Scientific Reports [https://www.nature.com/articles/s41598-018-37442-5]
- 89. The Divergent Pluripotent States in Mouse and Human Cells - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9408542/]
- 90. Principles of signaling pathway modulation for enhancing human naive pluripotency induction - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8423434/]
- 91. Wnt signaling in development and disease | Cell & Bioscience | Full Text [https://cellandbioscience.biomedcentral.com/articles/10.1186/2045-3701-2-14]
- 92. Roles of Wnt and ROR2 Receptor in Embryonic... Signaling Pathway [https://pmc.ncbi.nlm.nih.gov/articles/PMC8808015/]
- 93. | 41768 Publications | 343273 Citations Wnt signaling pathway [https://scispace.com/topics/wnt-signaling-pathway-17lctn1d]
- 94. /β-catenin mediated Wnt in cancer: recent... signaling pathways [https://pmc.ncbi.nlm.nih.gov/articles/PMC12150590/]
- 95. Wnt/β-catenin mediated signaling pathways in cancer: recent advances, and applications in cancer therapy | Molecular Cancer | Full Text [https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-025-02363-1]
- 96. Wnt signaling, NetPath (WP363) - Homo sapiens | WikiPathways [https://www.wikipathways.org/pathways/WP363.html]
- 97. Expression of Wnt-signaling pathway genes and their associations with miRNAs in colorectal cancer - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC5814196/]
- 98. Characteristic Markers of the WNT Signaling Pathways Are Differentially Expressed in Osteoarthritic Cartilage - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4297187/]
- 99. signaling in Wnt and development - PMC disease [https://pmc.ncbi.nlm.nih.gov/articles/PMC2854277/]
- 100. Frontiers | The Emerging Mechanisms of Wnt Secretion and Signaling... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2021.714746/full]
- 101. Role of the Wnt the complex microenvironment... signaling pathway in [https://www.spandidos-publications.com/10.3892/ijo.2025.5742]
- 102. Frontiers | MicroRNAs at the crossroad of cancer therapeutics: insights... [https://www.frontiersin.org/journals/molecular-biosciences/articles/10.3389/fmolb.2025.1616221/full]
- 103. Role of TCF/LEF Transcription Factors in Bone Development and Osteogenesis [https://www.medsci.org/v15p1415.htm]
- 104. -1 and LEF 4 TCF correlate inversely with survival in... expression [https://translational-medicine.biomedcentral.com/articles/10.1186/1479-5876-8-123]
- 105. Transcriptional activity mediated by β-CATENIN and TCF / LEF family... [https://www.nature.com/articles/s41598-022-27261-0?error=cookies_not_supported&code=2569efba-1ab5-4da1-bf57-a9cd9466240f]
- 106. A β-catenin-driven switch in TCF / LEF transcription factor binding to... [https://elifesciences.org/articles/64444]