ZEB2 in Somitogenesis: Unraveling its Critical Role in Mouse and Human Development
This article synthesizes current research on the transcription factor ZEB2 and its indispensable function in mouse and human somitogenesis.
ZEB2 in Somitogenesis: Unraveling its Critical Role in Mouse and Human Development
Abstract
This article synthesizes current research on the transcription factor ZEB2 and its indispensable function in mouse and human somitogenesis. We explore the foundational biology of ZEB2, from its discovery as a SMAD-interacting protein to its role as a key regulator of paraxial mesoderm patterning. The content details advanced methodological approaches, including the use of gastruloid models and degron-based perturbation systems combined with single-cell omics, which have been pivotal in identifying ZEB2's critical role. We further address common challenges in ZEB2 research and discuss the validation of its functions across species, highlighting implications for understanding human developmental disorders like Mowat-Wilson Syndrome and potential translational applications. This resource is tailored for researchers, scientists, and drug development professionals seeking a comprehensive overview of ZEB2 in embryonic segmentation.
ZEB2: From Discovery to Fundamental Roles in Embryonic Patterning
Discovery of ZEB2 as a SMAD-Binding Transcription Factor
The discovery of Zinc finger E-box binding homeobox 2 (ZEB2) as a SMAD-binding transcription factor marked a pivotal advancement in understanding embryonic development and disease pathogenesis. Initially identified in 1999 through yeast two-hybrid screening as a DNA-binding and SMAD-binding transcription factor that determines cell fate in amphibian embryos, ZEB2 emerged as a crucial nuclear fine-tuner of transcriptional responses to TGFβ/BMP signaling [1]. This discovery laid the foundation for comprehending ZEB2's multifaceted roles in neural development, neural crest cell formation, and its clinical significance in Mowat-Wilson syndrome [1] [2]. Subsequent research utilizing conditional knockout murine models and stem cell systems has further elucidated ZEB2's mechanism of action as a transcriptional repressor that integrates multiple signaling pathways, including BMP, Wnt, and Notch, during critical developmental processes such as somitogenesis [1] [3].
ZEB2, originally named SIP1 (SMAD Interacting Protein 1), represents one of two vertebrate members of the zinc-finger E-box binding homeodomain (ZEB) transcription factor family, characterized by their unique two-handed zinc finger/homeodomain structure [4] [3]. The discovery of its direct interaction with activated SMAD proteins positioned ZEB2 as a key nuclear effector in the transforming growth factor β (TGFβ) signaling pathway, which is essential during early fetal development [4]. This interaction mechanism provides a crucial link between extracellular signaling and transcriptional regulation, enabling precise control of developmental gene expression programs.
The structural configuration of ZEB2 includes eight zinc fingers and one homeodomain, which facilitate its DNA-binding capacity and protein-protein interactions [4]. As a transcription factor, ZEB2 predominantly functions as a transcriptional repressor but can also activate specific target genes depending on cellular context [2]. Its ability to interact with receptor-activated SMAD proteins allows ZEB2 to serve as a transcriptional corepressor that fine-tunes cellular responses to TGFβ/BMP signaling gradients, thereby influencing critical cell fate decisions during embryogenesis [1] [4].
Historical Context and Initial Discovery
The Original Experimental Approach
The initial identification of ZEB2 occurred through a yeast two-hybrid screening methodology using the transcription activation (MH2) domain of BMP-SMAD1 as bait against cDNA libraries derived from mid-gestation mouse embryos [1]. This approach was predicated on the hypothesis that receptor-activated SMADs required cooperation with DNA-binding transcription factors for their transcriptional regulatory functions in the nucleus. The screening identified several candidate SMAD-interacting proteins (SIPs), including a partial cDNA that exhibited significant sequence homology with the previously discovered transcription factor δEF1 (now known as ZEB1) [1].
Table 1: Key Experiments in ZEB2 Discovery
| Year | Experimental System | Key Finding | Significance |
|---|---|---|---|
| 1999 | Yeast two-hybrid screening | Identification of SIP1/ZEB2 as SMAD-binding protein | First demonstration of ZEB2-SMAD interaction [1] |
| 2001-2003 | Human genetic studies | ZEB2 mutations cause Mowat-Wilson syndrome | Established clinical relevance of ZEB2 [1] |
| 2003+ | Conditional KO mice | Embryonic lethality in full KO; tissue-specific functions | Revealed essential developmental roles [1] |
| 2016 | Zeb2 KO mouse ESCs | Defective exit from pluripotency | Identified role in stem cell differentiation [5] |
| 2020 | R26_Zeb2 mESCs | Positive regulation of myogenic differentiation | Demonstrated non-neural roles in mesodermal lineages [3] |
Technical Limitations and Methodological Considerations
The original yeast two-hybrid approach, while groundbreaking, presented several technical limitations that influenced the initial characterization of ZEB2. The screen identified partial cDNA clones rather than full-length transcripts, requiring subsequent complete cDNA isolation and sequencing. Additionally, the artificial environment of yeast cells may not have fully recapitulated the native nuclear context of mammalian cells, potentially missing certain post-translational modifications or protein co-factors essential for optimal SMAD-ZEB2 interactions. Despite these limitations, the initial discovery provided the crucial foundation for all subsequent functional studies of ZEB2 in development and disease.
Experimental Models and Methodologies
In Vivo Model Systems
Mouse models have been instrumental in elucidating ZEB2 functions. General homozygous Zeb2-knockout mice exhibit early post-gastrulation embryonic lethality around E8.5, with severe defects in somitogenesis, neural plate formation, and neural crest development [5] [2]. This early lethality necessitated the development of conditional knockout strategies using Zeb2+/fl(Δex7) mice, enabling tissue-specific deletion of ZEB2 functions in the nervous system, neural crest derivatives, and other tissues [1]. These models have revealed both cell-autonomous and non-autonomous functions of ZEB2, particularly in brain development where ZEB2 determines neurogenic-to-gliogenic switching timing through paracrine signaling mechanisms [1].
Zebrafish models with zeb2 knockdown display early axial and neural patterning defects alongside neural crest abnormalities, establishing evolutionary conservation of ZEB2 functions and providing a complementary system for rapid functional screening [1]. The transparency and external development of zebrafish embryos facilitate live imaging of developmental processes affected by ZEB2 deficiency.
In Vitro and Stem Cell Systems
Embryonic stem cell systems have provided critical insights into ZEB2's molecular mechanisms. Zeb2 knockout mouse ESCs can exit naive pluripotency but stall in an early epiblast-like state, failing to properly differentiate into neural or mesendodermal lineages [5]. Transcriptomic and DNA methylome analyses of these systems revealed that ZEB2 regulates the silencing of pluripotency networks and establishment of DNA methylation patterns necessary for lineage commitment [5].
Human ESC studies demonstrate that ZEB2 knockdown promotes mesendodermal differentiation while its overexpression enhances neuroectoderm formation, highlighting its role in early cell fate decisions [5]. More recent gastruloid models provide three-dimensional in vitro systems that recapitulate aspects of somitogenesis and enable precise manipulation of ZEB2 function during mesoderm patterning [6].
Table 2: Comparison of Major Experimental Systems for ZEB2 Research
| System | Key Applications | Advantages | Limitations |
|---|---|---|---|
| Mouse KO | Developmental functions, tissue-specific roles | Physiologically relevant, enables tissue-specific analysis | Embryonic lethal in full KO, time-consuming [1] |
| Zebrafish KD | Neural crest, early patterning | Rapid screening, easy visualization | May not fully recapitulate mammalian development [1] |
| Mouse ESCs | Differentiation mechanisms, molecular pathways | Genetic manipulation, in vitro differentiation | May not capture all in vivo complexities [5] |
| Human ESCs | Human-specific mechanisms, disease modeling | Human-relevant, disease modeling | Ethical considerations, technical challenges [5] |
| Gastruloids | Somitogenesis, 3D patterning | 3D architecture, high-throughput capability | Still developing, may lack some tissue interactions [6] |
Molecular Mechanisms of ZEB2 Action
SMAD Interaction and Transcriptional Regulation
ZEB2 functions as a nuclear effector of TGFβ/BMP signaling through its direct interaction with receptor-activated SMAD proteins (R-SMADs). This interaction occurs primarily through the N-terminal domain of ZEB2 and the MH2 domain of SMADs, allowing ZEB2 to be recruited to SMAD-binding elements in target gene promoters [1] [4]. Once bound to DNA, ZEB2 typically functions as a transcriptional repressor through its recruitment of corepressor complexes, including CtBP and NCOR, which mediate histone deacetylation and chromatin compaction at target loci [2].
The transcriptional regulatory activity of ZEB2 exhibits context-dependent specificity, influenced by cellular environment, developmental stage, and interacting protein partners. In some contexts, ZEB2 can function as a transcriptional activator, though this represents a less common mode of action [2]. This functional versatility enables ZEB2 to integrate multiple signaling inputs and execute appropriate transcriptional outputs during complex developmental processes.
Signaling Pathway Integration
Beyond its canonical role in TGFβ/BMP signaling, ZEB2 interacts with multiple additional signaling pathways to coordinate developmental processes:
Figure 1: ZEB2 Integration of Multiple Signaling Pathways. ZEB2 directly interacts with SMAD proteins and integrates inputs from Wnt and Notch pathways to regulate target gene expression.
The anti-BMP activity of ZEB2 was among its first identified signaling modulatory functions, particularly evident in neural plate patterning where ZEB2 opposes BMP-mediated ventralization [1]. Additionally, ZEB2 modulates Wnt signaling through indirect mechanisms that remain to be fully elucidated, and regulates Notch signaling outcomes through transcriptional repression of Notch pathway components or effectors [1] [3]. This multifaceted signaling integration capacity positions ZEB2 as a central node in the regulatory networks controlling embryonic patterning and cell fate decisions.
Comparative Analysis of ZEB2 Functions Across Biological Contexts
Role in Neurodevelopment and Neural Crest
ZEB2 plays indispensable roles in nervous system development, functioning at multiple stages from initial neural induction through terminal differentiation. During early neurodevelopment, ZEB2 regulates the timing of neurogenic-to-gliogenic transition in the forebrain, serving as the first identified transcription factor that controls this critical switch in a cell non-autonomous manner [1]. In the developing cortex, ZEB2 determines the size and layering of cortical regions by controlling the relative proportions of different neuronal subtypes.
In neural crest development, ZEB2 orchestrates the epithelial-to-mesenchymal transition (EMT) required for neural crest cell delamination, followed by regulation of migration and differentiation into diverse derivatives, including enteric nervous system neurons, melanocytes, and craniofacial structures [1] [2]. These functions explain why ZEB2 haploinsufficiency in Mowat-Wilson syndrome affects both central and peripheral nervous systems, with clinical manifestations including Hirschsprung disease, intellectual disability, and distinctive facial features [1] [4].
Role in Somitogenesis and Mesodermal Differentiation
While initially characterized for its roles in neural development, ZEB2 also plays critical functions in mesodermal lineages, particularly during somitogenesis. Mouse embryos with complete Zeb2 knockout exhibit severe defects in somite formation and patterning, revealing essential requirements for ZEB2 in this process [1]. Recent studies using gastruloid models have further delineated ZEB2's role in regulating mouse and human somitogenesis, with degradation-tag (degron) mediated perturbation of ZEB2 function causing specific defects in somite patterning [6].
In pluripotent stem cell differentiation, ZEB2 positively regulates myogenic differentiation, with Zeb2-overexpressing mouse ESCs showing enhanced expression of myogenic markers (Pax3, Pax7, MyoD, Myogenin) and myomiRs (miR-1, miR-133b, miR-206, miR-208) compared to Zeb2-null counterparts [3]. This pro-myogenic function involves ZEB2's ability to modulate TGFβ/BMP signaling, which normally opposes myogenic differentiation, thereby creating a permissive environment for muscle-specific gene expression programs.
Table 3: ZEB2 Functional Roles Across Developmental Contexts
| Developmental Context | Main Functions | Key Target Genes/Pathways | Phenotype of Loss-of-Function |
|---|---|---|---|
| Forebrain Development | Timing of neuro-gliogenic switch, cortical layering | TGFβ/BMP signaling, paracrine factors | Microcephaly, intellectual disability [1] |
| Neural Crest | EMT, migration, differentiation | E-cadherin, TGFβ pathway | Hirschsprung disease, craniofacial defects [1] [2] |
| Somitogenesis | Somite patterning, segmentation | Unknown targets | Somitogenesis defects, axial patterning defects [1] [6] |
| Myogenic Differentiation | Enhancement of muscle differentiation | MyoD, Myogenin, myomiRs | Impaired muscle differentiation [3] |
| Stem Cell Pluripotency | Exit from naive pluripotency | Tet1, pluripotency factors | Stalling in epiblast-like state [5] |
Technical Approaches and Research Toolkit
Essential Research Reagents and Models
Table 4: Essential Research Reagents for ZEB2 Investigation
| Reagent/Model | Type | Key Applications | Specific Function in ZEB2 Research |
|---|---|---|---|
| Zeb2+/fl(Δex7) mice | Animal model | Tissue-specific knockout studies | Enables Cre-mediated deletion of critical exon [1] |
| R26_Zeb2 mice | Animal model | Conditional overexpression | Rosa26 locus-driven cDNA expression [1] |
| Zeb2 KO mESCs | Cell line | Molecular mechanism studies | Elucidation of differentiation defects [5] |
| R26_Zeb2 mESCs | Cell line | Gain-of-function studies | Assessment of Zeb2 overexpression effects [3] |
| Anti-ZEB2 antibodies | Immunological reagent | Protein detection | Western blot, immunohistochemistry, ChIP [3] |
| Gastruloid systems | 3D in vitro model | Somitogenesis studies | Modeling early patterning events [6] |
| NIBR-17 | N6-(6-methoxypyridin-3-yl)-2-morpholino-[4,5'-bipyrimidine]-2',6-diamine | Get N6-(6-methoxypyridin-3-yl)-2-morpholino-[4,5'-bipyrimidine]-2',6-diamine (CAS 944396-88-7) for phosphoinositide 3-kinase (PI3K) research. This product is for Research Use Only (RUO). Not for human or veterinary use. | Bench Chemicals |
| McN5691 | McN5691, CAS:99254-95-2, MF:C30H35NO3, MW:457.6 g/mol | Chemical Reagent | Bench Chemicals |
Key Methodological Approaches
Transcriptomic profiling through RNA-sequencing of wild-type versus Zeb2 mutant cells has identified comprehensive sets of ZEB2-dependent genes across multiple developmental contexts [1] [5]. These studies reveal that ZEB2 regulates diverse target genes involved in cell differentiation, signaling pathways, and metabolic processes.
Epigenomic analyses including reduced representation bisulfite sequencing (RRBS) and chromatin immunoprecipitation (ChIP) have elucidated ZEB2's impact on the epigenome, particularly its role in maintaining DNA methylation patterns during differentiation [5]. Zeb2 knockout ESCs initially acquire appropriate DNA methylation marks but fail to maintain them during neural differentiation, instead reverting toward a more naive methylome state [5].
Lineage tracing and fate mapping approaches in conditional knockout models have revealed cell-autonomous versus non-autonomous functions of ZEB2, particularly important in understanding its roles in tissue patterning and cell-cell communication [1].
Figure 2: Experimental Workflow for ZEB2 Functional Characterization. Comprehensive approaches combining multiple model systems and analytical methods are required to fully elucidate ZEB2 functions.
Implications for Human Development and Disease
Mowat-Wilson Syndrome
The discovery that heterozygous ZEB2 mutations cause Mowat-Wilson syndrome (MOWS) provided crucial clinical relevance to basic research on ZEB2 function [1] [4]. This rare autosomal dominant disorder (incidence 1:50,000-70,000 live births) exemplifies the pleiotropic effects of ZEB2 haploinsufficiency, affecting multiple organ systems including the central nervous system, neural crest derivatives, heart, and genitourinary tract [1]. The majority of MOWS cases result from complete gene deletions, nonsense mutations, or frameshift mutations that trigger nonsense-mediated decay of ZEB2 mRNA, effectively creating null alleles [1]. Rare missense mutations and C-terminal truncations typically cause milder phenotypes, providing structure-function insights into critical protein domains [1].
Hirschsprung Disease
Isolated Hirschsprung disease (aganglionic megacolon) can result from specific ZEB2 mutations that preferentially affect enteric nervous system development without the full spectrum of MOWS features [4]. This reflects the particular sensitivity of neural crest-derived enteric neurons to ZEB2 gene dosage, with compromised EMT, migration, or differentiation of enteric neural crest progenitors leading to incomplete colonization of the gastrointestinal tract [1] [2].
Cancer and Fibrosis
While beyond the scope of this developmental focus, ZEB2 has emerging roles in cancer progression and fibrotic diseases, primarily through its regulation of epithelial-to-mesenchymal transition [1] [7]. In hepatocellular carcinoma, a specific single-nucleotide polymorphism (rs3806475) in the ZEB2 promoter region associates with increased cancer risk under a recessive model [4]. These pathological contexts highlight the continued relevance of ZEB2 mechanisms in adult tissue homeostasis and disease.
Future Directions and Unanswered Questions
Despite significant advances since its initial discovery as a SMAD-binding transcription factor, numerous questions regarding ZEB2 biology remain unresolved. The complete repertoire of direct ZEB2 target genes across different developmental contexts requires further elucidation through combinatorial ChIP-seq and transcriptomic approaches. The precise mechanisms by which ZEB2 interacts with various chromatin-modifying complexes and how these interactions determine transcriptional outcomes need additional characterization.
The functional significance of ZEB2 protein isoforms generated through alternative splicing represents another area of limited understanding, as does the regulation of ZEB2 expression itself by transcriptional and post-transcriptional mechanisms. From a therapeutic perspective, strategies to modulate ZEB2 function in disease contexts, particularly Mowat-Wilson syndrome, remain largely unexplored and would benefit from continued research into ZEB2 structure-function relationships and downstream effector pathways.
The integration of emerging technologies such as single-cell multi-omics, CRISPR-based screening, and organoid models will undoubtedly provide new insights into ZEB2 functions in development and disease, building upon the foundational discovery of ZEB2 as a SMAD-binding transcription factor that coordinates complex transcriptional programs during embryogenesis.
ZEB2 Expression Dynamics During Early Embryogenesis
Zinc finger E-box binding homeobox 2 (ZEB2) is a DNA-binding transcriptional repressor and critical developmental regulator essential for early embryonic patterning. As a nuclear fine-tuner of transcriptional responses to TGF-β/Nodal-Activin and BMP signaling, ZEB2 regulates cell fate decisions across multiple lineages during embryogenesis [3] [8]. Heterozygous mutations in ZEB2 cause Mowat-Wilson syndrome (MOWS), characterized by severe intellectual disability, Hirschsprung disease, epilepsy, and various structural anomalies including congenital heart defects and craniofacial dysmorphisms [9] [8]. This review synthesizes current understanding of ZEB2 expression dynamics and functional roles during early embryogenesis, with particular emphasis on its recently identified critical functions in mouse and human somitogenesis. We provide comprehensive experimental comparisons and methodological frameworks to guide future research into this pivotal developmental regulator.
Temporal and Spatial Expression Patterns of ZEB2
Early Embryonic Expression Dynamics
ZEB2 exhibits remarkably early and dynamic expression patterns during embryogenesis. In human embryonic stem cell (hESC) models of cranial neural crest induction, ZEB2 is among the earliest factors expressed in prospective human neural crest, with transcripts detectable as early as 12 hours after induction initiation and showing continuous increase over 5 days of differentiation [9]. This rapid activation positions ZEB2 as a primary responder to Wnt signaling during neural crest formation.
Comparative studies in chick embryos reveal conserved early expression patterns, with Zeb2 transcripts present throughout the entire epiblast of Hamburger-Hamilton stage 3 (HH3) gastrula embryos, prior to expression of the earliest neural crest specification marker Pax7 [9]. As development proceeds, Zeb2 becomes restricted to the prospective neural plate and neural crest at HH4+, and by HH8, expression localizes to neural folds with subsequent presence in neural tube and migrating neural crest streams at HH12 [9].
Table 1: Comparative ZEB2 Expression Dynamics Across Model Systems
| Developmental Stage | hESC Model | Chick Embryo | Mouse Model |
|---|---|---|---|
| Pre-gastrulation | Not applicable | Ubiquitous epiblast expression (HH3) | Not detected in naïve ESCs |
| Early specification | Detectable at 12h, increasing through day 5 | Restricted to neural plate/neural crest (HH4+) | Upregulated during exit from naïve pluripotency |
| Neural crest formation | Maintained throughout NC induction | Neural folds (HH8), migrating NC (HH12) | Critical for NC specification and differentiation |
| Somitogenesis | Not thoroughly investigated | Not thoroughly investigated | Essential for somite patterning and differentiation |
Expression in Pluripotency Transitions
In mouse embryonic stem cells (mESCs), Zeb2 is undetectable in the naïve pluripotent state but demonstrates significant upregulation during the transition from primed pluripotency, accompanying efficient conversion of naïve ESCs into epiblast-like stem cells [5] [10]. This pattern establishes ZEB2 as a critical factor for exit from primed pluripotency and entry into general and neural differentiation programs.
Comparative Functional Analysis of ZEB2 in Early Lineage Specification
Neural Crest Specification
ZEB2 plays indispensable roles in neural crest formation through multiple mechanisms. Knockdown studies in hESC-based neural crest models reveal that ZEB2 promotes neural crest identity while repressing non-neural/pre-placodal ectodermal genes [9]. At day 3 of neural crest induction, ZEB2 knockdown results in upregulation of neural plate border genes (PAX7, MSX1/2, DLX5) and NC specifiers (FOXD3, SOX9, SNAI2), followed by subsequent downregulation at day 5, indicating failure of proper NC cell formation [9].
ZEB2 interacts with the nucleosome remodeling and deacetylase (NuRD) complex to regulate the epigenetic landscape during neural crest development. Mutant ZEB2 lacking the N-terminal NuRD-interacting domain fails to properly recruit HDAC1 and displays derepressed enhancers during human neural crest induction, with compromised terminal differentiation into osteoblasts, peripheral neurons, and neuroglia [9].
Regulation of Pluripotency Exit
ZEB2 serves as a critical regulator of the transition from pluripotency to committed lineages. Zeb2 knockout mESCs can exit the naïve state but stall in an early epiblast-like state and demonstrate impaired neural and mesendodermal differentiation capacity [5]. These mutant cells maintain the ability to re-adapt to 2i+LIF conditions even after prolonged differentiation exposure, indicating failed irreversible commitment.
Mechanistically, Zeb2 knockout ESCs exhibit deregulated expression of genes involved in pluripotency, epithelial-to-mesenchymal transition (EMT), and DNA methylation, including elevated Tet1 levels [5]. The aberrant maintenance of pluripotency networks and DNA methylation patterns in Zeb2-deficient cells establishes ZEB2 as a link between pluripotency networks and irreversible differentiation commitment.
Table 2: ZEB2 Functional Roles Across Developmental Contexts
| Developmental Context | Primary Function | Key Regulatory Targets | Phenotype of Perturbation |
|---|---|---|---|
| Neural crest formation | Specifies NC fate, represses non-neural lineages | PAX7, MSX1/2, FOXD3, SOX9, BMP pathway components | Failed NC differentiation, BMP signaling dysregulation |
| Pluripotency exit | Promotes irreversible commitment | Tet1, pluripotency factors, EMT genes | Stalled epiblast state, maintained pluripotency |
| Somitogenesis | Regulates somite patterning | Unknown targets in presomitic mesoderm | Somitogenesis defects in mouse models |
| Myogenic differentiation | Enhances muscle commitment | MyoD, Myogenin, myomiRs | Impaired skeletal muscle differentiation |
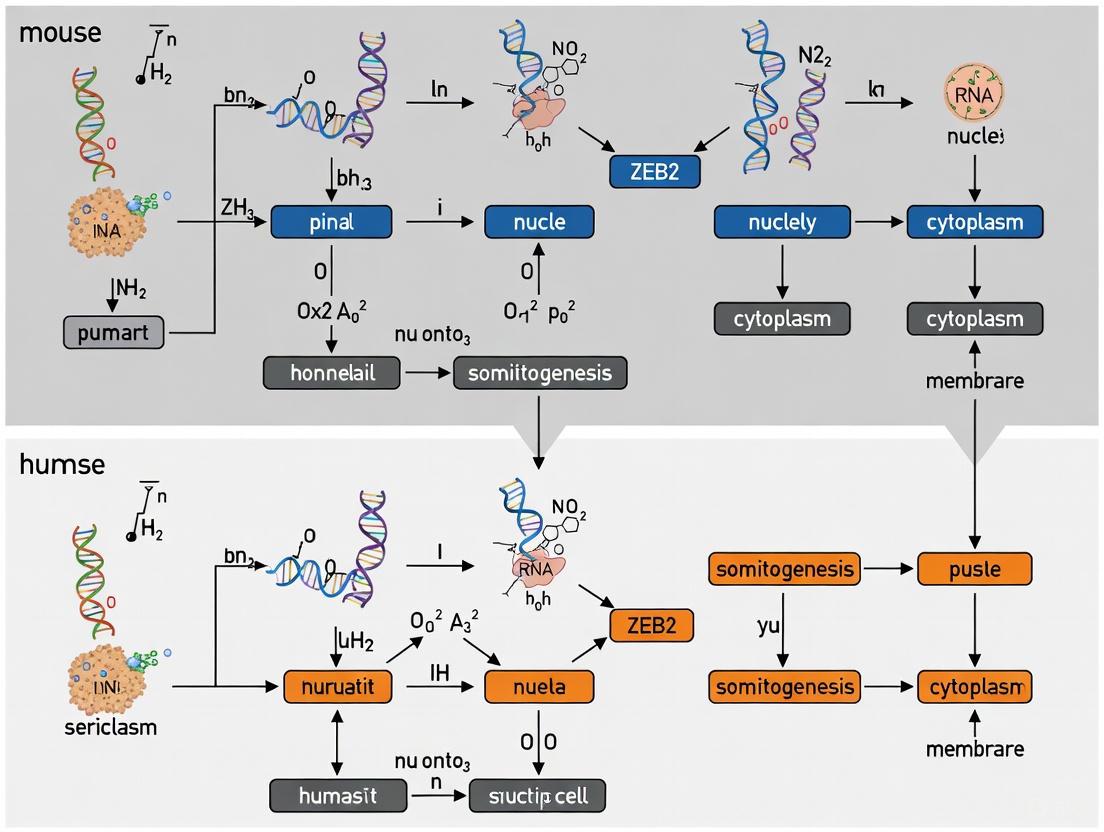
Emerging Role in Somitogenesis
Recent research utilizing mouse gastruloids and multilayered proteomics approaches has identified a critical role for ZEB2 in mouse and human somitogenesis [11] [12]. Gastruloid differentiation studies combined with degron-based perturbations and single-cell RNA sequencing demonstrate ZEB2's essential function in somite patterning, consistent with earlier observations of somitogenesis defects in Zeb2 knockout mice [11] [5] [8].
ZEB2 in Somitogenesis: Mouse and Human Conservation
Evidence from Gastruloid Models
Recent advances in synthetic embryo models have provided unprecedented insight into ZEB2 function during somitogenesis. Studies utilizing mouse gastruloids combined with multilayered mass spectrometry-based proteomics have revealed ZEB2 as a critical regulator of somite formation in both mouse and human systems [11] [12]. These models enable precise temporal resolution of protein expression dynamics during germ layer specification and their derivatives.
Gastruloid differentiation studies employing degron-based perturbations combined with single-cell RNA sequencing have specifically identified ZEB2's essential role in somitogenesis, demonstrating conserved function across mouse and human systems [11] [12]. This approach has established gastruloids as a powerful platform for investigating ZEB2 dynamics during paraxial mesoderm patterning.
Historical Evidence and Mechanistic Insights
Earlier studies of general Zeb2-knockout mice revealed embryonic lethality shortly after E8.5 with multiple defects, including impaired somitogenesis [5] [8]. Expression analyses in amphibian and mouse embryos further identified Zeb2 transcripts in presomitic mesoderm, suggesting direct involvement in somite patterning [8].
The mechanism of ZEB2 action in somitogenesis likely involves its characteristic function as a transcriptional repressor that fine-tunes cellular responses to morphogen signaling, potentially modulating BMP, TGF-β, or Wnt pathway activity in the presomitic mesoderm, similar to its actions in other developmental contexts [9] [3] [8].
Experimental Approaches for Investigating ZEB2 Function
Stem Cell Differentiation Models
Multiple stem cell-based systems have been developed to investigate ZEB2 function during early lineage specification:
Neural Crest Differentiation Protocol [9]:
- Base model: Human embryonic stem cells (hESCs)
- Induction method: 2 days of exogenous Wnt activation
- Key markers: PAX7 (NPB), FOXD3, SOX9 (NC specifiers)
- Timeline: ZEB2 expression onset at 12h, increasing through 5 days
- Perturbation approaches: siRNA knockdown, N-terminal mutant analysis
Neural Differentiation Protocol [5] [10]:
- Base model: Mouse embryonic stem cells (mESCs)
- Method: Embryoid body formation in KO DMEM + 15% FBS, then N2B27 + retinoic acid (500 nM) from day 4
- Timeline: 8-15 days to neuroprogenitor cells
- Key observations: Zeb2 upregulation essential for NPC formation
Gastruloid-Based Somitogenesis Analysis [11] [12]:
- Model system: Mouse gastruloids
- Analytical approach: Multilayered (phospho)proteomics + P300 proximity labeling
- Perturbation method: Degron-based Zeb2 depletion + scRNA-seq
- Key finding: Essential role in mouse and human somitogenesis
Signaling Pathway Modulation
ZEB2 functions as a critical modulator of multiple developmental signaling pathways. In neural crest development, ZEB2 regulates appropriate BMP signaling levels, with N-terminal truncated ZEB2 mutants causing early misexpression of BMP signaling ligands that can be rescued through BMP attenuation [9]. This positions ZEB2 as a key regulator of BMP-mediated cell fate decisions.
ZEB2 also interacts with activated SMADs of BMP and TGF-β pathways to inhibit expression of downstream targets, functioning as a nuclear fine-tuner of transcriptional responses to extracellular signals [3] [8]. This SMAD-interaction capability enables ZEB2 to integrate multiple signaling inputs during cell fate specification.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for ZEB2 Investigation
| Reagent Category | Specific Example | Application | Key Features/Considerations |
|---|---|---|---|
| Cell Models | Zeb2-V5 mESCs [10] | ChIP-seq mapping | Endogenous tagging for DNA-binding studies |
| ZEB2 KO hiPSC (KICRi002A-4) [13] | Differentiation studies | Homozygous 790bp deletion in ZEB2 | |
| R26_Zeb2 mESCs [3] | Gain-of-function studies | cDNA expression from Rosa26 safe harbor | |
| Perturbation Tools | siRNA ZEB2 knockdown [9] | Acute depletion | ~80% protein reduction in hNC model |
| Degron-based depletion [11] | Rapid protein degradation | Gastruloid studies with scRNA-seq readout | |
| Zeb2flox/flox mice [5] | Conditional knockout | Cell-type specific deletion approaches | |
| Analytical Methods | V5-tag ChIP-seq [10] | DNA-binding mapping | 2,432 binding sites identified in NPCs |
| Multilayered proteomics [11] | Protein expression | Phosphoprotein dynamics in gastruloids | |
| P300 proximity labeling [12] | Enhancer interaction mapping | Gastruloid-specific enhancer landscapes | |
| Differentiation Systems | hESC neural crest model [9] | NC specification studies | 2-day Wnt activation, 5-day differentiation |
| Mouse gastruloids [11] [12] | Somitogenesis studies | 3D model for paraxial mesoderm patterning | |
| AZD 4407 | AZD 4407, CAS:166882-70-8, MF:C19H21NO3S2, MW:375.5 g/mol | Chemical Reagent | Bench Chemicals |
| Org30958 | Org30958, CAS:99957-90-1, MF:C21H30O2S2, MW:378.6 g/mol | Chemical Reagent | Bench Chemicals |
ZEB2 represents a master regulatory transcription factor with critical functions at multiple stages of early embryogenesis. From its early expression in gastrulating embryos to its essential roles in neural crest formation, pluripotency exit, and somitogenesis, ZEB2 coordinates complex developmental processes through transcriptional repression and signaling pathway modulation. The conservation of ZEB2 functions between mouse and human systems, particularly in recently identified roles in somitogenesis, underscores its fundamental importance in embryonic patterning. Continued investigation using the rapidly expanding toolkit of stem cell models, genome editing approaches, and multi-omics technologies will further elucidate ZEB2's diverse mechanisms of action and potential therapeutic applications for related developmental disorders.
The transcription factor ZEB2 (Zinc Finger E-Box Binding Homeobox 2) represents a critical regulator in mammalian embryonic development, with its functions elucidated primarily through sophisticated knockout mouse models. These models demonstrate that ZEB2 is indispensable for proper embryogenesis, particularly in somitogenesis, neural development, and hematopoietic differentiation. This guide systematically compares phenotypic outcomes across various Zeb2 knockout systems, providing researchers with consolidated experimental data and methodologies relevant to developmental biology and therapeutic discovery. The essential nature of ZEB2 is further highlighted by its association with Mowat-Wilson syndrome (MOWS) in humans, a neurodevelopmental disorder caused by ZEB2 haploinsufficiency [1].
Comparative Phenotypic Analysis of ZEB2 Knockout Models
Table 1: Developmental Defects in ZEB2 Knockout Models
| Knockout Model/System | Key Developmental Defects | Developmental Stage | Primary Experimental Evidence |
|---|---|---|---|
| Conventional Homozygous KO [1] | Early embryonic lethality, multiple defects in somitogenesis, neural plate, and neural crest cells [5] | Lethal shortly after E8.5 [5] | Histological analysis, mRNA expression studies |
| Conditional KO (Tie2-Cre, Vav-iCre) [14] | Defective HSC/HPC differentiation and fetal liver colonization; enhanced cell adhesion; perinatal lethality with cephalic hemorrhaging [14] | Embryonic/perinatal lethality [14] | FACS analysis, histological staining, gene expression (Angpt1, β1 integrin, Cxcr4) |
| Zeb2 KO Embryonic Stem Cells (ESCs) [5] | Stalled in epiblast-like state; impaired neural and mesendodermal differentiation; failed pluripotency exit [5] | In vitro differentiation (Embryoid Bodies) | RNA-sequencing, RRBS (DNA-methylation analysis), immunostaining |
| Zeb2 KO in Somitogenesis [12] | Critical role in mouse and human somitogenesis [12] | Gastrulation and early organogenesis [12] | scRNA-seq, proteomics, enhancer interactome profiling via P300 proximity labeling |
The data reveal that complete Zeb2 knockout results in early embryonic lethality (around E8.5), precluding the study of its roles in later organogenesis [5] [1]. Consequently, conditional knockout (cKO) models using Cre-loxP technology have been indispensable for elucidating ZEB2 functions in specific tissues and developmental stages, such as hematopoiesis [14]. In vitro models, particularly Zeb2 knockout embryonic stem cells (ESCs), have provided deep mechanistic insights, demonstrating the transcription factor's necessity for exiting the pluripotent state and committing to differentiated lineages [5].
Experimental Workflows in Key Knockout Studies
Workflow for Analyzing Zeb2 KO ESCs and Differentiation Potential
The following diagram outlines the core experimental workflow used to establish ZEB2's critical role in exit from pluripotency and lineage commitment.
Figure 1: Experimental Workflow for Zeb2 KO ESC Differentiation
Detailed Methodology:
- Generation of Zeb2 KO mESC Lines: Control lines are derived from
Zeb2flox/floxmice. Knockout is achieved via nucleofection of a Cre recombinase vector into low-passage ESCs, followed by blasticidin selection [5]. - ESC Maintenance and Differentiation Initiation: ESCs are maintained feeder-free in 2i + LIF medium (containing 1 μM PD0325901, 3 μM CHIR99021, and 1000 U/mL LIF) to sustain the naive pluripotent state. Differentiation is triggered by withdrawing 2i + LIF [5].
- Embryoid Body (EB) Formation and Directed Differentiation: For neural differentiation, 3 million ESCs are plated in bacterial petri dishes in EB medium (KO DMEM, 15% FBS, NEAA, etc.). On day 4, the medium is switched to N2B27 supplemented with 500 nM retinoic acid to promote neural fate [5].
- Phenotypic Analysis:
- Transcriptomics: Temporal RNA-sequencing identifies deregulated gene networks [5].
- DNA Methylation Analysis: Reduced Representation Bisulfite Sequencing (RRBS) assesses genome-wide methylation changes [5].
- Functional Rescue: Knockdown of
Tet1(a gene deregulated in KO cells) is performed to test partial rescue of differentiation impairment [5].
Workflow for Conditional KO in Hematopoietic System
The methodology for defining ZEB2 function in embryonic hematopoiesis using conditional knockout models is summarized below.
Figure 2: Workflow for Hematopoietic cKO Analysis
Detailed Methodology:
- Mouse Model Generation: Conditional deletion of Zeb2 is achieved using Tie2-Cre or Vav-iCre driver lines, which induce recombination in endothelial and hematopoietic lineages [14].
- Cellular Analysis of Hematopoietic Tissues: Detailed cellular analysis is performed on the aorta-gonad-mesonephros (AGM) region, fetal liver, and bone marrow. Techniques include fluorescence-activated cell sorting (FACS) to isolate HSCs/HPCs and functional assays to test their differentiation and colonization capacity [14].
- Molecular and Histological Examination:
Decoding ZEB2-Controlled Signaling Pathways
ZEB2 operates as a nodal point, integrating signals from multiple key developmental pathways. The following diagram synthesizes its central regulatory role.
Figure 3: ZEB2 as an Integrator of Core Developmental Pathways
ZEB2 functions primarily as a transcriptional repressor but can also activate some genes [5] [1]. Its protein structure allows it to bind activated SMADs (downstream of TGFβ/BMP receptors), thereby fine-tuning the transcriptional response to these morphogens [1]. This interaction underpins its anti-BMP activity, which is crucial for neural patterning and neural crest development [1]. Furthermore, ZEB2 is implicated in modulating cellular responses to Wnt and Notch signaling, positioning it as a central integrator of extracellular cues that determine cell fate [1].
A critical function is its regulation of the epithelial-to-mesenchymal transition (EMT) by repressing epithelial genes like E-cadherin (Cdh1) [5]. This is vital for the migration of neural crest cells and hematopoietic progenitors [14]. During differentiation, ZEB2 is required for the irreversible silencing of the core pluripotency network (e.g., Oct4, Nanog), thereby facilitating commitment [5]. Mechanistically, ZEB2 links pluripotency exit with epigenetic remodeling, as its loss leads to deregulated expression of DNA methylation regulators like TET1, resulting in failure to maintain differentiation-associated DNA methylation patterns [5].
The Scientist's Toolkit: Essential Reagents for ZEB2 Research
Table 2: Key Research Reagents for ZEB2 Functional Studies
| Reagent / Resource | Function and Application in ZEB2 Research | Example Use Case |
|---|---|---|
| Zeb2flox/flox Mice [5] | Enables tissue-specific knockout of Zeb2 via Cre-loxP recombination. | Generation of conditional KO models for studying tissue-specific functions (e.g., hematopoiesis [14]). |
| Cre Recombinase Lines (Tie2-Cre, Vav-iCre) [14] | Drives deletion of floxed Zeb2 alleles in specific cell lineages (endothelial, hematopoietic). | Studying Zeb2 function in embryonic HSC/HPC differentiation and mobilization [14]. |
| 2i + LIF Medium [5] | Chemical inhibitors (PD0325901, CHIR99021) + LIF cytokine maintain ESCs in naive pluripotent state. | Culture of control and Zeb2 KO mESCs prior to differentiation induction [5]. |
| R26Zeb2 ESC Line [5] | ESCs with Flag-tagged Zeb2 cDNA inserted into the Rosa26 safe-harbor locus for overexpression. | Rescue experiments; testing cell-autonomous effects of Zeb2 re-expression [5]. |
| N2B27 Medium + Retinoic Acid [5] | Defined medium for efficient neural differentiation of ESCs/EBs. | Directed neural differentiation of control and Zeb2 KO ESCs [5]. |
| P300 Proximity Labeling [12] | Maps enhancer interaction landscapes and identifies key TFs in developing systems. | Identifying ZEB2's role and interactions in mouse and human somitogenesis [12]. |
| FR 167653 | FR 167653, CAS:158876-66-5, MF:C24H20FN5O6S, MW:525.5 g/mol | Chemical Reagent |
| Asparagusic acid | Asparagusic acid, CAS:2224-02-4, MF:C4H6O2S2, MW:150.2 g/mol | Chemical Reagent |
This toolkit highlights the critical reagents that enable precise manipulation and analysis of ZEB2 function. The combination of conditional mouse models and well-defined ESC culture systems provides a powerful platform for dissecting the pleiotropic roles of ZEB2 from early lineage commitment to organ-specific development.
Regulation of Germ Layer Specification and Exit from Pluripotency
The exit from pluripotency and the subsequent specification into the three primary germ layers (ectoderm, mesoderm, and endoderm) are fundamental processes in embryonic development. This transition involves a complex interplay of transcription factors, signaling pathways, and epigenetic regulators that dismantle the pluripotent state and initiate lineage-specific programs. The transcription factor ZEB2 (Zinc Finger E-Box Binding Homeobox 2) has emerged as a critical node in this regulatory network, acting as a pivotal regulator of cell fate decisions in both mouse and human models. This guide provides a comparative analysis of the mechanisms governing exit from pluripotency and germ layer specification, with a specific focus on the role of ZEB2, integrating findings from key experimental models and methodologies to serve as a resource for researchers and drug development professionals.
Core Regulatory Circuits Governing Pluripotency Exit
The Pluripotency Network and Its Dismantling
The core pluripotency circuit, maintained by transcription factors including Oct4 (Pou5f1), Sox2, and Nanog, is intrinsically linked to the machinery that drives lineage selection [15]. These factors do not merely sustain pluripotency but also integrate external signals to guide fate decisions. During differentiation, their levels are asymmetrically modulated: Oct4 is upregulated in mesendoderm (ME) progenitors but repressed in neural ectoderm (NE) progenitors, while Sox2 shows the inverse pattern, being upregulated in NE and repressed in ME fates [15]. This reciprocal relationship helps steer cells toward distinct developmental paths.
The exit from the naïve pluripotent state is a rapid, reproducible, and irreversible process that can be modeled in vitro using mouse Embryonic Stem Cells (mESCs) [16]. Upon withdrawal of pluripotency-maintaining signals (such as 2i/LIF), cells undergo a metachronous but unidirectional transition toward a primed epiblast-like state, making this system ideal for dissecting the dynamics of enhancer activation, transcription factor binding, and transcriptional reorganization [16].
ZEB2 as a Master Regulator of Cell Fate Transition
ZEB2 is indispensable for the exit from pluripotency and the initiation of differentiation programs. In mESCs, ZEB2 protein is undetectable in the naïve state but is strongly upregulated during neural differentiation, accompanying the efficient conversion into epiblast-like cells and subsequent neuroprogenitor cells (NPCs) [10] [5].
Perturbation studies demonstrate its necessity; Zeb2 knockout (KO) mESCs fail to properly differentiate and instead stall in an early epiblast-like state, maintaining the ability to re-adapt to pluripotency conditions even after prolonged exposure to differentiation signals [5]. This stall is characterized by deregulation of the pluripotency network, epithelial-to-mesenchymal transition (EMT) genes, and DNA methylation machinery, including elevated levels of the demethylase TET1 [5]. Knockdown of Tet1 in Zeb2 KO cells partially rescues the differentiation impairment, indicating that ZEB2 links the pluripotency network with DNA methylation to ensure irreversible commitment [5].
Table 1: Key Regulators of Pluripotency Exit and Their Roles
| Regulator | Function in Pluripotency | Role in Lineage Specification | Consequence of Perturbation |
|---|---|---|---|
| ZEB2 | Low/undetectable in naïve state [10] | Essential for exit from epiblast state; promotes neural differentiation [5] | KO causes stall in epiblast-like state; impaired neural and mesendodermal differentiation [5] |
| OCT4 | Core pluripotency factor [15] | Upregulated in mesendoderm (ME) fate [15] | Necessary for ME fate choice; repression required for NE fate [15] |
| SOX2 | Core pluripotency factor [15] | Upregulated in neural ectoderm (NE) fate [15] | Necessary for NE fate choice; repression required for ME fate [15] |
| NANOG | Core pluripotency factor [15] | Downregulated for both ME and NE lineage selection [15] | Downregulation is necessary for cells to respond to differentiation signals [15] |
Figure 1: Fate Decisions at the Exit from Pluripotency. The core pluripotency factors OCT4 and SOX2 are asymmetrically modulated by external signals to direct cells toward mesendoderm (ME) or neural ectoderm (NE) fates.
ZEB2 in Vitro Models: From Pluripotency to Neural and Somitic Fates
ZEB2 in Neural Differentiation
Studies in mESCs have detailed ZEB2's function in neural differentiation. A key methodology involves the in vitro differentiation of mESCs into neuroprogenitor cells (NPCs) via embryoid body (EB) formation [10] [5]. In one protocol, ESCs are aggregated in non-adherent dishes, treated with retinoic acid from day 4 to promote neural differentiation, and then plated on coated surfaces at day 8 to obtain NPCs [10].
To identify direct genomic targets, researchers have used CRISPR-edited mESCs carrying an epitope-tagged Zeb2 allele (Flag-V5) [10]. Chromatin Immunoprecipitation followed by sequencing (ChIP-seq) in derived NPCs mapped 2,432 high-confidence ZEB2 DNA-binding sites, with a major site located in the Zeb2 promoter itself, indicating a critical autoregulatory loop [10]. Homozygous deletion of this site demonstrated that Zeb2 autoregulation is necessary for its proper function in ESC-to-NPC differentiation [10].
ZEB2 and the Path to Somitogenesis
The in vitro modeling of paraxial mesoderm and somite differentiation from human Pluripotent Stem Cells (hPSCs) provides a system to study ZEB2-relevant processes. A key protocol leverages transcriptomic data from human embryos, which revealed that downregulation of BMP and TGFβ signaling in the presomitic mesoderm (PSM) is a major regulator unique to human somitogenesis [17].
The in vitro differentiation protocol, therefore, involves:
- Initial WNT activation using a GSK3β inhibitor (CHIR99021) to specify primitive streak and posterior PSM, marked by T (Brachyury) and TBX6 [17].
- Subsequent inhibition of BMP and TGFβ signaling following WNT activation to robustly steer pPSM cells toward anterior PSM and somite fates [17]. These hPSC-derived somite cells are multipotent and can generate skeletal myocytes, osteocytes, and chondrocytes under lineage-specific conditions [17].
Table 2: Key In Vitro Differentiation Models for Studying ZEB2-Related Processes
| Model System | Key Inductive Signals | Critical Markers | Utility for ZEB2 Research |
|---|---|---|---|
| mESC to NPC [10] [5] | Retinoic Acid (RA); Aggregation (EB formation) | Nestin, Sox1; Loss of Oct4/Nanog | Models ZEB2's role in neurogenesis and autoregulation; platform for ChIP-seq. |
| hPSC to Somite [17] | WNT activation (CHIR99021) → BMP/TGFβ inhibition | T (Brachyury), TBX6 (PSM); PAX3, PAX7 (somite) | Models paraxial mesoderm development; context for ZEB2's role in somitogenesis. |
ZEB2 in Mouse and Human Somitogenesis
Conserved and Divergent Somitogenesis Pathways
Somitogenesis is a rhythmic process that segments the paraxial mesoderm into somites, the precursors of vertebrae, skeletal muscle, and dermis. The "clock and wavefront" model, involving oscillating gene expression (Notch, Wnt, FGF pathways) and opposing signaling gradients, is a conserved regulator across vertebrates [18]. In mouse, high-resolution transcriptional and chromatin maps (RNA-seq and ATAC-seq) of maturing somites have revealed a conserved molecular program followed by all somites, which includes the downregulation of Wnt and Notch signaling pathways and the upregulation of cell adhesion and migration programs associated with an epithelial-to-mesenchymal transition (EMT) [18].
However, transcriptomic profiling of human embryos revealed that BMP and TGFβ signaling are major regulators unique to human somitogenesis [17]. While these pathways are downregulated in nascent human somites, TGFβ signaling is upregulated in mouse somites, indicating a potential species-specific difference in the regulatory logic [17].
ZEB2's Role in Somitogenesis and Mowat-Wilson Syndrome
ZEB2 is expressed in the presomitic mesoderm and somites during mouse embryogenesis [8]. The crucial role of ZEB2 in this process is highlighted by the early embryonic lethality of general homozygous Zeb2-knockout mice, which display multiple defects, including in somitogenesis [8] [5]. Mowat-Wilson Syndrome (MOWS), a rare congenital disorder caused by haploinsufficiency of ZEB2, further underscores its importance in human development [10] [8]. MOWS patients display a wide array of clinical features, including intellectual disability, epilepsy, Hirschsprung disease, and various congenital heart defects, some of which may originate from disturbances in early mesodermal patterning [8].
ZEB2's function is context-dependent, mediated through its interaction with various partner proteins and signaling pathways, including TGFβ/BMP, Wnt, and Notch [8]. Its role in modulating cellular responses to these pathways positions it as a key integrator of the signals that guide somite formation and maturation.
Figure 2: Simplified Molecular Regulation of Somitogenesis. Somites form from the PSM under the control of the clock and wavefront mechanism. ZEB2 is a critical transcription factor in this process. Notably, BMP/TGFβ signaling shows a human-specific regulation pattern.
The Scientist's Toolkit: Essential Reagents and Models
Table 3: Research Reagent Solutions for Studying Pluripotency Exit and ZEB2 Function
| Reagent / Model | Specific Example | Function and Application |
|---|---|---|
| Gene-Edited ESC Lines | Zeb2-Flag-V5 mESCs [10] | Enables precise ChIP-seq mapping of endogenous ZEB2 binding sites in derived cell types (e.g., NPCs). |
| Conditional KO Models | Zeb2flox/flox mice [5] | Allows cell-type or temporal-specific deletion of Zeb2 to study its function in specific lineages (e.g., somites, neural crest). |
| Small Molecule Inhibitors/Activators | CHIR99021 (WNT agonist), PD0325901 (MEK inhibitor) [17] [5] | Used to direct hPSC/mESC differentiation by precisely modulating key signaling pathways (WNT, FGF). |
| Differentiation Media | N2B27 medium [5] [15] | A defined, serum-free medium used to maintain ESCs and to make them competent to respond to lineage-specific differentiation signals. |
| Lineage Reporters | Sox1-GFP mESC line [15] | Enables live tracking and isolation of neural ectoderm progenitors during fate choice experiments. |
| (Rac)-Valsartan-d9 | (Rac)-Valsartan-d9, CAS:1089736-73-1, MF:C24H29N5O3, MW:444.6 g/mol | Chemical Reagent |
| Faropenem daloxate | Faropenem Daloxate|Oral Penem Antibiotic|CAS 141702-36-5 |
The journey from a pluripotent stem cell to a committed progenitor is orchestrated by a tightly regulated network. The transcription factor ZEB2 acts as a critical conduit in this process, integrating signals from core pluripotency factors like OCT4 and SOX2 with those from major developmental pathways such as TGFβ/BMP and Wnt. Its non-redundant functions, evidenced by severe developmental defects in its absence, underscore its importance as a master regulator. The continued refinement of in vitro models, including neural and somitic differentiation from PSCs, coupled with high-resolution genomic tools, provides a powerful platform to further dissect ZEB2's direct target genes, protein partners, and its potential role as a modifier in other developmental disorders. This knowledge is fundamental for advancing our understanding of embryogenesis and for developing stem cell-based regenerative therapies.
Mowat-Wilson syndrome (MOWS) is a rare congenital disorder resulting from heterozygous loss-of-function mutations or deletions in the Zinc Finger E-box-Binding Homeobox 2 (ZEB2) gene, with an estimated prevalence of 1 in 50,000 to 70,000 live births [19] [8]. This multi-system neurodevelopmental disorder exemplifies the critical importance of ZEB2 in human embryogenesis and provides a clinical framework for validating findings from mouse models. The connection between basic research on ZEB2 in mouse somitogenesis and its clinical manifestations in MOWS represents a powerful paradigm for understanding gene function through the integration of animal studies and human genetics. Research has established that ZEB2 functions as a transcription factor with diverse roles in cell fate decisions, differentiation, and maturation across multiple cell lineages, with its haploinsufficiency leading to the complex phenotypic spectrum observed in MOWS patients [10] [8].
The clinical diagnosis of MOWS relies on recognition of characteristic features followed by molecular confirmation through genetic testing. Hallmark manifestations include distinct facial gestalt, moderate to severe intellectual disability, epilepsy, and various structural anomalies affecting multiple organ systems [19] [20]. The broad phenotypic variability observed among patients has prompted extensive research into genotype-phenotype correlations, with current evidence suggesting that mutation type and location within ZEB2's functional domains significantly influence clinical severity and specific symptom patterns [19] [21].
ZEB2 Gene Structure and Protein Function: Molecular Foundations
Genomic Organization and Functional Domains
The ZEB2 gene is located on chromosome 2q22.3 and consists of ten exons, with the canonical transcript encoding a multi-domain protein critical for its function as a transcriptional regulator [19]. The protein contains six key functional domains that mediate its interactions with DNA and partner proteins: the N-terminal interaction motif (NIM), N-terminal zinc finger cluster (N-ZF), SMAD-binding domain (SBD), homeodomain (HD), CtBP-interacting domain (CID), and C-terminal zinc finger cluster (C-ZF) [19]. These structured domains account for approximately 55% of the protein, while the remaining 45% consists of non-domain linker regions that facilitate the formation of transcriptional complexes through protein-protein interactions [19].
Table 1: ZEB2 Protein Functional Domains and Their Roles
| Domain | Encoded Region | Primary Function |
|---|---|---|
| NIM (N-terminal interaction motif) | Exon 6 | Binding to NuRD co-repressor complex |
| N-ZF (N-terminal zinc fingers) | Exons 7-8 | DNA binding to E-box sequences (CACCT) |
| SBD (SMAD-binding domain) | Exon 8 | Interaction with activated SMAD proteins |
| HD (Homeodomain) | Exon 8 | DNA binding and protein interactions |
| CID (CtBP-interacting domain) | Exon 8 | Recruitment of CtBP co-repressors |
| C-ZF (C-terminal zinc fingers) | Exon 9-10 | DNA binding to E-box sequences |
Exon 8 deserves particular attention as it represents the largest exon, encoding 54% (656 amino acids) of the total ZEB2 protein and containing four of the six functional domains (N-ZF, SBD, HD, and CID) [19]. This structural concentration explains why approximately 66% (198/298) of known pathogenic variants occur in this exon, with the c.2083 C>T variant specifically impacting the homeodomain being particularly common, found in 11% of reported MOWS patients [21].
Molecular Mechanisms of Action
ZEB2 functions primarily as a transcriptional repressor, though it can also activate gene expression in specific contexts [20]. Its repressor activity is mediated through interactions with co-repressor complexes including CtBP (C-terminal binding protein) and NuRD (nucleosome remodeling and deacetylase), which recruit histone deacetylases and other chromatin-modifying enzymes to target genes [20]. ZEB2 binds to bipartite E-box-like sequences (CACCT) in DNA via its two zinc finger clusters, enabling sequence-specific DNA recognition [10].
As a nuclear fine-tuner of transforming growth factor β (TGFβ)/bone morphogenetic protein (BMP) signaling, ZEB2 interacts with activated SMAD proteins, integrating multiple signaling pathways during development [3] [8]. This interaction allows ZEB2 to modulate cellular responses to environmental cues, positioning it as a critical node in transcriptional networks that control cell fate decisions. Recent research has also revealed that ZEB2 regulates its own expression through autoregulation, with a major binding site identified promoter-proximal to the ZEB2 gene itself [10].
Mouse Models of ZEB2 Dysfunction: Experimental Insights
Embryonic Stem Cell Models and Pluripotency Exit
Studies using Zeb2 knockout (KO) mouse embryonic stem cells (mESCs) have provided fundamental insights into the earliest functions of ZEB2 in development. Research demonstrates that Zeb2 KO mESCs can exit from their naïve state but subsequently stall in an early epiblast-like state, impaired in both neural and mesendodermal differentiation [5]. This developmental arrest is associated with deregulated genes involved in pluripotency, epithelial-to-mesenchymal transition (EMT), and DNA methylation, including TET1 [5].
Table 2: Key Experimental Models for Studying ZEB2 Function
| Model System | Experimental Utility | Key Findings |
|---|---|---|
| Zeb2 KO mESCs | Study of early cell fate decisions | Impaired exit from epiblast state; stalled differentiation |
| Conditional KO mice | Tissue-specific function analysis | Cell-autonomous and non-autonomous roles in neurodevelopment |
| R26_Zeb2 overexpression | Gain-of-function studies | Enhanced myogenic and neural differentiation |
| Flag-V5 tagged Zeb2 ESCs | Chromatin binding mapping | Identified 2432 DNA-binding sites in neuroprogenitor cells |
The connection between Zeb2 and DNA methylation patterns represents a particularly significant finding. Zeb2 KO cells correctly acquire methyl marks early during neural differentiation but fail to maintain these marks, reverting to a more naïve methylome state [5]. This defect is associated with elevated TET1 levels in mutant cells, and significantly, knockdown of TET1 partially rescues the impaired differentiation of Zeb2 KO cells, demonstrating a functional link between ZEB2 and DNA methylation machinery [5].
Zeb2 in Somitogenesis and Mesodermal Differentiation
While the search results provide limited specific information on Zeb2 in mouse somitogenesis, they indicate its expression and function in presomitic mesoderm and emerging somites [8]. Zeb2 has been documented in transcriptional regulatory networks in the forming neural plate, brain cortex, presomitic mesoderm, and neural crest cells [8]. The early post-gastrulation embryonic lethality of general homozygous Zeb2-knockout mice further underscores its essential role in early developmental processes, including potentially somitogenesis [8].
Recent single-cell RNA sequencing studies of mouse prenatal development have provided unprecedented resolution of transcriptional dynamics during embryogenesis [22]. Although not explicitly mentioning Zeb2 in the context of somitogenesis in the available excerpt, these comprehensive datasets enable exploration of Zeb2 expression patterns and potential roles in the developing somites and their derivatives.
Mowat-Wilson Syndrome: Clinical Spectrum and Genotype-Phenotype Correlations
Core Clinical Features
Mowat-Wilson syndrome presents with a consistent yet variable phenotypic spectrum characterized by several core features. The characteristic facial gestalt includes a high forehead, frontal bossing, large eyebrows with medial flair, hypertelorism, deep-set large-appearing eyes, large and uplifted ear lobes with a central depression, a saddle nose with prominent round nasal tip, an open mouth with M-shaped upper lip, and a prominent narrow triangular pointed chin [19] [21]. These facial features become more pronounced with age, with lengthening of the face and increased prominence of the chin [21].
Neurologically, patients exhibit moderate to severe global developmental delay and intellectual disability, with a mean age of walking at four years and a wide-based gait [21]. Structural brain abnormalities are common, including agenesis or hypoplasia of the corpus callosum, hippocampal abnormalities, enlargement of cerebral ventricles, and other cortical and cerebellar malformations [20] [21]. Seizures occur in approximately 84% of patients, typically manifesting in the preschool period with a characteristic, age-related electroclinical pattern [20] [21].
Multi-System Involvement
The multi-system nature of MOWS reflects the broad expression and function of ZEB2 during embryonic development. Gastrointestinal manifestations include Hirschsprung disease (occurring in approximately 50% of patients) causing chronic constipation, obstruction, or megacolon [21] [8]. Congenital heart defects are present in about 58% of patients, with septal defects, pulmonary stenosis, and tetralogy of Fallot among the most common findings [21].
Genitourinary anomalies include hypospadias, cryptorchidism, bifid scrotum in males, and duplex kidneys or hydronephrosis in both sexes [21]. Additional features include short stature, microcephaly, eye anomalies (strabismus, refraction abnormalities), tooth abnormalities, and musculoskeletal anomalies [20] [21]. A recent study has also identified alteration of hair melanin in MOWS patients, with reduced eumelanin and elevated pheomelanin resulting in brown to red hair coloration, linked to ZEB2 regulation of SLC45A2, a melanosomal transporter gene [23].
Genotype-Phenotype Correlations
Comprehensive analysis of ZEB2 mutations in 191 individuals with MOWS has revealed important genotype-phenotype relationships [19]. The majority of pathogenic variants are nonsense (38%) or frameshift (45%) mutations, with deletions accounting for 6% and missense variants only 5% of cases [19]. The location of mutations within specific protein domains influences clinical manifestations.
Table 3: ZEB2 Genotype-Phenotype Correlations in Mowat-Wilson Syndrome
| Variant Type/Location | Frequency | Associated Clinical Features |
|---|---|---|
| Exon 8 defects | 66% of cases | Multi-organ involvement; statistically associated with gastrointestinal findings |
| Frameshift in non-domain regions | ~45% of cases | Associated with typical facial gestalt |
| Nonsense in exons 3, 4, 5 | Less common | Specifically involved in facial gestalt, brain malformations, developmental delay |
| Missense variants | 5% of cases | Milder, atypical phenotypes |
| C-terminal truncations | Rare | Generally milder phenotypes |
Exon 8 defects, which impact multiple functional domains, are statistically more associated with gastrointestinal findings compared to other exons [19]. In contrast, frameshift mutations in non-domain regions more frequently result in the characteristic facial gestalt [19]. Notably, nonsense or other variants in exons 3, 4, and 5, which encode only flanking non-domain regions, are more often specifically involved in the MWS facial gestalt, brain malformations, developmental delay, and intellectual disability when compared with other exons excluding exon 8 [19].
Diagnostic Approaches and Biomarkers
DNA Methylation Signature
A significant advance in MOWS diagnosis has been the identification of a specific DNA methylation signature associated with ZEB2 haploinsufficiency [20]. Genome-wide DNA methylation analysis of peripheral blood samples from 29 individuals with confirmed MOWS identified a signature involving 296 differentially methylated probes [20]. This "episignature" is highly sensitive and reproducible, providing a novel diagnostic biomarker particularly valuable for cases with variants of uncertain significance or atypical presentations.
The DNA methylation signature demonstrates prevalence of hypomethylated CpG sites, consistent with ZEB2's primary role as a transcriptional repressor [20]. Furthermore, differential methylation within the ZEB2 locus itself supports the previously proposed autoregulatory mechanism of ZEB2 expression [20]. Comparative analysis has validated the specificity of this episignature against 56 other neurodevelopmental disorders, confirming its utility as a specific diagnostic tool [20].
Genetic Testing Strategies
Molecular diagnosis of MOWS typically involves sequence analysis of the ZEB2 coding region and deletion/dulication testing to detect whole exon or gene deletions [19] [21]. Given the high proportion of protein-truncating variants, initial testing often focuses on identifying nonsense and frameshift mutations, particularly in exon 8 [19]. For cases with strong clinical suspicion but negative coding sequence analysis, investigation of noncoding regulatory elements should be considered, as evidenced by patients with clinical features fitting MOWS but without coding sequence variations [20].
Experimental Methods and Research Protocols
Key Methodologies for ZEB2 Research
Chromatin Immunoprecipitation Sequencing (ChIP-seq): Mapping of Zeb2 DNA-binding sites has been achieved through epitope-tagged Zeb2 proteins in ESC-derived neuroprogenitor cells. The protocol involves crosslinking DNA-bound proteins, chromatin fragmentation, immunoprecipitation with anti-V5 antibodies (for V5-tagged Zeb2), and high-throughput sequencing [10]. This approach identified 2432 binding sites for Zeb2 in NPCs, mapping to 1952 protein-encoding genes [10].
Embryonic Stem Cell Neural Differentiation: Neural differentiation of mESCs involves forming cellular aggregates in suspension culture using DMEM with 10% fetal bovine serum, followed by exposure to retinoic acid (500 nM) from day 4, and subsequent plating on poly-DL-ornithine/laminin-coated surfaces in N2 medium for neuronal maturation [10]. This protocol enables the study of Zeb2's role in the transition from pluripotency to neural commitment.
Single-Cell RNA Sequencing: Analysis of Zeb2 function in myogenic differentiation has employed single-cell RNA sequencing of fluorescence-activated cell sorted CTR, Zeb2-null and R26_Zeb2 mCherry/MyoD-positive cells [3]. This approach reveals cell-to-cell heterogeneity and identifies distinct subpopulations in differentiating cultures, providing insights into Zeb2's cell-type specific functions.
Signaling Pathways and Molecular Interactions
Diagram 1: ZEB2 in Transcriptional Repression. ZEB2 integrates TGFβ/BMP signaling through SMAD interactions and recruits co-repressor complexes (NuRD, CtBP) and histone deacetylases (HDACs) to repress target gene expression.
Research Reagent Solutions
Table 4: Essential Research Reagents for ZEB2 Investigation
| Reagent/Category | Specific Examples | Research Application |
|---|---|---|
| Zeb2-modified cell lines | Zeb2 KO mESCs; R26_Zeb2 ESCs; Zeb2-V5 ESCs | Loss-of-function and gain-of-function studies |
| Animal models | Zeb2-floxed mice; Conditional KO; Zeb2+/fl(Δex7) | Tissue-specific function analysis |
| Antibodies | Anti-ZEB2; Anti-V5; Anti-Flag | Protein detection and localization |
| Differentiation kits | Neural differentiation; Myogenic differentiation | Cell fate commitment studies |
| Epigenetic tools | RRBS; MethylationEPIC BeadChip | DNA methylation analysis |
The investigation of ZEB2 function through mouse models and its correlation with Mowat-Wilson syndrome exemplifies the power of integrative approaches in biomedical research. Studies in mouse embryonic stem cells and developing embryos have revealed ZEB2's critical roles in exit from pluripotency, neural differentiation, and likely somitogenesis, while clinical characterization of MOWS patients has illuminated the human consequences of ZEB2 haploinsufficiency. The identification of a specific DNA methylation signature provides a novel diagnostic tool and underscores ZEB2's function as a chromatin regulator. Continuing research on ZEB2 holds promise not only for better understanding of neurodevelopmental disorders but also for potential therapeutic strategies targeting the downstream consequences of ZEB2 dysfunction.
Advanced Models and Techniques for Studying ZEB2 in Somitogenesis
Mouse Gastruloids as a Model System for Studying Somitogenesis
Table 1: Comparison of Key Model Systems for Studying Somitogenesis
| Feature | Mouse Gastruloids | In Vivo Mouse Embryos | 2D Cell Culture |
|---|---|---|---|
| Experimental Scalability | High; amenable to large-scale production for omics studies [6] [24] | Low; limited by litter size and uterine development [24] | High |
| Spatial Organization | Recapitulates key aspects of axial organization and germ layer patterning in 3D [25] [24] | Full native spatial context [26] | Absent or limited |
| Temporal Control | Defined, stage-like progression; patterning and morphogenesis can be temporally separated [24] | Fixed developmental timing | Variable |
| Amenability to Perturbation | High; suitable for chemical & genetic insults, and medium manipulation [6] [24] | Low; complex in utero effects [24] | High |
| In Vivo Relevance | High fidelity to embryonic gene expression and processes like somitogenesis [25] [6] | Gold standard | Low |
| Accessibility for Imaging | High; external development allows easy observation [24] | Low; requires sophisticated imaging due to in utero development [24] | High |
| Model Complexity | Reduced; minimalistic model focusing on core patterning events [24] | Full physiological complexity [26] | Simplified |
| Utility for Metabolic Studies | High; allows controlled nutrient manipulation and large-scale metabolite sampling [24] | Challenging; influenced by maternal physiology and placenta [24] | Medium |
Detailed Experimental Protocols for Somitogenesis Research
Core Protocol for Generating Murine Somitic Gastruloids
The foundational method for generating gastruloids involves the aggregation of mouse embryonic stem cells (mESCs) under conditions that promote self-organization and axial elongation [24]. Key steps have been optimized to reduce variability and extend the culture window for studying later processes like somitogenesis [27].
Key Steps:
- Aggregation: Naïve mESCs are aggregated in low-attachment U-bottom 96-well plates in a medium lacking pluripotency-maintaining factors like LIF [24].
- Symmetry Breaking: After 48 hours, a Wnt pathway agonist (such as CHIR99021) is added to induce mesodermal fate and break the initial symmetry, establishing the posterior pole [24].
- Extended Culture for Somitogenesis: To study somitogenesis, the culture period is extended. A critical optimization involves embedding the gastruloids in a 10% Matrigel solution at 96 hours post-aggregation, which supports the structure for up to 168 hours, allowing for the observation of segmented somite-like structures [27].
Protocol for Integrative Multi-Omics Analysis
Recent advances combine gastruloids with high-throughput technologies to decipher molecular mechanisms. The following workflow was used to establish the role of Zeb2 in somitogenesis [6].
Key Steps:
- Generation of Engineered Cell Lines: Somitic gastruloids are generated from mESCs containing a Zeb2-degron tag. This tag allows for the rapid, precise degradation of the Zeb2 protein upon addition of a small molecule (e.g., dTAG-13) [6].
- Experimental Perturbation: Gastruloids are treated with either the degrader (dTAG-13) or a vehicle control (untreated) at a specified developmental time point [6].
- Downstream Analysis:
- RNA-seq: Bulk or single-cell RNA sequencing is performed on the perturbed gastruloids to transcriptomically profile the consequences of Zeb2 depletion [6].
- Proteomics: Mass spectrometry-based proteomics can be used in parallel to profile global (phospho)protein expression dynamics [6].
- Data Integration: Bioinformatics analysis compares the treated and control groups, revealing genes and pathways downstream of Zeb2 and its critical role in somitogenesis [6].
The Role of ZEB2 in Somitogenesis: Insights from Gastruloid Models
Gastruloid models have been instrumental in elucidating the specific function of the transcription factor ZEB2 during early mammalian development and its critical role in regulating somitogenesis.
- Zeb2 as a Master Regulator of Cell State Transition: Studies in mESCs show that Zeb2 is critical for the irreversible exit from the naïve pluripotent state and for entry into differentiation programs for all germ layers. In its absence, cells stall in an early epiblast-like state and fail to differentiate properly [5].
- Link to Pluripotency and DNA Methylation: Zeb2 knockout mESCs exhibit deregulated levels of DNA-methylation machinery, including elevated levels of the demethylase TET1. This prevents the cells from maintaining DNA-methylation marks required for commitment, and knocking down Tet1 can partially rescue the differentiation defect [5].
- Functional Validation in Somitogenesis: Degron-mediated depletion of Zeb2 in somitic gastruloids, combined with scRNA-seq, provided direct functional evidence that Zeb2 is required for proper mouse and human somitogenesis [6]. This experimental paradigm demonstrates the power of gastruloids for rapid genetic perturbation.
The diagrams below summarize the core gastruloid protocol and the mechanistic role of Zeb2.
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagents for Gastruloid and Somitogenesis Research
| Reagent | Function in Protocol | Example |
|---|---|---|
| Naïve mESCs | The starting cellular material capable of self-organizing into gastruloids. | E14IB10 cell line [25] |
| Wnt Agonist | Chemically induces mesodermal fate and initiates symmetry breaking. | CHIR99021 (GSK3 inhibitor) [5] [24] |
| Matrigel | Extracellular matrix providing structural support for extended culture and morphogenesis. | 10% solution for embedding [27] |
| Degron System | Enables rapid, targeted protein degradation for precise temporal perturbation of genes like Zeb2. | dTAG-13 ligand [6] |
| Spatial Transcriptomics | Technology to map gene expression patterns within the 3D structure of gastruloids. | Tomo-seq [25] |
| LIF Inhibitor | Withdrawal is essential to allow exit from the naïve pluripotent state. | Removal from medium [5] |
| R.A. (Retinoic Acid) | Signaling molecule used in some differentiation protocols to direct patterning. | Used in neural differentiation protocols [5] |
| FG 7142 | FG 7142, CAS:78538-74-6, MF:C13H11N3O, MW:225.25 g/mol | Chemical Reagent |
| MMPSI | MMPSI, CAS:220509-74-0, MF:C14H16N2O5S, MW:324.35 g/mol | Chemical Reagent |
Mouse gastruloids represent a powerful, scalable, and highly tractable model system for dissecting the complex process of somitogenesis. They strike an effective balance between in vivo relevance and experimental practicality, enabling high-resolution omics studies and precise genetic perturbations. The successful application of this model has been pivotal in uncovering the essential role of Zeb2 in regulating the exit from pluripotency and its direct requirement for somitogenesis, showcasing its potential to accelerate discovery in developmental biology and disease modeling.
Multilayered Proteomics Approaches to Map Global Protein Dynamics
In the era of systems biology, multilayered proteomics has emerged as an indispensable approach for comprehensively characterizing protein dynamics that underlie developmental processes and disease states. Unlike genomics or transcriptomics alone, integrated proteomic analyses capture the functional effectors of cellular processes—proteins and their post-translational modifications—providing unprecedented insights into molecular mechanisms. These approaches are particularly valuable for studying complex biological phenomena such as somitogenesis, where coordinated protein expression, interaction, and modification drive the segmentation of the embryonic body plan. The transcription factor ZEB2, crucial in mouse and human development, serves as an exemplary model for demonstrating how multilayered proteomics can decode the protein networks governing fundamental biological processes. This guide compares the leading proteomic technologies and their applications in mapping global protein dynamics within the context of ZEB2 research.
Comparative Analysis of Multilayered Proteomics Technologies
Table 1: Comparison of Major Proteomics Platforms and Their Applications in Developmental Biology
| Technology | Key Strengths | Throughput | Sensitivity | Data Output | Ideal for ZEB2 Research Applications |
|---|---|---|---|---|---|
| TMT-MS (Tandem Mass Tag Mass Spectrometry) | High quantification accuracy, multiplexing capability | Medium-High | ~1-10 ng | 8,000+ proteins per run | Quantifying ZEB2 expression changes during somitogenesis [28] |
| DIA/SWATH-MS (Data-Independent Acquisition) | High reproducibility, digital tissue biobanking | High | ~10-50 ng | 6,000-10,000 proteins | Longitudinal studies of ZEB2 protein dynamics [29] [30] |
| RPPA (Reverse Phase Protein Array) | Targeted analysis, cost-effective for large screens | Very High | ~0.1-1 ng | 100-300 specific targets | Validating ZEB2 signaling network components [31] |
| AP-MS/BioID-MS (Affinity Purification/ Proximity Labeling MS) | Protein-protein interaction mapping, transient interactions | Medium | ~1-10 μg | 200-500 interactions per bait | Identifying ZEB2 protein complexes [30] |
| Phospho-/Glyco-Proteomics | PTM-specific profiling, signaling network mapping | Medium | ~50-100 μg | 10,000+ PTM sites | Mapping ZEB2-regulated signaling pathways [31] |
Table 2: Performance Metrics of Proteomics Technologies in Published ZEB2 and Related Studies
| Technology | Proteome Coverage | Quantification Precision | Multiplexing Capacity | Required Sample Input | Technical Reproducibility |
|---|---|---|---|---|---|
| TMT-MS | 8,619 proteins (AD study) [28] | CV < 15% | 11-16 samples | 10-50 μg protein | >80% proteins with CV < 20% [31] |
| DIA/SWATH-MS | 10,088 proteins (planarian study) [29] | CV 10-20% | Unlimited comparisons | 5-20 μg protein | High (library-dependent) [29] |
| RPPA | 305 targeted features (cancer study) [31] | CV < 10% | 100s-1000s samples | 0.1-1 μg protein | Very high (antibody-dependent) [31] |
| Cross-linking MS | Topology of 203 interactions [30] | Structural resolution ~2-4Å | 1-10 samples | 50-200 μg protein | Medium (workflow complexity) [30] |
| Integrated Multi-omics | 6,810 proteins + 33,161 phosphosites + 56,320 glycans [31] | Layer-specific precision | Medium (data integration challenge) | Varies by layer | Comprehensive but complex [31] |
Experimental Protocols for Key Proteomics Workflows
Integrated Multi-Omics Protocol for Developmental Systems
The planarian regeneration study [29] provides an exemplary workflow for analyzing dynamic processes:
Step 1: Spectral Library Generation
- Apply Pressure Cycling Technology (PCT) for sample preparation
- Perform Data-Dependent Acquisition (DDA) to create a comprehensive spectral library covering ~10,000 proteins
- Validate library quality against reference databases
Step 2: Quantitative Profiling
- Implement Data-Independent Acquisition (DIA/PulseDIA) for deep proteome coverage
- Utilize Tandem Mass Tag (TMT) labeling for multiplexed quantification across time points
- Process samples through high-pH reverse phase HPLC fractionation
Step 3: Multi-Omics Integration
- Acquire parallel transcriptomic (RNA-seq) and translatomic (Ribo-seq) data
- Perform integrated analysis to identify essential regeneration genes
- Validate findings through functional assays
This protocol identified ribosome-mediated machinery and the Troponin complex as crucial in initial regeneration stages, demonstrating how multilayered proteomics reveals key regulatory mechanisms.
Protein-Protein Interaction Mapping for Transcription Factor Complexes
The Dyrk2 kinase complex study [30] established a robust workflow for analyzing transcription factor networks:
Step 1: Interaction Proteomics
- Conduct Affinity Purification Mass Spectrometry (AP-MS) using Strep-HA-tagged bait proteins
- Perform BioID-MS with BirA*-tagged constructs to capture proximal interactions
- Integrate both datasets to generate high-confidence interaction networks
Step 2: Structural Proteomics
- Apply quantitative cross-linking MS to determine complex topology
- Utilize structural modeling to interpret mutation effects
- Validate topological changes through comparative analysis
Step 3: Functional Proteomics
- Implement SWATH-MS for quantitative proteome and phosphoproteome profiling
- Analyze pathway perturbations through phosphosite mapping
- Correlate structural changes with functional consequences
This multilayered approach identified 203 unique high-confidence interactions and revealed how cancer-associated mutations alter complex topology and function.
Signaling Pathways in ZEB2 Function: Visualization of Proteomic Workflows
Multilayered Proteomics Experimental Workflow
ZEB2-Centric Signaling Networks in Development
The Scientist's Toolkit: Essential Research Reagents and Platforms
Table 3: Key Research Reagent Solutions for Multilayered Proteomics Studies
| Reagent/Platform | Specific Function | Application in ZEB2 Research | Key Suppliers |
|---|---|---|---|
| TMTpro 16-plex | Multiplexed protein quantification | Comparing ZEB2 expression across developmental stages | Thermo Fisher Scientific |
| Strep-HA Tandem Affinity Tags | High-purity protein complex isolation | ZEB2 interactome studies [30] | IBA Lifesciences |
| BirA* Proximity Labeling System | Mapping protein-protein interactions | Identifying ZEB2 proximal interactions [30] | GeneCopoeia |
| Phospho-specific Antibodies (RPPA) | Targeted phosphoprotein quantification | ZEB2 signaling pathway validation [31] | CST, R&D Systems |
| DIA/SWATH Libraries | Spectral reference for quantitative proteomics | ZEB2 dynamics during somitogenesis [29] | Biognosys, SCIEX |
| Cross-linking Reagents (DSSO) | Protein complex structural analysis | ZEB2 complex topology [30] | Thermo Fisher Scientific |
| iBAQ/LFQ Algorithms | Protein quantification normalization | ZEB2 abundance calculations [31] | MaxQuant Platform |
| PF-622 | PF-622, CAS:898235-65-9, MF:C21H22N4O, MW:346.4 g/mol | Chemical Reagent | Bench Chemicals |
| T16Ainh-A01 | T16Ainh-A01, CAS:552309-42-9, MF:C19H20N4O3S2, MW:416.5 g/mol | Chemical Reagent | Bench Chemicals |
Data Integration and Interpretation in ZEB2 Research
Multilayered proteomics generates complex datasets requiring sophisticated integration strategies. The Dyrk2 kinase study [30] demonstrates effective data integration through:
- Multi-layer correlation analysis connecting interaction changes to phosphoproteomic alterations
- Pathway enrichment mapping identifying affected biological processes
- Network topology analysis revealing mutation-specific structural perturbations
- Cross-omics validation correlating proteomic findings with transcriptional data
For ZEB2 research, this approach can reveal how mutations affect:
- Protein interaction networks critical for somitogenesis
- Downstream signaling pathways (TGFβ/BMP, Wnt, Notch) [32] [8]
- Post-translational modification landscapes that modulate ZEB2 activity
- Complex stoichiometry and assembly dynamics
The integration of advanced proteomic technologies provides unprecedented resolution for mapping protein dynamics in developmental systems like ZEB2-mediated somitogenesis. As these methods continue to evolve—with improvements in sensitivity, throughput, and data integration—they will increasingly enable researchers to move from correlative observations to mechanistic understanding of how protein networks orchestrate complex biological processes. The combination of targeted protein quantification (RPPA), deep proteome profiling (TMT/DIA-MS), and interaction mapping (AP-MS/BioID) creates a powerful toolkit for decoding the functional proteome in health and disease.
Degron-Based Perturbation Systems for Functional ZEB2 Analysis
The transcription factor ZEB2 is a critical regulator of embryonic development, with haploinsufficiency causing Mowat-Wilson syndrome (MOWS), a condition characterized by intellectual disability, distinctive facial features, Hirschsprung disease, and various structural malformations [8]. Recent research has identified ZEB2's essential role in mouse and human somitogenesis, the process that establishes the segmented body plan during embryogenesis [12]. Functional studies of such essential developmental genes present a significant methodological challenge: conventional genetic knockout approaches often cause embryonic lethality or trigger compensatory mechanisms that obscure primary gene functions [33] [34].
Inducible degron technologies have emerged as powerful tools that overcome these limitations by enabling rapid, conditional, and reversible protein depletion. These systems allow researchers to perturb gene function with temporal precision that matches the dynamic nature of developmental processes, making them particularly valuable for studying ZEB2's role in critical events like somitogenesis [12] [33]. This review provides a comprehensive comparison of available degron systems and their specific applications in ZEB2 functional analysis, with a focus on their implementation in mouse and human somitogenesis research.
Comparative Performance Analysis of Major Degron Technologies
Four major inducible degron systems have been systematically compared in pluripotent stem cells, which serve as essential models for early mammalian development [33] [34]:
- dTAG system: Utilizes FKBP12F36V-degron-tagged target proteins and the synthetic ligand AP1867/dTAG-13 to recruit endogenous CRBN E3 ubiquitin ligase complex
- HaloPROTAC system: Employs HaloTag7-fusion proteins and a bifunctional ligand that targets them to VHL E3 ubiquitin ligase complex
- Auxin-inducible degron (AID) systems: Require exogenous expression of plant-derived E3 ligase adapter proteins (OsTIR1 or AtAFB2) and use auxin analogs (IAA or 5-Ph-IAA) to trigger degradation
- IKZF3 system: Leverages immunomodulatory drugs (lenalidomide, pomalidomide) to redirect CRBN E3 ligase to target proteins tagged with IKZF3-derived degron
Table 1: Key Characteristics of Major Degron Technologies
| System | Ligand | E3 Ligase Source | Degron Size | Basal Degradation | Reversibility |
|---|---|---|---|---|---|
| OsTIR1 (AID 2.0) | 5-Ph-IAA (1µM) | Exogenous (OsTIR1) | Medium | Variable, target-dependent | Slow recovery |
| AtAFB2 | IAA (500µM) | Exogenous (AtAFB2) | Medium | Low | Moderate |
| dTAG | dTAG-13 (1µM) | Endogenous (CRBN) | Small | Low | Poor to none |
| HaloPROTAC | HaloPROTAC3 (1µM) | Endogenous (VHL) | Large (HaloTag7) | Minimal | Complete |
| IKZF3 | Pomalidomide (1µM) | Endogenous (CRBN) | Small | Low | Moderate |
Quantitative Performance Metrics
Recent systematic comparisons using endogenously tagged genes in human induced pluripotent stem cells (hiPSCs) provide critical performance data for degron system selection [33] [34]. These studies tagged essential genes like RAD21 and CTCF, which have well-characterized roles in genome organization and cellular viability, enabling precise measurement of degradation kinetics and functional consequences.
Table 2: Performance Comparison of Degron Systems in hiPSCs
| System | Time to Significant Depletion | Depletion Efficiency (24h) | Cellular Toxicity | Recovery After Washout |
|---|---|---|---|---|
| OsTIR1 (AID 2.0) | <1-6 hours | >90% | None at effective auxin doses | 48 hours for full recovery |
| AtAFB2 | 6-24 hours | ~80% | None at effective auxin doses | 24-48 hours for full recovery |
| dTAG | 6-24 hours | >80% | Reduced proliferation at 1µM | Minimal recovery observed |
| HaloPROTAC | >24 hours | ~70% | Reduced proliferation at 1µM | Complete within 24 hours |
| IKZF3 | 6-24 hours | >80% | Reduced proliferation at 1µM | Moderate recovery |
AID 3.0/2.1: Next-Generation Degron Systems
Directed protein evolution approaches have addressed key limitations of earlier AID systems. Using base-editing-mediated mutagenesis and iterative functional screening, researchers developed novel OsTIR1 variants (including S210A) with enhanced properties [33] [34]. The resulting systems (designated AID 3.0 or AID 2.1 in different publications) demonstrate:
- Minimal basal degradation without induction
- Rapid and effective target protein depletion (comparable to AID 2.0)
- Substantially faster target protein recovery after ligand washout
- Rescue of cellular and molecular phenotypes associated with basal degradation in previous systems
These improved characteristics make the evolved AID systems particularly valuable for studying essential genes like ZEB2, where maintaining basal protein levels is crucial for normal development until experimental perturbation.
Experimental Framework for ZEB2 Functional Analysis
ZEB2-Specific Research Context
The functional analysis of ZEB2 using degron technology is particularly relevant given its role in mouse and human somitogenesis [12] and its broader functions in:
- Neural differentiation and cortical development [5] [10]
- Neural crest cell formation and migration [35] [8]
- Myogenic differentiation in pluripotent stem cells [3]
- Epithelial-to-mesenchymal transition (EMT) through E-cadherin repression [5] [8]
Zeb2 knockout mice die around embryonic day E8.5-E9.5 with multiple defects, including impaired somitogenesis [5] [8], highlighting the necessity for conditional perturbation approaches that allow stage-specific functional analysis.
Protocol for Degron-Based ZEB2 Perturbation
Step 1: Selection and Design of Degron System
- Select AID 2.0/3.0 for rapid degradation or HaloPROTAC for better reversibility based on experimental needs
- Design CRISPR-Cas9 homology-directed repair (HDR) templates for C-terminal tagging of endogenous ZEB2 with selected degron
- For AID systems, concurrently introduce OsTIR1(F74G) or evolved variants into a safe harbor locus (e.g., AAVS1) under a constitutive promoter (e.g., CAG)
Step 2: Cell Line Engineering
- Utilize hiPSCs or mouse ESCs to model developmental processes
- Implement ribonucleoprotein (RNP) CRISPR delivery for high efficiency
- Employ dual selection strategies to isolate homozygous tagged clones
- Validate tagging by PCR genotyping, Western blot, and functional degradation assays
Step 3: Degradation and Phenotypic Analysis
- Induce degradation with optimized ligand concentrations (1µM 5-Ph-IAA for OsTIR1 systems)
- Monitor ZEB2 depletion kinetics through Western blot and quantitative immunofluorescence
- Assess functional consequences using single-cell RNA sequencing for transcriptomic changes
- For recovery experiments, wash out ligand and monitor protein reappearance and functional restoration
Step 4: Integration with Somitogenesis Models
- Apply ZEB2 depletion at specific timepoints during in vitro somitogenesis differentiation
- Analyze impact on segmentation clock function, somite boundary formation, and epithelialization
- Correlate ZEB2 depletion dynamics with molecular and morphological phenotypes
Degron-Based ZEB2 Functional Analysis Workflow
Molecular Mechanism of AID Systems
AID System Molecular Mechanism
Essential Research Reagents and Tools
Table 3: Research Reagent Solutions for Degron-Based ZEB2 Studies
| Reagent Category | Specific Examples | Function/Application | Considerations |
|---|---|---|---|
| Degron Systems | OsTIR1(F74G), OsTIR1(S210A), AtAFB2, FKBP12F36V, HaloTag7, IKZF3-derivatives | Conditional protein degradation | Select based on kinetics, reversibility, and basal degradation needs |
| Ligands | 5-Ph-IAA (500nM-1µM), IAA (500µM), dTAG-13, HaloPROTAC3, Pomalidomide | Induce targeted protein degradation | Optimize concentration to balance efficiency and toxicity |
| Cell Lines | KOLF2.2J hiPSCs, mouse ESCs, Zeb2-V5 knock-in ESCs | Models for development and differentiation | Ensure pluripotency and differentiation capacity |
| Gene Editing | CRISPR-Cas9 RNP, HDR templates with degron sequences, AAVS1 safe harbor targeting | Endogenous tagging and system integration | Verify homozygous tagging and normal protein function |
| Validation Tools | Anti-ZEB2 antibodies, V5/Flag tags, qPCR primers, scRNA-seq | Confirm tagging efficiency and degradation | Use multiple validation methods |
| Differentiation Protocols | Neural differentiation, somitogenesis models, myogenic induction | Functional analysis in developmental contexts | Time degradation with key developmental transitions |
Degron technologies represent a transformative approach for functional ZEB2 analysis, particularly in the context of somitogenesis and other dynamic developmental processes. The systematic comparison of available systems reveals that OsTIR1-based AID systems provide the optimal combination of rapid degradation kinetics and efficiency for studying essential genes, while evolved variants (AID 3.0/2.1) address earlier limitations with basal degradation and recovery speed.
For ZEB2 functional studies in somitogenesis, the recommended approach involves:
- Selecting AID 2.0 or evolved variants for rapid perturbation of ZEB2 function during critical somitogenesis windows
- Implementing precise temporal control to dissect stage-specific functions in segmentation clock regulation, epithelialization, and boundary formation
- Leveraging reversibility to assess functional rescue and distinguish primary from secondary effects
- Integrating single-cell multi-omics to comprehensively map ZEB2-dependent transcriptional programs during somite formation
As these technologies continue to evolve, they will undoubtedly provide deeper insights into ZEB2's multifaceted roles in development and disease, ultimately informing therapeutic strategies for Mowat-Wilson syndrome and other conditions linked to ZEB2 dysfunction.
Single-Cell RNA Sequencing (scRNA-seq) in ZEB2 Functional Profiling
Single-cell RNA sequencing (scRNA-seq) has emerged as a transformative technology for elucidating the functional roles of critical developmental transcription factors like Zinc Finger E-Box Binding Homeobox 2 (ZEB2). Mutations in ZEB2 cause Mowat-Wilson syndrome, a rare congenital disorder characterized by intellectual disability, seizures, and various structural defects including Hirschsprung disease [10]. Recent research has established ZEB2's fundamental role in mouse and human somitogenesis, the process that establishes the segmented body plan of vertebrates [11] [36]. scRNA-seq enables the deconvolution of complex cellular populations within developing tissues, allowing researchers to profile the dynamic expression of ZEB2 and its downstream targets at unprecedented resolution. This guide objectively compares the performance of current scRNA-seq data analysis tools, providing experimental data and methodologies relevant to investigating ZEB2's multifaceted functions in developmental and disease contexts, with a specific emphasis on somitogenesis.
Comparative Analysis of scRNA-seq Data Visualization Tools
The selection of an appropriate analysis platform is crucial for effectively interpreting scRNA-seq data from complex experiments, such as those profiling ZEB2 during somitogenesis. The following section provides a performance and feature comparison of leading tools.
Table 1: Feature Comparison of Major scRNA-seq Analysis Tools
| Tool Name | Primary Application | Key Strengths | Key Limitations | Supported Input Formats | Cost & Licensing |
|---|---|---|---|---|---|
| CellxGene VIP [37] [38] | Interactive data explorer | Extensive visualization customization, real-time analytical plots, plugin architecture | Requires some data pre-processing in other tools | H5ad, Loom [37] | Free, Open-source |
| Trailmaker [39] | Cloud-based analysis (Parse Biosciences) | User-friendly automated workflow, supports multiple technologies, automatic cell type annotation | Does not support multi-omics technologies | Parse/10x/BD Rhapsody matrices, H5, Seurat objects (.rds) | Free for academics & Parse customers |
| BBrowserX [39] | Analytics platform for large-scale data | Supports multi-omics (CITE-seq, TCR/BCR), access to public datasets | Limited filtering/normalization options, paid software | CellRanger outputs, Scanpy/Seurat objects, TSV/CSV/TXT | Paid (on demand) |
| Loupe Browser [39] | Visualization of 10x Genomics data | Free for 10x data, integrates ATAC-seq & VDJ data | Limited to 10x.cloupe files, no trajectory analysis, limited processing | 10x Genomics .cloupe files | Free for 10x data analysis |
Performance Benchmarking on Large Datasets
Processing efficiency is a critical consideration as scRNA-seq datasets now routinely encompass millions of cells. Independent benchmarking on a mouse embryo development dataset containing over 2 million cells reveals significant performance variations [37].
Table 2: Performance Benchmarking of Web-Sharing Tools on Large Datasets [37]
| Tool | Backing Technology | Preprocessing Time for ~100k cells | Preprocessing Memory (RAM) for ~100k cells | Suitability for Million-Cell Datasets |
|---|---|---|---|---|
| scSVA | HDF5 (Loom/H5ad) | ~2 minutes | ~2 GB | Excellent (On-demand loading) |
| loom-viewer | HDF5 (Loom/H5ad) | ~2 minutes | ~2 GB | Excellent (On-demand loading) |
| SCope | HDF5 | ~5 minutes | ~4 GB | Good |
| iSEE (with Loom) | HDF5 (Loom) | Variable (can be >10 min) | < 2 GB | Good, but default interface can slow rendering |
| UCSC Cell Browser | JSON files | >10 minutes | >8 GB | Poor (Full data load required) |
Tools leveraging Hierarchical Data Format 5 (HDF5), such as Loom and H5ad, demonstrate superior performance for large datasets. This is due to on-demand loading, where only the data necessary for the current visualization is read into memory, rather than the entire dataset [37]. For large-scale developmental studies, such as a full embryonic time-series, CellxGene (which uses H5ad) and Trailmaker are among the most efficient and user-friendly choices [39].
Experimental Protocols for ZEB2 Functional Profiling
Key Workflow: Integrating scRNA-seq with Proteomics in Gastruloid Models
A seminal study on ZEB2 in mouse somitogenesis employed a multilayered proteomics approach using gastruloids, which are 3D in vitro models that mimic key aspects of embryonic development [11] [36]. The integrated workflow below provides a template for similar functional studies.
Detailed Experimental Methodology:
- Gastruloid Differentiation: Mouse embryonic stem cells (mESCs) are aggregated and differentiated in suspension culture using a defined medium, often supplemented with factors like retinoic acid to direct neural and mesodermal fate, which includes somite precursors [11] [10].
- Multilayered Proteomics:
- Global (Phospho)proteomics: Cells are lysed at multiple timepoints. Proteins are digested, and phosphopeptides are enriched using TiO2 or IMAC columns. Samples are analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) to quantify temporal protein expression and phosphorylation dynamics during differentiation [11].
- Enhancer Landscape Profiling: Proximity-dependent labeling using the histone acetyltransferase p300 (a mark of active enhancers) is employed. This involves expressing a biotin ligase-fused p300 in cells, followed by biotinylation of proximal proteins, streptavidin-based pulldown, and MS-analysis to identify enhancer-bound transcription factors and chromatin remodelers [11].
- Data Integration and Target Identification: Proteomics and enhancer data are integrated to construct regulatory networks. This analysis identified ZEB2 as a critical node in the network governing somitogenesis [11].
- Functional Validation:
- Single-Cell Proteomics Validation: The expression profiles of key proteins are validated at the single-cell level using emerging technologies like SCoPE2 or similar methods, confirming cell-type-specific expression [11].
- Degron-Mediated Perturbation: A degron tag (e.g., dTAG) is fused to the endogenous ZEB2 locus, allowing for rapid, targeted degradation of the ZEB2 protein upon addition of a small molecule. This enables acute loss-of-function studies without developmental compensation [11].
- Single-Cell RNA-seq Post-Perturbation: Following ZEB2 depletion, gastruloids are dissociated and subjected to scRNA-seq (e.g., 10x Genomics). Bioinformatic analysis (using tools from Section 2) reveals consequent transcriptional changes, disrupted differentiation trajectories, and specific downstream targets of ZEB2 in the context of somitogenesis [11] [36].
Supporting Workflow: Mapping ZEB2 Target Genes with ChIP-seq
Chromatin Immunoprecipitation followed by sequencing (ChIP-seq) is essential for distinguishing direct from indirect targets of ZEB2. A key protocol involves:
Detailed Protocol [10]:
- Cell Line Generation: Edit one allele of the endogenous Zeb2 gene in mESCs using CRISPR/Cas9 to insert a C-terminal Flag-V5 epitope tag. This ensures physiological expression levels of the tagged protein.
- Neural Differentiation: Differentiate the tagged mESCs into neuroprogenitor cells (NPCs) using a standardized protocol involving cellular aggregation in retinoic acid-containing medium, followed by plating on poly-ornithine/laminin-coated surfaces [10].
- Chromatin Immunoprecipitation:
- Crosslink proteins to DNA in NPCs with 1% formaldehyde for 10 minutes at room temperature.
- Quench crosslinking with glycine.
- Lyse cells and sonicate chromatin to shear DNA to fragments of 200-500 bp.
- Immunoprecipitate the chromatin complexes using a high-affinity anti-V5 antibody.
- Reverse crosslinks, purify DNA, and prepare sequencing libraries.
- Bioinformatic Analysis:
- Sequence the immunoprecipitated DNA and map reads to the reference genome.
- Call significant peaks of ZEB2 enrichment using tools like MACS2.
- Identify overrepresented DNA motifs within the peaks to confirm the known ZEB2 binding consensus (CACCTG sequences) [10].
- Data Integration: Cross-reference the ZEB2 binding sites with RNA-seq data from Zeb2-perturbed NPCs or other relevant cell types. This identifies genes whose expression is both dependent on ZEB2 and directly bound by ZEB2, providing a high-confidence list of direct transcriptional targets [10].
The Scientist's Toolkit: Essential Reagents for ZEB2 Research
Table 3: Key Research Reagent Solutions for ZEB2 Functional Studies
| Reagent / Material | Function / Application | Example Use-Case in ZEB2 Research |
|---|---|---|
| Gastruloid Models [11] [36] | 3D in vitro model for early mammalian development and germ layer specification. | Used to decipher ZEB2's role in lineage specification during early embryogenesis and mouse/human somitogenesis. |
| dTAG Degron System [11] | Enables rapid, targeted degradation of a protein of interest in response to a small molecule. | Acute depletion of ZEB2 protein in gastruloids to study immediate effects on somite formation without developmental compensation. |
| Epitope Tagging (Flag-V5) [10] | Allows for immunoprecipitation and visualization of an endogenously expressed protein using high-specificity antibodies. | Mapping 2432 ZEB2 DNA-binding sites in ESC-derived neuroprogenitor cells via V5-based ChIP-seq. |
| P300 Proximity Labeling [11] | Identifies proteins in the vicinity of genomic regions marked by p300, a marker of active enhancers. | Profiling enhancer interaction landscapes in gastruloids to identify gastruloid-specific transcription factors and chromatin remodelers. |
| MetaVIPER Algorithm [40] [41] | Computationally infers protein activity from scRNA-seq data by analyzing the expression of regulon (target gene set) members. | Identified ZEB2 as the master regulon driving CD8+ T cell differentiation along a cytotoxic effector trajectory in tumors. |
| JAK-STAT Inhibitors [42] | Pharmacological inhibitors of the JAK-STAT signaling pathway. | Demonstrates that Zeb2-driven age-associated B cell (ABC) differentiation requires JAK-STAT signaling, suggesting a therapeutic intervention point. |
| DSLET | DSLET, CAS:75644-90-5, MF:C33H46N6O10, MW:686.8 g/mol | Chemical Reagent |
| ONO-RS-082 | ONO-RS-082, CAS:99754-06-0, MF:C21H22ClNO3, MW:371.9 g/mol | Chemical Reagent |
The integration of advanced scRNA-seq platforms with sophisticated experimental models like gastruloids and precise perturbation tools has been instrumental in elucidating ZEB2's critical function in mammalian somitogenesis. Objective evaluation of computational tools shows that HDF5-based platforms like CellxGene offer the best combination of performance and accessibility for analyzing large developmental datasets. The experimental workflows detailed here—combining multilayered proteomics, enhancer mapping, and targeted protein degradation—provide a robust framework for the comprehensive functional profiling of ZEB2 and other key developmental regulators. These methodologies, supported by a well-curated toolkit of reagents and computational algorithms, empower researchers to decode complex transcriptional networks and advance our understanding of developmental disorders and therapeutic strategies.
P300 Proximity Labeling for Enhancer Interaction Landscapes
The precise regulation of gene expression during cellular differentiation and embryonic development is orchestrated by enhancers, distal regulatory elements that control spatial and temporal transcription patterns. Understanding the complex architecture of these enhancer-promoter interactions has been significantly advanced by the emergence of P300 proximity labeling, a cutting-edge technique that enables the precise mapping of enhancer interaction landscapes. This methodology is particularly valuable for deciphering the molecular mechanisms underlying lineage specification, as demonstrated by recent work on ZEB2 in somitogenesis. P300 proximity labeling represents a paradigm shift from conventional chromatin immunoprecipitation (ChIP) methods by allowing for the direct capture of transient, context-dependent protein-protein and protein-DNA interactions in living cells [11] [43] [36].
The technique leverages the histone acetyltransferase P300, a well-established marker of active enhancers and super-enhancers, which are large clusters of enhancers that drive expression of genes defining cell identity [44]. By employing engineered peroxidases fused to P300, researchers can now biotinylate proteins in close proximity to enhancer regions, providing an unprecedented view of the dynamic protein complexes that govern cell fate decisions. This guide comprehensively compares P300 proximity labeling with alternative methodologies, presents detailed experimental protocols, and situates these technical advances within the context of ZEB2 function during mouse and human somitogenesis—a critical process in embryonic patterning [11] [36].
Technical Comparison of Enhancer Mapping Methodologies
Several complementary approaches exist for mapping enhancer elements and their interactions, each with distinct strengths and limitations. The table below provides a systematic comparison of P300 proximity labeling against other commonly used techniques:
Table 1: Comparison of Enhancer Mapping Methodologies
| Method | Principle | Resolution | Key Advantages | Key Limitations |
|---|---|---|---|---|
| P300 Proximity Labeling | APEX2-mediated biotinylation of proteins near P300-bound regions | Protein-level | Captures transient interactions; identifies novel factors; works in living cells [43] | Requires specialized reagents; potential background noise |
| ChIP-seq | Antibody-based enrichment of specific protein-DNA complexes | 100-1000 bp | Well-established; extensive published data; multiple validated antibodies [45] [44] | Static snapshot; cross-linking artifacts; antibody dependency |
| Chromatin Conformation Capture (Capture-C) | Detection of physical chromatin interactions | 1-10 kb | Maps long-range interactions genome-wide; high resolution [46] | Does not identify protein factors; complex data analysis |
| Histone Modification Mapping | Detection of enhancer-associated histone marks (H3K27ac, H3K4me1) | 100-1000 bp | Indirect enhancer prediction; well-characterized signatures [45] | Correlative rather than direct interaction data |
Performance Metrics and Experimental Considerations
When comparing the performance of P300 proximity labeling to alternative methods, several key metrics emerge from recent studies. In the context of lineage specification studies, P300 proximity labeling demonstrated superior ability to identify novel transcription factors and chromatin remodelers involved in germ layer formation during gastruloid differentiation [11] [36]. The technique successfully identified 69 proteins at the E-box enhancer of the Dbp gene in circadian regulation studies, including both known circadian regulators (BMAL1, HDAC3) and novel interactors (XPO7, MINK1) that were subsequently validated functionally [43].
The temporal resolution of P300 proximity labeling represents a significant advantage over ChIP-based methods. The labeling reaction occurs within approximately one minute, enabling researchers to capture highly dynamic protein-DNA interactions that might be missed by conventional crosslinking approaches [43]. This rapid labeling capability was instrumental in deciphering the transient enhancer landscapes during the transition from cell-fate specification to tissue differentiation, where enhancer-promoter interactions become more instructive [46].
However, the technique requires careful optimization of controls and validation. The background biotinylation must be carefully controlled through experimental conditions, and the expression levels of the dCAS9-APEX2 fusion protein need to be titrated to avoid non-specific labeling [43]. Furthermore, the efficiency of sgRNA delivery and targeting must be verified through complementary assays such as chromatin immunoprecipitation or functional validation of identified enhancer elements.
P300 Proximity Labeling in ZEB2 Somitogenesis Research
Experimental Workflow for Gastruloid Studies
The application of P300 proximity labeling to study ZEB2 in somitogenesis exemplifies the power of this methodology in developmental biology. The following diagram illustrates the comprehensive workflow used in recent pioneering studies:
Figure 1: Experimental workflow for studying ZEB2 in somitogenesis using P300 proximity labeling in gastruloids.
This integrated approach revealed the critical role of ZEB2 in mouse and human somitogenesis, demonstrating how enhancer interaction landscapes are rewired during germ layer specification [11] [36]. The study combined P300 proximity labeling with single-cell proteomics and degron-based perturbation experiments to establish a causal relationship between ZEB2-dependent enhancer interactions and somitogenesis defects observed upon ZEB2 depletion.
Key Research Reagents and Solutions
Successful implementation of P300 proximity labeling requires specific reagents and tools. The following table details essential components used in recent ZEB2 somitogenesis research:
Table 2: Essential Research Reagents for P300 Proximity Labeling Studies
| Reagent Category | Specific Examples | Function/Application |
|---|---|---|
| Cell Models | Mouse embryonic stem cells (mESCs), Gastruloids, Dbp(luc) array NIH3T3 cells [43] [36] | Provide biologically relevant systems for studying enhancer dynamics in development |
| Enzyme Systems | dCAS9-APEX2 fusion protein, P300-based labeling constructs [43] | Catalyzes proximity-dependent biotinylation of proteins near targeted genomic loci |
| Molecular Tools | Target-specific sgRNAs (e.g., for Dbp E-box or ZEB2 locus), Biotin-phenol, H2O2 [43] | Enables targeted localization and activation of the labeling system |
| Affinity Reagents | Streptavidin-coated beads, Antibodies for validation (P300, H3K27ac) [43] | Isolation and validation of biotinylated proteins and associated complexes |
| Analytical Platforms | Liquid chromatography-mass spectrometry (LC-MS/MS), Single-cell RNA sequencing [11] [43] | Identification and quantification of captured proteins; transcriptional profiling |
Detailed Experimental Protocol for P300 Proximity Labeling
Cell Preparation and Targeting
The initial phase involves establishing the appropriate cellular system and targeting machinery. For studies of somitogenesis, researchers typically employ mouse gastruloids as a model system that recapitulates key aspects of in vivo development [11] [36]. Cells are engineered to express inducible dCAS9-APEX2 fusion protein, with expression controlled by a doxycycline-responsive promoter. Simultaneously, guide RNAs (sgRNAs) are designed to target specific genomic regions of interest—for ZEB2 studies, these would include regions within the ZEB2 locus and potential regulatory elements. The sgRNAs should be positioned within 500 base pairs of the target site to ensure efficient biotinylation, based on the estimated range of APEX2 activity [43].
Critical optimization steps include titrating doxycycline concentration to achieve optimal dCAS9-APEX2 expression (typically 0.25 µg/mL in NIH3T3 systems) and validating sgRNA efficiency through complementary assays such as DNA FISH or ChIP-qPCR [43]. For gastruloid studies, proper differentiation protocols must be established with clear staging markers to ensure reproducibility across experiments. The entire process from gastruloid formation to analysis typically spans 5-7 days, with P300 proximity labeling performed at specific developmental timepoints corresponding to key transitions in mesoderm patterning and somite formation [36].
Biotinylation, Enrichment, and Analysis
The core labeling procedure involves precise timing and condition optimization. The following diagram illustrates the molecular mechanism of proximity labeling at enhancer regions:
Figure 2: Molecular mechanism of P300 proximity labeling at enhancer regions using dCAS9-APEX2.
Cells are incubated with biotin-phenol for 30 minutes to allow penetration into cellular compartments, followed by precisely timed H2O2 stimulation (1 minute) to activate the APEX2-mediated biotinylation reaction [43]. The reaction is promptly quenched with specific quenching solutions containing Trolox and sodium ascorbate to minimize background labeling. Cells are then harvested and lysed under denaturing conditions to preserve protein complexes and prevent post-lysis interactions.
Biotinylated proteins are captured using streptavidin-coated beads with rigorous washing steps to remove non-specifically bound proteins. The enriched proteins are then subjected to on-bead tryptic digestion and prepared for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis [43]. For integrated analyses, parallel samples should be processed for transcriptomic profiling (RNA-seq) to correlate enhancer interactions with gene expression changes, particularly for ZEB2 and its downstream targets during somitogenesis [11] [6].
Data Interpretation and Integration with Complementary Approaches
Analytical Frameworks for Enhancer Landscape Data
The raw data from P300 proximity labeling experiments require specialized bioinformatic processing to yield biologically meaningful insights. Mass spectrometry data are processed using standard proteomic pipelines (MaxQuant, Proteome Discoverer) against appropriate protein databases, with specific attention to the elimination of common contaminants and statistical assessment of enrichment (e.g., using Significance Analysis of INTeractome [SAINT] methodology) [43]. Proteins consistently identified in negative control samples (lacking H2O2 stimulation or expressing catalytically dead APEX2) should be stringently filtered out.
Integration with complementary genomic datasets greatly enhances the biological interpretation of P300 proximity labeling results. For the ZEB2 somitogenesis studies, researchers integrated proteomic data with single-cell RNA sequencing from Zeb2-degron tagged gastruloids, revealing how ZEB2-dependent enhancer interactions translate to transcriptional outputs during mesoderm differentiation [11] [6]. This multi-layered approach demonstrated that ZEB2 coordinates the expression of key somitogenesis regulators through specific enhancer hubs, and that disruption of these interactions leads to defects in somite formation in both mouse and human model systems.
Functional Validation Strategies
Candidate proteins identified through P300 proximity labeling require rigorous functional validation. In the ZEB2 study, this involved degron-based perturbations combined with single-cell RNA sequencing to assess the functional consequences of ZEB2 depletion on the transcriptional programs driving somitogenesis [11] [36]. Additional validation methods include CRISPR-based editing of identified enhancer elements, followed by assessment of gene expression changes and phenotypic characterization in gastruloids.
For the novel interactors identified at enhancer regions, validation typically includes reciprocal immunoprecipitation to confirm physical interactions, followed by functional assays assessing the impact of depletion or overexpression on enhancer activity (using reporter assays) and target gene expression [43]. In the circadian regulation study, novel E-box interactors XPO7 and MINK1 were validated using siRNA-mediated depletion in human U-2 OS cells, which resulted in disrupted circadian rhythms, confirming their functional relevance to enhancer-mediated regulation [43].
P300 proximity labeling represents a transformative methodology for mapping enhancer interaction landscapes with unprecedented resolution and context specificity. When compared to traditional enhancer mapping techniques, this approach provides unique advantages in capturing transient interactions, identifying novel regulatory factors, and working in living cellular systems. The application of this technology to ZEB2 function in somitogenesis has illuminated how specific enhancer networks are rewired during germ layer specification and has established causal relationships between enhancer interactions and developmental outcomes.
As the field advances, the integration of P300 proximity labeling with single-cell multi-omics and computational modeling will further enhance our understanding of the dynamic protein complexes that govern enhancer function across developmental timelines. The reagents and protocols detailed in this guide provide a roadmap for researchers seeking to implement this powerful technology in their own investigations of gene regulation in development and disease.
Addressing Technical Challenges and Optimizing ZEB2 Research
Overcoming Early Embryonic Lethality in ZEB2 Knockout Studies
Zinc finger E-box binding homeobox 2 (ZEB2) is a crucial transcription factor with multifaceted roles in embryonic development, stem cell differentiation, and disease pathogenesis. General homozygous knockout of Zeb2 in mice results in embryonic lethality around E8.5, preventing the study of its functions in later developmental stages and adulthood. This review comprehensively compares the experimental strategies that overcome this lethality, detailing their methodologies, applications, and limitations within the context of ZEB2's role in somitogenesis and broader developmental processes. We provide structured comparisons of quantitative data, detailed protocols, and visual frameworks to guide researchers in selecting appropriate models for investigating ZEB2 functions.
ZEB2 (also known as SIP1 or ZFHX1B) is a DNA-binding and SMAD-binding transcription factor that determines cell fate across multiple lineages. Its critical importance is underscored by the fact that general homozygous knockout (KO) mice die around embryonic day 8.5 (E8.5), exhibiting severe developmental defects including impaired somitogenesis, neural plate formation, and neural crest cell development [1] [5]. This early lethality presents a significant barrier to studying ZEB2 functions in later organogenesis, postnatal development, and adult tissue homeostasis.
The biological rationale for this lethality stems from ZEB2's fundamental role in early developmental processes. ZEB2 governs the exit from pluripotency in embryonic stem cells (ESCs) and is essential for initiating differentiation programs [5]. In the absence of ZEB2, cells fail to properly transition from the naive pluripotent state, impairing subsequent lineage commitment. Furthermore, ZEB2 regulates key signaling pathways including TGF-β/BMP, Wnt, and Notch, which are crucial for patterning and morphogenesis during early embryogenesis [1].
ZEB2 in Development and Disease Contexts
Biological Functions of ZEB2
ZEB2 orchestrates numerous developmental processes through its function as a transcriptional regulator. During neurodevelopment, ZEB2 determines the timing of neuro-/gliogenesis and controls the layering of the cortex in a cell non-autonomous fashion [1]. In neural crest cells, ZEB2 is indispensable for proper migration and differentiation, particularly in the enteric nervous system, explaining why its deficiency causes Hirschsprung disease in Mowat-Wilson syndrome (MOWS) patients [1] [47]. ZEB2 also regulates epithelial-to-mesenchymal transition (EMT) through E-cadherin repression and influences cell fate decisions in various lineages including hematopoietic, immune, and muscle cells [1] [3].
Clinical Significance: Mowat-Wilson Syndrome
Heterozygous mutations in ZEB2 cause Mowat-Wilson syndrome (MOWS), a rare autosomal dominant disorder characterized by severe intellectual disability, distinctive facial features, Hirschsprung disease, epilepsy, and multiple congenital anomalies [1]. Most pathogenic variants are nonsense or frameshift mutations leading to premature stop codons and haploinsufficiency through nonsense-mediated decay. The spectrum of clinical features underscores ZEB2's pleiotropic functions during embryonic development and provides clinical rationale for investigating its mechanisms of action using appropriate model systems [1].
Methodological Approaches to Overcome Embryonic Lethality
Conditional Knockout Models
Conditional knockout (cKO) strategies utilizing Cre-loxP technology represent the primary approach for circumventing early embryonic lethality in ZEB2 research:
Table 1: Conditional ZEB2 Knockout Mouse Models
| Model System | Genetic Design | Key Applications | Major Findings |
|---|---|---|---|
| Zeb2+/fl(Δex7) mice [1] | Exon 7 flanked by loxP sites | Neural development, Neural crest studies | Cell-autonomous & non-autonomous functions in CNS; ENS defects linking to MOWS |
| Cell-type specific cKO [1] | Cre recombinase driven by cell-specific promoters | Hematopoiesis, Immune function, Postnatal brain development | Role in adult hematopoietic differentiation; Myeloproliferative disorder phenotypes |
| Embryonic stem cell models [5] | Zeb2-null ESCs | Pluripotency exit, Neural differentiation | ZEB2 essential for exit from epiblast state; Links pluripotency network with DNA methylation |
The fundamental principle involves generating mice with loxP sites flanking critical exons of Zeb2 (typically exon 7), which are then crossed with various Cre-driver lines to achieve spatial and temporal control of gene deletion [1]. This enables investigation of ZEB2 function in specific cell types and developmental stages without triggering systemic lethality.
Experimental Protocol: Generation and Validation of Conditional Knockout Mice
- Targeting Vector Construction: Design a targeting vector with loxP sites flanking exon 7 of Zeb2, incorporating positive (e.g., neomycin resistance) and negative (e.g., thymidine kinase) selection markers.
- ESC Electroporation and Selection: Introduce the targeting vector into mouse embryonic stem cells (mESCs) via electroporation, followed by selection with appropriate antibiotics.
- Blastocyst Injection and Germline Transmission: Inject validated targeted mESCs into blastocysts, implant into pseudopregnant females, and breed chimeric offspring for germline transmission.
- Cre Crosses: Cross Zeb2-floxed mice with tissue-specific Cre-driver lines (e.g., Nestin-Cre for neural lineages, Wnt1-Cre for neural crest).
- Phenotypic Validation: Confirm efficient deletion via PCR genotyping, Western blotting, and immunohistochemistry. Assess tissue-specific phenotypes through histological analysis and functional assays relevant to the target tissue [1].
Embryonic Stem Cell Models
Zeb2 knockout ESCs provide a powerful in vitro system for investigating ZEB2 functions without embryonic lethality constraints:
Table 2: ZEB2 Manipulation in Stem Cell Models
| ESC Model | Genetic Manipulation | Differentiation Potential | Key Regulatory Roles Identified |
|---|---|---|---|
| Zeb2 KO ESCs [5] | Complete Zeb2 knockout | Stalled in epiblast-like state; impaired neural & mesendodermal differentiation | Controls pluripotency exit; Connects DNA methylation with differentiation commitment |
| R26_Zeb2 ESCs [3] | cDNA expression from Rosa26 locus | Enhanced myogenic differentiation | Positive regulator of skeletal muscle differentiation; Upregulates myogenic markers |
| Zeb2-V5 ESCs [10] | Endogenous tagging with Flag-V5 epitope | Normal differentiation to NPCs | Enables ChIP-seq mapping of DNA-binding sites in neuroprogenitor cells |
Zeb2 KO ESCs exhibit a distinctive phenotype: they can exit the naive state but stall in an early epiblast-like state and are impaired in both neural and mesendodermal differentiation [5]. These cells maintain the ability to re-adapt to pluripotency conditions even after prolonged differentiation exposure, highlighting ZEB2's role in irreversible commitment.
Experimental Protocol: Zeb2 KO ESC Derivation and Differentiation
- ESC Culture Maintenance: Maintain ESCs feeder-free in 2i + LIF medium (1 μM PD0325901, 3 μM CHIR99021, and 1000 U LIF/mL in N2B27 base medium) to preserve ground state pluripotency [5].
- Genetic Manipulation: For KO generation, nucleofect Zeb2-floxed ESCs with Cre-recombinase vector (e.g., pcDNA6-His-eGFP:Cre) using Amaxa A-23 program with blasticidin selection (48 hours). For rescue models, introduce Flag-tagged Zeb2 cDNA into the Rosa26 locus via RMCE technology [5] [3].
- Neural Differentiation: Plate 3×10^6 ESCs in bacterial petri dishes in EB medium (KO DMEM, 15% FBS, NEAA, sodium pyruvate, β-mercaptoethanol). On day 2, refresh EB medium; on day 4, change to N2B27 + 500 nM retinoic acid; refresh on day 6. From days 8-15, culture in N2B27 refreshed every other day [5].
- Analysis: Assess differentiation efficiency via immunostaining for lineage markers, RNA-seq transcriptomics, and RRBS for DNA methylation analysis.
Alternative Model Organisms
Zebrafish models with zeb2 knockdown provide additional insights while avoiding mammalian lethality:
Experimental Protocol: Zebrafish Morpholino Knockdown
- Morpholino Design: Design antisense morpholino oligonucleotides targeting the translation start site or splice junctions of zeb2.
- Microinjection: Inject 1-4 cell stage zebrafish embryos with 1-2 nL of morpholino solution (0.1-0.5 mM).
- Phenotypic Analysis: Assess axial patterning, neural crest defects, and enteric nervous system development at 24-72 hours post-fertilization. Zebrafish zeb2 knockdown recapitulates key MOWS features including neural crest defects and enteric nervous system deficiencies, providing a complementary in vivo model [1].
Analytical Frameworks for ZEB2 Functional Studies
Signaling Pathways Regulated by ZEB2
ZEB2 interacts with multiple signaling pathways essential for somitogenesis and overall embryonic development. The following diagram illustrates these key regulatory relationships:
Figure 1: ZEB2 regulates multiple signaling pathways. ZEB2 integrates TGF-β/BMP signaling via SMAD interactions, represses pluripotency factors, promotes differentiation programs, controls EMT, and modulates DNA methylation.
ZEB2 functions as a nuclear fine-tuner of transcriptional responses to TGFβ/Nodal-Activin and BMP signaling [3]. It exhibits both anti-BMP and anti-Wnt activities depending on cellular context, and regulates key developmental transitions through epigenetic mechanisms including DNA methylation [5] [3].
Experimental Workflow for Conditional KO Analysis
The following diagram outlines a comprehensive strategy for studying ZEB2 functions using conditional knockout models:
Figure 2: Workflow for overcoming ZEB2 embryonic lethality. The conditional knockout strategy enables tissue-specific and temporal control of ZEB2 deletion through intersection of floxed alleles with Cre drivers, facilitating both in vivo phenotypic analysis and in vitro mechanistic studies.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for ZEB2 Studies
| Reagent/Cell Line | Type | Key Applications | Function/Features |
|---|---|---|---|
| Zeb2+/fl(Δex7) mice [1] | Mouse model | Conditional knockout studies | Exon 7 flanked by loxP sites; Base for tissue-specific deletions |
| Zeb2 KO ESCs [5] | Embryonic stem cell | Pluripotency exit studies | Stalls in epiblast-like state; Differentiation impairment |
| R26_Zeb2 ESCs [3] | Rescue model | Myogenic differentiation studies | Zeb2 cDNA from Rosa26 locus; Enhanced muscle differentiation |
| Zeb2-V5 ESCs [10] | Endogenously tagged | ChIP-seq mapping | Flag-V5 tag before stop codon; Identifies DNA-binding sites |
| C2C12-Zeb2 [3] | Myoblast model | Muscle differentiation | Zeb2 transfection; Enhanced myogenic marker expression |
| Anti-ZEB2 antibodies | Immunodetection | WB, IHC, IF | Protein detection; Limited availability of ChIP-grade versions |
| Cre-recombinase lines | Genetic tool | Tissue-specific deletion | Drivers: Nestin-Cre (neural), Wnt1-Cre (neural crest) |
| NS1-IN-1 | REDD1 Inducer|For Cell Stress Research (RUO) | Explore cellular stress responses with our REDD1 Inducer. This reagent is for Research Use Only (RUO) and is not intended for diagnostic or personal use. | Bench Chemicals |
Discussion and Future Perspectives
The methodological advances summarized herein have enabled remarkable progress in understanding ZEB2 functions beyond early embryogenesis. Conditional knockout models have revealed ZEB2's critical roles in adult hematopoiesis, immune function, and tissue repair [1] [48]. Stem cell models have illuminated its fundamental mechanisms in pluripotency exit and lineage commitment [5] [3]. The recent mapping of ZEB2 DNA-binding sites in neuroprogenitor cells provides a foundation for identifying direct target genes and understanding its transcriptional networks [10].
Future directions include developing inducible knockout systems for precise temporal control, generating human iPSC models of Mowat-Wilson syndrome, and creating double conditional alleles for studying ZEB2 interactions with other developmental regulators. The continuing refinement of these experimental approaches will further elucidate ZEB2's multifaceted functions in development, homeostasis, and disease, potentially identifying therapeutic targets for conditions associated with ZEB2 dysfunction.
Overcoming the early embryonic lethality of ZEB2 knockout requires sophisticated genetic strategies including conditional mouse models, stem cell systems, and alternative organism approaches. Each method offers distinct advantages and limitations, enabling investigators to address specific biological questions about ZEB2 function in different contexts. The experimental frameworks and reagent tools summarized in this review provide a roadmap for researchers investigating this crucial transcription factor in development, disease, and regenerative processes.
Conditional Knockout Strategies for Tissue-Specific Analysis
Conditional knockout (cKO) strategies represent a cornerstone of modern genetic research, enabling the precise dissection of gene function in a spatiotemporal manner. Within the specific context of ZEB2 research, these technologies are indispensable for unraveling the gene's multifaceted roles in mammalian development, particularly in complex processes like somitogenesis—the segmentation of the embryonic body into somites that give rise to vertebrae, skeletal muscle, and dermis. Mutations in the ZEB2 gene cause Mowat-Wilson syndrome (MOWS), a neurodevelopmental disorder often accompanied by axial skeletal defects, underscoring the critical nature of properly regulated ZEB2 function during embryogenesis [1]. Conventional, full-gene knockout of Zeb2 in mice results in early embryonic lethality around E8.5, precluding the study of its function in later developmental stages and in specific tissues [5] [1]. The advent of cKO technologies has therefore been pivotal, allowing researchers to bypass this lethality and investigate ZEB2's cell-autonomous and non-autonomous roles in somitogenesis and the development of the nervous system, neural crest, and other MOWS-relevant tissues.
Fundamental Principles
At its core, a cKO strategy allows for the deletion of a target gene in a specific cell type or tissue at a chosen time. The most widely used system is the Cre-loxP system. This involves generating a mouse line in which an essential exon(s) of the gene of interest is "floxed"—flanked by loxP sites, which are 34-base pair recognition sequences for the Cre recombinase enzyme. These mice are then crossed with a second line expressing Cre recombinase under the control of a tissue-specific promoter. In the resulting double-positive offspring, Cre mediates the recombination between the loxP sites, excising the critical genomic region and knocking out the gene exclusively in Cre-expressing cells [49] [50]. The key advantage is the ability to study genes essential for early development or viability in a highly controlled manner.
Established cKO Workflow
The following diagram illustrates the foundational process for generating and using conditional knockout mice, a methodology that has been central to ZEB2 research.
Comparison of Modern cKO Methodologies
The field of genetic engineering has evolved significantly, moving beyond traditional embryonic stem (ES) cell-based methods. The table below provides a comparative overview of established and contemporary cKO strategies.
Table 1: Comparison of Conditional Knockout Methodologies
| Method | Core Principle | Key Advantages | Key Limitations | Typical Efficiency/Time | Suitability for ZEB2 Studies |
|---|---|---|---|---|---|
| Traditional Cre-loxP [49] | Homologous recombination in ES cells to insert loxP sites. | High precision; well-established; numerous tissue-specific Cre lines available. | Time-consuming (multiple breeding steps, ~1 year); technically demanding; low efficiency in non-ES cells. | ~10% targeting efficiency in ES cells; requires extensive screening. | Foundation of early ZEB2 research (e.g., neural crest, forebrain cKOs) [1]. |
| Rapid Cre-loxP (Sequential Electroporation) [50] | CRISPR/Cas9-assisted sequential insertion of loxP sites into zygotes. | Faster (~4 months); uses desired genetic background (C57BL/6); no ES cells needed. | Requires two rounds of embryo manipulation; optimization needed for different targets. | High embryo survival (>97%); successful floxing in 1/10 pups in a study. | Useful for rapid model generation to study ZEB2 in somitogenesis. |
| Inducible Cas9 cKO [49] [51] | Doxycycline-induced Cas9 expression with gRNAs to disrupt the gene. | Fast, one-step cloning; simultaneous multi-gene targeting; no need for floxed alleles. | Potential for in-frame mutations (incomplete KO); requires stable integration of Cas9/gRNA. | ~70% biallelic mutation efficiency for one gene; ~40-50% for two genes [51]. | Ideal for acute KO in adult stages or for combinatorial gene studies post-somitogenesis. |
| CRISPR/Cas9 with LSL-Cas9 [51] | Cross floxed-stop-Cas9 mice with tissue-specific Cre and ubiquitous gRNA mice. | Spatiotemporal control of Cas9; one gRNA line works with any Cre line; good for multi-gene KO. | Off-target potential; requires breeding of three components. | ~74% mutation rate in hepatocytes shown in a model [51]. | Suitable for tissue-specific, multi-gene analysis of ZEB2 networks. |
| FLIP-Cassette cKO [49] | CRISPR-assisted insertion of an invertible gene-trap intron (FLIP cassette). | Effective in aneuploid cells; one targeting event can disrupt both alleles post-Cre. | Complex vector; may affect gene expression before Cre induction; moderate efficiency. | ~6% of clones were correct FLIP/- for Ctnnb1 cKO [49]. | Potential for in vitro ZEB2 studies in hard-to-transfect primary cells. |
Detailed Experimental Protocols
Protocol 1: Rapid Generation of Floxed Mice via Sequential Electroporation
This protocol, adapted from a 2021 study, outlines a method to generate floxed mice in approximately four months using CRISPR/Cas9 and electroporation, significantly faster than traditional ES cell-based methods [50].
Key Research Reagents:
- ssODNs: Single-stranded oligodeoxynucleotides containing the loxP sequence and homology arms (~200 nt). Function as donor DNA for homology-directed repair.
- gRNA (crRNA + tracrRNA): Guides the Cas9 nuclease to the specific genomic loci upstream and downstream of the exons to be floxed.
- HiFi Cas9 Protein: A high-fidelity version of the Cas9 nuclease that creates double-strand breaks at the target sites with reduced off-target effects.
- C57BL/6 N Mice: Source of zygotes, allowing direct work on the desired genetic background.
Procedure:
- First Electroporation (5' loxP insertion): Perform in vitro fertilization (IVF) using C57BL/6 N mice. Electroporate the resulting 1-cell stage zygotes with a mixture containing gRNA targeting the upstream intron, HiFi Cas9 protein, and the 5' loxP ssODN. Transfer viable 2-cell embryos into pseudopregnant female mice.
- Founder Generation and Validation: Genotype the resulting offspring to identify founders carrying the correct 5' loxP insertion.
- Second Electroporation (3' loxP insertion): Collect sperm from a 5' loxP founder and perform a second round of IVF with wild-type oocytes. Electroporate these fertilized eggs with the gRNA for the downstream intron and the 3' loxP ssODN. Transfer embryos and generate offspring.
- Identification of Floxed Mice: Genotype the pups from the second round to identify those with both loxP sites correctly inserted on the same allele. Confirm loxP orientation and sequence by PCR and sequencing.
- Functional Validation: Test the functionality of the floxed allele in vitro by incubating a PCR product spanning the loxP sites with recombinant Cre protein. Successful recombination, observed as a band shift on a gel, confirms the loxP sites are functional [50].
Protocol 2: Inducible Cas9-Mediated cKO in Macrophages
This protocol demonstrates a method for achieving cell-type-specific, simultaneous knockout of multiple genes without the need for floxed alleles, using a loxP-stop-loxP (LSL)-Cas9 system [51].
Key Research Reagents:
- LSL-Cas9 Transgenic Mice: Mice harboring a transgene where Cas9 expression is blocked by a floxed transcriptional STOP cassette.
- Tissue-Specific Cre Mice: Mice expressing Cre recombinase under a cell-specific promoter (e.g., LysM-Cre for macrophages).
- Ubiquitous sgRNA Transgenic Mice: Mice expressing sgRNAs targeting the gene(s) of interest (e.g., c-Maf and MafB) from a ubiquitous promoter.
Procedure:
- Mouse Crossbreeding: Cross the LSL-Cas9 mouse with the tissue-specific Cre mouse (e.g., LysM-Cre) to generate LSL-Cas9;Cre offspring. Then, cross these mice with the ubiquitous sgRNA transgenic mouse.
- Induction of cKO: In the target tissue of the triple-transgenic offspring (LSL-Cas9;Cre;sgRNA), the Cre recombinase excises the STOP cassette, allowing expression of Cas9. The expressed Cas9, guided by the sgRNAs, introduces double-strand breaks in the target genes, leading to functional knockout.
- Validation:
- Molecular Analysis: Isolate genomic DNA from the target cells and perform T7 endonuclease I (T7EN1) assay or sequencing to detect insertion/deletion (indel) mutations at the target loci.
- Protein Analysis: Perform Western blotting on protein lysates from the target cells to confirm the loss of the target protein(s) [51].
Advanced Applications and Visualization: cKO in ZEB2 Research
The diagrams below illustrate how modern cKO strategies are applied to investigate the role of ZEB2 in specific biological contexts, such as brain development and somitogenesis.
Application in Neuronal and Somitogenesis Studies
Molecular Pathway of ZEB2 in Somitogenesis
Understanding the molecular function of ZEB2 is key to interpreting cKO phenotypes. ZEB2 is a transcription factor that can both activate and repress gene expression. Recent proteomic and functional studies in gastruloids have highlighted its critical role in somitogenesis [12].
Diagram 2 Explanation: This diagram summarizes the molecular role of ZEB2, informed by studies in ESCs and gastruloids [5] [12]. ZEB2 is regulated by key developmental signaling pathways. It executes its function by binding to E-box sequences in DNA, leading to the repression of specific target genes like Tet1 (a demethylase) and Cdh1 (E-cadherin). Repression of Tet1 helps establish a stable DNA methylation landscape, while repression of other targets facilitates the exit from the pluripotent epiblast state. Through a combination of repression and activation of distinct target genes, ZEB2 ensures the proper execution of the somitogenesis program. Its deletion via cKO disrupts this network, leading to stalled development and segmentation defects.
The landscape of conditional knockout technology has expanded dramatically, offering researchers a powerful toolkit for tissue-specific genetic analysis. For the study of a critical developmental regulator like ZEB2, the choice of strategy is paramount. Traditional Cre-loxP models have laid the groundwork, revealing ZEB2's essential functions in the brain and neural crest. However, newer methods such as inducible Cas9 systems and rapid electroporation-based floxing provide enhanced speed, flexibility, and the capacity for multi-gene analysis. These advanced methodologies are particularly suited for probing the dynamic and complex role of ZEB2 in processes like somitogenesis, allowing researchers to move beyond embryonic lethality and model the tissue-specific defects seen in Mowat-Wilson syndrome with ever-greater precision. As these technologies continue to evolve, they will undoubtedly yield deeper insights into the gene regulatory networks controlled by ZEB2 and their contribution to development and disease.
Validating Protein Expression and Phosphoproteome Dynamics
Within the broader study of ZEB2's essential role in mouse and human somitogenesis, the precise validation of protein expression and the mapping of phosphoproteome dynamics are critical. Somitogenesis, the process of body segmentation during embryonic development, relies on intricate signaling networks and precise protein-level regulation. Research has demonstrated that ZEB2 plays a key role in mouse and human somitogenesis, and its function is deeply intertwined with phosphorylation-driven signaling pathways that coordinate this complex morphogenetic event [12]. This guide objectively compares the primary technologies and computational workflows used to dissect these molecular events, providing a foundation for research and drug development aimed at developmental disorders.
Protein Expression Validation in Model Systems
Validating protein expression for a developmental transcription factor like ZEB2 often requires the use of recombinant protein production for functional studies, antibodies, or standards. The choice of expression system is dictated by the protein's biochemical properties and required post-translational modifications.
Table 1: Key Characteristics of Common Gene Expression Systems
| Expression System | Best For Protein Types | Folding & Assembly Capacity | Post-Translational Modifications | Typical Yield | Cost & Speed |
|---|---|---|---|---|---|
| E. coli | Prokaryotic proteins; simple eukaryotic proteins without complex PTMs [52] | Limited for multi-domain/complex proteins [52] | None beyond disulfide bonds in periplasm [52] | High for soluble targets [52] | Low cost; very fast [52] |
| Yeast | Secreted eukaryotic proteins; some membrane proteins [52] | Good for proteins with disulfide bonds [52] | Hyper-mannose glycosylation (non-human) [52] | Moderate to High [52] | Low cost; fast [52] |
| Insect Cells | Complex eukaryotic proteins; multi-subunit complexes; many membrane proteins like GPCRs [52] | High capacity for complex assembly [52] | Paucimannose glycosylation (non-human) [52] | High [52] | Moderate cost and speed [52] |
| Mammalian Cells | Proteins requiring authentic human PTMs (e.g., complex glycosylation); complex membrane proteins [52] | Highest fidelity for human protein complexes [52] | Complex, human-like glycosylation and other PTMs [52] | Low to Moderate [52] | High cost; slower [52] |
For challenging proteins critical in development, a new generation of machine learning models, such as MPB-EXP, can predict heterologous expression levels from protein sequences across 88 different species with high accuracy, helping to prioritize constructs and systems for experimental validation [53].
Comparative Analysis of Phosphoproteomic Technologies
Phosphoproteomics enables the system-wide study of signaling dynamics, which is essential for understanding the role of ZEB2 in the phosphorylation-regulated processes of somitogenesis. The two primary mass spectrometry acquisition methods, Data-Dependent Acquisition (DDA) and Data-Independent Acquisition (DIA), offer different trade-offs.
Table 2: DDA vs. DIA Phosphoproteomics at a Glance
| Feature | Data-Dependent Acquisition (DDA) | Data-Independent Acquisition (DIA) |
|---|---|---|
| Acquisition Principle | Selects top-N most abundant precursors for fragmentation in each cycle [54] | Fragments all precursors within pre-defined, sequential m/z windows [54] |
| Dynamic Range | Limited by precursor intensity [54] | Order of magnitude broader [54] |
| Reproducibility | Lower identification reproducibility between replicates [54] | High reproducibility of identification and quantification [54] |
| Quantitative Accuracy | Good (R² ~0.89 in benchmark studies) [54] | Excellent (R² ~0.93 in benchmark studies) [54] |
| Throughput & Depth | Deeper analysis requires longer instrument times; ~7,000 phosphopeptides in 15 min [54] | Higher throughput without sacrificing depth; >10,000 phosphopeptides in 15 min [54] |
| Data Analysis | Relies on project-specific or public spectral libraries [54] | Can use spectral libraries or direct, library-free analysis (dDIA) [54] |
A benchmark study comparing quantitative accuracy using yeast phosphopeptides spiked into a HeLa background at known ratios (0.25:1 to 2:1) found that both DIA and dDIA quantified twice as many phosphopeptides as DDA, while all methods accurately estimated the expected ratios on median [54].
Optimized Experimental Workflows
Workflow for Deep Phosphoproteome Profiling
An optimized, rapid workflow for phosphoproteomics using DIA has been established for high-throughput signaling studies [54]:
- Sample Preparation: Digest 200 μg of protein lysate to peptides.
- Phosphopeptide Enrichment: Use high-throughput magnetic Ti-IMAC beads to selectively bind phosphopeptides.
- LC-MS/MS Analysis: Perform single-shot 15-minute liquid chromatography runs coupled to a Q Exactive HF-X mass spectrometer.
- DIA Acquisition: Employ an optimized method with a 2-second cycle time, 48 mass windows of 14 Da width, and 1 Th overlap between windows, using a normalized collision energy of 25 [54].
- Data Processing: Analyze data with Spectronaut software incorporating a PTM-specific site localization score, either using a project-specific library or via direct DIA (dDIA) which generates pseudo-MS/MS spectra from the DIA data [54].
Optimal Differential Expression Analysis Workflow
For proteomics data, an extensive benchmarking study of 34,576 combinatoric workflows on spike-in datasets revealed that optimal performance is predictable and depends on the quantification setting (e.g., DDA, DIA, TMT) [55]. Key high-performing rules were identified:
- For Label-Free Data (DDA/DIA): Workflows are enriched for directLFQ intensity, no normalization (referring to distribution correction methods not embedded in quantification), and imputation methods like SeqKNN, Impseq, or MinProb [55].
- Influential Steps: The choice of normalization and statistical methods for differential expression analysis are particularly influential for label-free DDA and TMT data, while for DIA, the matrix type is also critical [55].
- Ensemble Inference: Integrating results from multiple top-performing individual workflows (e.g., combining results from top0, directLFQ, and MaxLFQ intensities) can expand differential proteome coverage, improving the partial area under the curve (pAUC) by up to 4.61% and the G-mean score by up to 11.14% [55].
The ZEB2 Signaling Nexus in Development
ZEB2 is a critical transcriptional regulator in early development. Studies in mouse embryonic stem cells (ESCs) show that ZEB2 is essential for exiting the naïve pluripotent state and for progression into differentiation lineages, including neural and mesendodermal fates [5]. Without ZEB2, cells stall in an early epiblast-like state and cannot commit to differentiation, linking ZEB2 directly to the early stages of cell fate decisions that precede somitogenesis [5]. This function involves Zeb2's role as a transcriptional repressor that helps silence the pluripotency network (including Oct4 and Nanog) and regulates key processes like epithelial-to-mesenchymal transition (EMT) and DNA methylation dynamics through factors like TET1 [5].
A multi-omics study on mouse gastruloids—a model for early development—revealed that germ layer specification is associated with a global rewiring of the (phospho)proteome, further underscoring the link between proteome dynamics, phosphorylation, and lineage decisions in which ZEB2 is a key player [12].
Successfully navigating protein expression and phosphoproteomics requires a curated set of reagents, databases, and computational tools.
Table 3: Essential Research Reagents and Resources
| Category / Item | Specific Examples | Function and Application |
|---|---|---|
| Expression Systems | E. coli (BL21), HEK293, Sf9 insect cells [52] | Heterologous protein production for antibodies, functional assays, and standardization. |
| Phospho-Enrichment | Ti-IMAC magnetic beads [54] | Selective enrichment of phosphopeptides from complex peptide digests for MS analysis. |
| Mass Spectrometry | Q Exactive HF-X [54] | High-resolution, high-sensitivity LC-MS/MS for DDA and DIA acquisition of phosphoproteomes. |
| Critical Software | Spectronaut, MaxQuant, FragPipe, DIA-NN [55] [54] | Processing raw MS data for identification, quantification, and site localization of phosphopeptides. |
| Proteomics Databases | UniProt, PRIDE, Peptide Atlas [56] | Reference protein sequences (UniProt) and public repositories for raw MS data (PRIDE). |
| Interaction Databases | STRING-db, IntAct [56] | Contextualizing phosphosignaling within known and predicted protein-protein interaction networks. |
The journey from validating the expression of a key developmental regulator like ZEB2 to mapping its effects on the dynamic phosphoproteome relies on a carefully selected suite of technologies. While E. coli offers a rapid route for protein production, mammalian systems are indispensable for authentic PTM studies. For phosphoproteomics, DIA is increasingly the method of choice for high-throughput, reproducible, and accurate system-wide analyses. The integration of optimized wet-lab protocols with robust, benchmarked computational workflows—and potentially ensemble inference methods—provides the most powerful approach for uncovering the nuanced signaling dynamics that govern critical processes like somitogenesis. This objective comparison of tools and data underscores the technological foundation upon which a deeper understanding of ZEB2 in development and disease can be built.
Optimizing Gastruloid Differentiation Protocols for Somite Modeling
The transcription factor ZEB2 is a critical regulator of embryonic development, with mutations in humans causing Mowat-Wilson syndrome, which features congenital anomalies including skeletal defects [1]. Research using Zeb2 knockout (KO) mouse embryonic stem cells (mESCs) has demonstrated its essential role in exit from the epiblast state and successful differentiation into all germ layers [5]. Without ZEB2, cells stall in an early epiblast-like state and are impaired in both neural and mesendodermal differentiation, directly linking its function to the formation of derivatives like somites [5]. Gastruloids—three-dimensional in vitro models derived from pluripotent stem cells—have emerged as powerful tools to study these early developmental events, including somitogenesis. This guide compares current gastruloid differentiation protocols, evaluating their effectiveness for modeling somite formation and their integration with ZEB2 research, providing experimental data to inform protocol selection.
Comparative Analysis of Gastruloid Differentiation Systems
Key Performance Metrics for Somite Modeling
Table 1: Comparison of Gastruloid Protocol Performance for Somite and Posterior Structure Formation
| Protocol Type | Efficiency of Somite Formation | Key Induced Cell Types | Morphological Structures | Developmental Stage Equivalent | Technical Variability |
|---|---|---|---|---|---|
| Conventional Mouse Gastruloids (with Matrigel) [57] [58] | Variable, often low and inconsistent | Presomitic mesoderm, some somite derivatives | Elongated structures, often lacks organized somites | Mouse E8.5 | High inter-gastruloid heterogeneity |
| Mouse TLS Protocol (Trunk-Like Structures) [57] | High, robust | Differentiated somites (FST+, PAX3+), neural tube, gut tube | Neural tube flanked by segmented somites | Mouse E8.5-E9.0 | More consistent, lower variability |
| Conventional Human Gastruloids [57] | Low or absent | Presomitic mesoderm (TBX6+), NMPs; lacks substantial neural tube | Elongated body axis without segmented somites | Carnegie Stage 7-8 human | Moderate heterogeneity |
| Human RA-Gastruloids (with Retinoic Acid pulse) [57] | Very high (89% of elongated gastruloids) | Segmented somites, neural tube, neural crest, renal progenitors, myocytes | Neural tube flanked by multiple, segmented somites | ~E9.5 mouse / CS11 cynomolgus monkey | Low inter-individual variation |
Quantitative Molecular Patterning Data
Table 2: Molecular and Gene Expression Profiles Across Gastruloid Models
| Protocol | NMP/Patterning Marker Expression | Mesoderm/Somite Marker Expression | Neural Marker Expression | Key Signaling Pathway Activities |
|---|---|---|---|---|
| Conventional Mouse [58] | TBXT+, CDX2+ | TBX6+ (PSM), MESP2+ (differentiation front) | IRX3+, SOX1+, PAX6+ (neural tube) | WNT, FGF signaling present |
| Mouse TLS [57] | Robust TBXT+, SOX2+, CDX2+ | FST+, PAX3+ (differentiated somites) | Strong IRX3+, SOX1+ neural tube patterning | WNT, BMP, FGF pathways active |
| Conventional Human [57] | TBXT+, SOX2+, NKX1-2+, CDX2+ (NMPs) | TBX6+ (PSM), MESP2+, RIPPLY2+ (differentiation front) | Lacks significant neural tube markers (IRX3+, SOX1+, PAX6+) | High WNT, low endogenous RA signaling |
| Human RA-Gastruloids [57] | Balanced NMP population (TBXT+, SOX2+) | Robust TBX6+, MESP2+, RIPPLY2+, PAX3+ somite segmentation | Strong posterior neural tube (SOX2+, SOX1+), neural crest (PAX3+) | Corrected WNT/RA balance, BMP for somite patterning |
Detailed Experimental Protocols and Methodologies
Retinoic Acid-Induced Human Gastruloids with Somites
The most advanced protocol for generating human gastruloids with segmented somites utilizes a temporally discontinuous retinoic acid (RA) pulse regimen [57].
Cell Culture and Patterning Workflow:
Critical Steps and Optimization:
- Initial cell seeding density must be optimized (3,000-6,000 cells per aggregate) for consistent somite formation [57].
- Early RA pulse (0-24 hours) is essential for maintaining bipotentiality of neuromesodermal progenitors (NMPs); neither retinol nor retinal can substitute for RA [57].
- Matrigel supplementation beginning at 48 hours promotes epithelialization and structural organization of somites and neural tube [57].
- CHIR99021 concentration during pre-treatment requires optimization, as WNT signaling levels critically influence mesodermal versus neural differentiation from NMPs [57].
Mouse Trunk-Like Structure (TLS) Gastruloids
The mouse TLS protocol generates the most advanced somite structures in mouse gastruloids, with clear segmentation and neural tube formation [57] [58].
Key Protocol Modifications:
- Pre-culture conditions significantly impact differentiation consistency. Transitioning mESCs from 2i to ESLIF medium before aggregation reduces heterogeneity and enhances mesodermal contributions [58].
- Matrigel embedding before elongation stage promotes somite epithelialization and neural tube formation [57] [58].
- WNT activation timing using CHIR99021 must be precisely controlled (typically 48-72 hours post-aggregation) to establish proper anteroposterior patterning [58].
Stem Cell Pluripotency State Modulation
The starting pluripotency state of stem cells dramatically influences gastruloid patterning consistency and somite differentiation potential [58].
Epigenetic Regulation:
- 2i-grown mESCs display genome-wide DNA methylation of ~30% and general H3K27me3 distribution [58].
- ESLIF-grown mESCs show 80% genome-wide DNA methylation with focused H3K27me3 at promoters [58].
- Transition protocols (2i to ESLIF pre-culture) optimize the epigenetic state for gastruloid formation, resulting in more complex mesodermal contributions and reduced heterogeneity [58].
Signaling Pathways Governing Somitogenesis in Gastruloids
The formation of somites in gastruloids recapitulates in vivo signaling principles, with specific pathway requirements that must be balanced.
Core Somitogenesis Signaling Network:
Pathway Interactions:
- WNT signaling establishes the primitive streak and induces NMP population [57] [59].
- Retinoic acid balances NMP differentiation toward neural versus mesodermal fates; human gastruloids naturally exhibit low ALDH1A2 (RA synthesis) and high CYP26 (RA degradation) expression, requiring supplementation [57].
- BMP signaling initiates at gastruloid edges and is antagonized by NOG at the center, creating patterning gradients essential for germ layer organization [60].
- FGF signaling maintains the presomitic mesoderm in an immature state, working with the segmentation clock to generate periodic somite formation [57].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Gastruloid Somite Differentiation and Their Applications
| Reagent/Category | Specific Examples | Function in Protocol | Application Notes |
|---|---|---|---|
| Small Molecule Inducers | CHIR99021 (GSK3β inhibitor) | Activates WNT signaling, induces primitive streak and NMPs | Concentration and timing critical; typically 3-6µM [57] [58] |
| Retinoic Acid (RA) | Balances NMP differentiation toward neural vs. mesodermal fates | Early pulse (0-24h) essential in human systems; 100nM-1µM [57] | |
| Extracellular Matrix | Matrigel | Promotes epithelialization, structural organization of somites and neural tube | Added at 48h (10%) for human RA-gastruloids; embedding for mouse TLS [57] |
| Cell Culture Media | N2B27 medium | Defined medium for neural and mesodermal differentiation | Base medium for many gastruloid protocols [5] [58] |
| 2i medium (MEK + GSK3β inhibitors) | Maintains ground-state pluripotency for consistent starting population | Transition to ESLIF before aggregation improves outcomes [58] | |
| Lineage Reporters | SOX2-mCitrine, TBXT-GFP | Live monitoring of neural and mesodermal differentiation | Enables real-time tracking of patterning efficiency [57] |
| Aneuploidy Inducers | Reversine (MPS1 kinase inhibitor) | Models chromosomal instability effects on lineage specification | Induces heterogeneous aneuploidy for disease modeling [60] |
Advanced Technologies for Gastruloid Analysis and Screening
High-Throughput Screening Platforms
Microraft Array Technology enables large-scale screening and sorting of individual gastruloids based on phenotypic features [60]:
- Array design: 529 indexed magnetic microrafts (789µm side length) with photopatterned central ECM islands (500µm diameter) for standardized gastruloid formation [60].
- Sorting efficiency: 98±4% release and 99±2% collection efficiency for individual gastruloids [60].
- Application: Enables correlation of morphological phenotypes (e.g., DNA content/area) with gene expression patterns (e.g., NOG, KRT7) in aneuploid versus euploid gastruloids [60].
Advanced Imaging and Computational Analysis
Two-Photon Imaging Pipeline permits deep-tissue visualization of multilayered gastruloids [61]:
- Sample preparation: Glycerol clearing (80%) provides 3-fold/8-fold reduction in intensity decay at 100µm/200µm depth compared to PBS [61].
- Computational analysis: Open-source Python package (Tapenade) with napari plugins for 3D nuclei segmentation, gene expression quantification, and tissue-scale organization analysis [61].
- Application: Reveals 3D spatial patterns of gene expression and nuclear morphology relationships to tissue-scale organization in developing somites [61].
Integration with ZEB2 Research Applications
Gastruloid systems provide ideal platforms for investigating ZEB2 functions in somitogenesis, with specific methodological considerations:
ZEB2 Loss-of-Function Studies:
- Zeb2 KO mESCs stall in an epiblast-like state and fail to properly silence pluripotency networks, impairing differentiation into mesodermal lineages including somites [5].
- Rescue approaches: Tet1 knockdown in Zeb2 KO cells partially restores differentiation capacity, linking ZEB2 to DNA methylation regulation during lineage commitment [5].
- Disease modeling: Mowat-Wilson syndrome patient-derived iPSCs can be differentiated in gastruloid systems to study somite development defects associated with ZEB2 haploinsufficiency [1].
Experimental Design Considerations:
- Temporal control: Inducible Cre systems enable stage-specific ZEB2 deletion to dissect its roles in NMP specification, presomitic mesoderm formation, and somite differentiation [5] [1].
- Single-cell multiomics: Combined scRNA-seq and scATAC-seq reveals ZEB2-dependent gene regulatory networks in skeletal development, applicable to gastruloid systems [62].
- Aneuploidy screening: Microraft arrays facilitate isolation of gastruloids with specific patterning defects for molecular analysis of ZEB2-dependent processes [60].
Zinc finger E-box binding homeobox 2 (ZEB2) is a transcription factor fundamental to embryonic development, cell fate decisions, and differentiation across multiple lineages. Research spanning mouse models and human biological systems has revealed its indispensable functions, particularly in the process of somitogenesis—the segmentation of the embryonic body into somites that give rise to vertebrae, skeletal muscle, and other tissues. Mutations in ZEB2 cause Mowat-Wilson syndrome (MOWS), a rare congenital disorder characterized by intellectual disability, epilepsy, Hirschsprung disease, and distinctive facial features, underscoring the gene's critical developmental role [10]. This guide objectively compares the experimental approaches, findings, and limitations of mouse and human models in ZEB2 research, providing a framework for validating developmental mechanisms across species.
Section 1: ZEB2 in Mouse Models of Somitogenesis
Phenotypic Evidence from Genetic Knockouts
Mouse models with targeted disruption of Zeb2 provide the most direct evidence of its role in somitogenesis. Homozygous null mutants exhibit severe developmental defects and die between embryonic days 9.5 and 10.5 [63] [5]. These defects include profound abnormalities in somite formation, highlighting a cell-autonomous requirement for Zeb2 during this critical window of segmentation and axis elongation [5].
Mechanistic Insights from In Vitro Stem Cell Models
Mouse Embryonic Stem Cells (mESCs) have been instrumental in dissecting Zeb2's molecular function. Zeb2 knockout (KO) mESCs can exit the naïve pluripotent state but subsequently stall in an early epiblast-like state [5]. They are impaired in executing both neural and mesendodermal differentiation programs, indicating Zeb2 is critical for progression beyond this primed state into committed lineages, including the presomitic mesoderm.
Key molecular disruptions in Zeb2 KO mESCs include:
- Deregulated Pluripotency Network: Failure to efficiently silence core pluripotency genes like Oct4 and Nanog.
- Altered DNA Methylation Dynamics: Deregulation of DNA (de)methylation machinery, including elevated levels of the demethylase Tet1. Cells initially acquire methylation marks but fail to maintain them, reverting to a more naïve methylome state [5].
- Rescue Potential: Knockdown of Tet1 in Zeb2 KO cells partially restores their ability to differentiate, directly linking Zeb2 to the epigenetic control of cell fate [5].
Table 1: Key Phenotypes of Zeb2 Mouse Models
| Model Type | Key Phenotypic Features | Developmental Stage | Primary References |
|---|---|---|---|
| Homozygous Null Mutant | Lethality; multiple defects including aberrant somitogenesis | Death between E9.5-E10.5 | [63] [5] |
| Conditional Knockouts | Defects in specific lineages (CNS, PNS, neural crest) | Embryonic to postnatal | [5] |
| Zeb2 KO mESCs | Stalled in epiblast-like state; impaired neural and mesendodermal differentiation | In vitro differentiation | [5] |
Section 2: Advancing to Human Model Systems
The Advent of Human Somitoids
Recent breakthroughs in stem cell biology have enabled the generation of somitoids from human pluripotent stem cells (hPSCs) [64]. These 3D cellular aggregates mimic key aspects of human axis formation, including elongation and the sequential formation of paired somite-like structures. This model provides a tractable, ethically accessible system for studying human-specific aspects of development that are difficult to interrogate in mouse models or embryos.
Targeting the Human Segmentation Clock
The segmentation clock is a molecular oscillator that dictates the rhythmic formation of somites. A 2025 study by Meijer et al. utilized somitoids to dissect the role of Notch signaling in pacing this clock in humans [64]. They identified that the stability of the Notch1 intracellular domain (NICD), controlled by its interaction with the E3 ligase FBXW7, is essential for setting the clock's tempo.
The critical experimental finding was that a single point mutation in NICD (S2513A), which stabilizes the protein, led to somitoids with accelerated and rapidly dampened oscillations of the core clock gene HES7, accompanied by failure to properly elongate [64]. This demonstrates that precise control of protein stability is a tunable mechanism for developmental timing in humans.
Section 3: Direct Comparative Analysis of Mouse and Human Models
The following tables synthesize quantitative data and methodological details to facilitate a direct comparison between mouse and human models in ZEB2 and somitogenesis research.
Table 2: Cross-Species Comparison of Key Experimental Findings
| Research Aspect | Findings in Mouse Models | Findings in Human Models |
|---|---|---|
| ZEB2 Loss-of-Function | Early embryonic lethality (E9.5-E10.5); somitogenesis defects [63] [5] | Linked to Mowat-Wilson syndrome; human neurodevelopmental defects [10] |
| Primary Model System | Genetic knockout mice; Zeb2 KO mESCs [5] | Somitoids derived from human PSCs [64] |
| Key Molecular Pathway | Exit from pluripotency; EMT (e.g., Cdh1 repression); DNA methylation (Tet1) [10] [5] | Segmentation clock (HES7 oscillation); NICD/FBXW7 regulation of protein stability [64] |
| Technical Advantage | Well-established genetics; in vivo physiological context | Studies human-specific biology; amenable to live imaging of oscillations |
Table 3: Detailed Experimental Protocols from Key Studies
| Study Component | Methodology in Mouse mESC Model [5] | Methodology in Human Somitoid Model [64] |
|---|---|---|
| Model Generation | Derivation of Zeb2 KO mESCs from Zeb2flox/flox blastocysts; gene targeting via Cre-lox system. | Differentiation of human PSCs into presomitic mesoderm; 3D aggregation to form somitoids. |
| Genetic Manipulation | Conditional knockout; cDNA rescue via Rosa26 locus; Tet1 knockdown. | Tunable manipulation of endogenous FBXW7; introduction of NICD-S2513A point mutation. |
| Differentiation Protocol | Embryoid Body (EB) formation in bacterial petri dishes; neural differentiation with retinoic acid. | Directed differentiation using specific signaling factors to pattern PSCs into paraxial mesoderm. |
| Key Readouts | RNA-sequencing; RRBS for DNA methylation; immunostaining for pluripotency/differentiation markers. | Live imaging of HES7-ACHILLES reporter oscillations; biochemical assays for protein stability; morphology scoring. |
Section 4: Visualizing Signaling Pathways and Workflows
The following diagrams illustrate the core signaling pathways and experimental workflows derived from the research, providing a visual summary of the complex relationships and processes.
ZEB2 and Notch in Development
Diagram Title: ZEB2 and Notch Signaling Interplay in Development
Experimental Workflow for Cross-Species Validation
Diagram Title: Cross-Species Validation Workflow
Section 5: The Scientist's Toolkit
This section catalogs essential research reagents and solutions that form the methodological backbone of contemporary ZEB2 and somitogenesis research.
Table 4: Essential Research Reagents and Solutions
| Reagent / Solution | Function in Research | Example Application |
|---|---|---|
| Zeb2-V5 Tagged mESCs | Enables mapping of DNA-binding sites for endogenous Zeb2 via V5-based ChIP-seq. | Identification of 2432 Zeb2 binding sites in neuroprogenitor cells [10]. |
| Zeb2 Knockout (KO) mESCs | Reveals gene networks and processes dependent on Zeb2 function. | Studying exit from pluripotency and epigenetic regulation (e.g., Tet1) [5]. |
| HES7-ACHILLES Reporter | A fluorescent reporter providing a live readout of segmentation clock oscillations. | Quantifying clock periodicity and damping in live human somitoids [64]. |
| Somitoid Differentiation Protocol | A defined system to generate human presomitic mesoderm and 3D somite-forming aggregates. | Modeling human somitogenesis and its disorders in a dish [64]. |
| Flag-V5 Epitope Tag | A dual epitope tag inserted into endogenous loci for highly specific ChIP-grade immunoprecipitation. | Overcoming the lack of ChIP-seq grade Zeb2 antibodies [10]. |
| 2i + LIF Medium | A chemical-defined medium that maintains mESCs in a naïve pluripotent "ground state." | Culture of mESCs for consistent baseline in differentiation studies [5]. |
The cross-species validation journey from mouse models to human biology underscores both the conserved fundamental roles of ZEB2 and the critical, species-specific nuances in its mechanisms. Mouse models have been indispensable in establishing ZEB2's non-negotiable role in somitogenesis and early development, while human stem cell-based models, particularly somitoids, are now revealing the precise tuning of protein stability and oscillator dynamics that govern human developmental timing. Together, these complementary approaches provide a more complete picture of ZEB2 in development and disease, offering robust platforms for future investigation into related human congenital disorders and therapeutic strategies.
Validating ZEB2 Function Across Species and Disease Contexts
Conserved Role of ZEB2 in Mouse and Human Somitogenesis
Somitogenesis, the process of segmental body formation during embryonic development, is fundamentally conserved across mammals. The transcription factor Zinc Finger E-Box Binding Homeobox 2 (ZEB2) has emerged as a critical regulator of this process, with its functions elucidated through both mouse models and human disease studies. This guide objectively compares experimental findings from multiple systems to delineate ZEB2's conserved role, providing researchers with a synthesis of current data, methodological approaches, and technical resources. Mutations in human ZEB2 cause Mowat-Wilson syndrome (MOWS), a neurodevelopmental disorder often featuring congenital defects indicative of somitogenesis abnormalities, thereby highlighting the gene's essential developmental function [8]. Recent single-cell transcriptomic atlases of mouse prenatal development have further illuminated the dynamic expression patterns of Zeb2 across somitogenesis stages [22]. This guide integrates phenotypic observations with molecular data to present a comprehensive comparison of ZEB2 function in mouse and human somitogenesis.
Comparative Analysis of ZEB2 Function in Somitogenesis
Table 1: Phenotypic Consequences of ZEB2 Perturbation in Model Systems
| Model System | Phenotypic Outcome | Key Molecular Alterations | Experimental Evidence |
|---|---|---|---|
| Mouse (General KO) [8] | Embryonic lethality by ~E8.5; multiple defects including impaired somitogenesis. | Disrupted transcriptional networks governing pluripotency exit and EMT. | Phenotypic analysis of Zeb2 knockout embryos. |
| Mouse (ESC Model) [5] | Stalling in early epiblast-like state; impaired neural and mesendodermal differentiation. | Deregulated pluripotency genes (e.g., Nanog, Sox2); elevated Tet1; faulty DNA methylation dynamics. | RNA-seq, RRBS, and rescue via Tet1 knockdown in differentiating EBs. |
| Human (MOWS) [8] | Congenital anomalies (e.g., skeletal, cardiac) suggestive of somitogenesis defects. | Haploinsufficiency due to deletions, nonsense, or frameshift mutations. | Genetic analysis of patient cohorts (~350 patients). |
| Mouse Gastruloids [6] | Critical role in regulating mouse and human somitogenesis. | N/A | Degron-based perturbation combined with scRNA-seq. |
Table 2: Key Direct Target Genes and Dependent Processes of ZEB2
| Gene / Process | Regulation by ZEB2 | Functional Context | Experimental Support |
|---|---|---|---|
| ZEB2 (itself) [10] | Positive Autoregulation | Necessary for efficient ESC-to-NPC differentiation. | ChIP-seq in NPCs and deletion of promoter-proximal binding site. |
| Cdh1 (E-cadherin) [5] [8] | Repression | Epithelial-to-Mesenchymal Transition (EMT). | Target gene identification and expression analysis. |
| Id1, Smad7 [8] | Dependent Expression | Cell differentiation. | Transcriptomic phenotyping of Zeb2 mutant cells. |
| Nanog, Sox2 [5] [8] | Repression/Silencing | Exit from pluripotency network. | RNA-seq analysis of Zeb2 KO ESCs. |
| Tet1 [5] | Repression | DNA methylation control; exit from naive state. | RRBS and expression analysis in KO ESCs; rescue by KD. |
Detailed Experimental Protocols and Workflows
Protocol 1: Analyzing ZEB2 Function in Mouse Embryonic Stem Cell (ESC) Differentiation
This protocol is used to investigate ZEB2's role in the exit from pluripotency and the initiation of differentiation, processes fundamental to lineage specification during somitogenesis [5].
1. Cell Culture and Maintenance:
- Mouse ESC Lines: Use wild-type (WT), Zeb2 knockout (KO), and rescue (e.g., R26_Zeb2) lines [5] [3].
- Maintenance Medium: Culture ESCs feeder-free in "2i + LIF" medium to maintain a naive pluripotent state. This consists of N2B27 base medium supplemented with [5]:
- 1 μM PD0325901: MEK/ERK pathway inhibitor.
- 3 μM CHIR99021: GSK3 inhibitor.
- 1000 U/mL LIF: Leukemia Inhibitory Factor.
2. Directed Differentiation via Embryoid Body (EB) Formation:
- Day 0: Plate 3-4 million ESCs in a 10-cm bacterial-grade Petri dish in EB medium (e.g., KO-DMEM with 15% FBS, non-essential amino acids, and β-mercaptoethanol). The non-adherent dish promotes aggregate formation [5] [10].
- Day 2 & 4: Refresh the EB medium [5].
- Day 4 Onwards (For Neural Differentiation): Change medium to N2B27 supplemented with 500 nM retinoic acid (RA) to induce neural differentiation. Refresh medium every two days [5] [10].
- Day 8-15: Culture EBs in base N2B27 medium, refreshing it every other day [5].
3. Sample Collection and Analysis:
- Molecular Phenotyping: Harvest cells at specific time points (e.g., day 0, 4, 8, 15) for downstream analysis [5] [10].
- RNA-seq: For transcriptomic profiling.
- Reduced Representation Bisulfite Sequencing (RRBS): For analyzing DNA methylation dynamics.
- Western Blot/Immunofluorescence: To confirm protein-level changes.
4. Perturbation and Rescue Experiments:
- Genetic Knockdown: Transfect cells with shRNAs (e.g., against Tet1) to test for functional rescue of differentiation defects in Zeb2 KO cells [5].
- For Myogenic Differentiation: Transfer ESCs to specific induction media and/or transiently transfect with a MyoD expression construct to drive skeletal muscle differentiation [3].
Figure 1: Workflow for Analyzing ZEB2 in ESC Differentiation. The core differentiation protocol (yellow) can be perturbed using genetic models (red, blue, green) to probe ZEB2 function. Abbreviations: KO (Knockout), OE (Overexpression), KD (Knockdown), RA (Retinoic Acid), IF (Immunofluorescence), WB (Western Blot).
Protocol 2: Mapping ZEB2 DNA-Binding Sites via ChIP-seq
This protocol outlines the method for identifying genome-wide direct target genes of ZEB2, which is crucial for understanding its transcriptional regulatory network in development [10].
1. Generation of Endogenously Tagged ZEB2 Cell Line:
- Gene Editing: Use CRISPR/Cas9 to insert a dual epitope tag (e.g., Flag-V5) in-frame at the C-terminus of the endogenous Zeb2 gene in mouse ESCs. This avoids the artifacts associated with antibody cross-reactivity or overexpression [10].
2. Differentiation and Cell Collection:
- Differentiate the tagged ESCs into the desired cell type, such as neuroprogenitor cells (NPCs), using a protocol similar to the one described in Section 3.1 [10].
- Harvest cells at the desired stage (e.g., day 8 or 10 of neural differentiation) [10].
3. Chromatin Immunoprecipitation (ChIP):
- Cross-link proteins to DNA in living cells (typically with formaldehyde).
- Lyse cells and shear chromatin by sonication to fragment DNA into sizes of 200-500 bp.
- Immunoprecipitate the protein-DNA complexes using an antibody against the epitope tag (e.g., anti-V5) [10].
- Reverse cross-links, purify the DNA, and prepare sequencing libraries.
4. Sequencing and Data Analysis:
- Perform high-throughput sequencing (ChIP-seq).
- Map sequencing reads to the reference genome and call significant peaks of ZEB2 binding.
- Cross-reference binding sites with transcriptome data (e.g., from Zeb2 KO cells) to identify direct, functional target genes [10].
ZEB2 Signaling Pathways and Gene Regulatory Network
ZEB2 functions as a central node in a complex regulatory network that integrates multiple signaling pathways to control cell fate during somitogenesis and other developmental processes. Its action as a transcriptional repressor is pivotal for exiting pluripotency and committing to differentiation [5] [8].
Figure 2: ZEB2's Integrative Role in Gene Regulation. ZEB2 (blue) is upregulated by key signaling pathways (green) and the pluripotency network (yellow). It then represses key targets (red) to silence the pluripotency network, promote EMT (via Cdh1), and control DNA methylation (via Tet1). A positive autoregulatory loop stabilizes its own expression.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Studying ZEB2 in Development
| Reagent / Resource | Function and Application | Example Use Case |
|---|---|---|
| Zeb2 KO Mouse ESCs [5] | Model for ZEB2 loss-of-function; study of pluripotency exit and differentiation stall. | Transcriptomic (RNA-seq) and epigenomic (RRBS) phenotyping [5]. |
| R26_Zeb2 cDNA OE ESCs [3] | Model for ZEB2 gain-of-function; rescue and overexpression studies. | Enhancing myogenic differentiation in ESCs and C2C12 cells [3]. |
| Zeb2-Flag-V5 ESCs [10] | Endogenously tagged ZEB2 for mapping direct DNA binding sites. | Chromatin Immunoprecipitation followed by sequencing (ChIP-seq) [10]. |
| 2i + LIF Medium [5] | Maintains mouse ESCs in a naive, ground state of pluripotency. | Culture and expansion of ESCs prior to differentiation induction [5]. |
| Gastruloid Models [6] | 3D in vitro model that recapitulates aspects of gastrulation and somitogenesis. | Studying the role of ZEB2 in a scalable, defined system mimicking early development [6]. |
| dTAG Degron System [6] | Allows rapid, precise degradation of a tagged protein of interest. | Acute perturbation of ZEB2 function to study immediate effects on somitogenesis. |
The conserved role of ZEB2 in mouse and human somitogenesis is underscored by its non-negotiable function in exiting pluripotency and committing to mesodermal lineages. Data from mouse KO models, ESC differentiation systems, and human genetics converge to define ZEB2 as a critical transcriptional repressor that silences the pluripotency network and regulates the epigenetic landscape, thereby enabling proper lineage specification. The experimental protocols and reagents detailed herein provide a roadmap for continued investigation into the mechanisms of ZEB2 action, with implications for understanding congenital disorders and guiding stem cell-based differentiation protocols for regenerative medicine.
Comparative Analysis of ZEB2-Dependent Gene Regulatory Networks
Zinc finger E-box binding homeobox 2 (ZEB2) is a pivotal transcription factor governing embryonic development, neural induction, and cell fate determination. This review provides a systematic comparison of ZEB2-dependent gene regulatory networks (GRNs) in mouse and human models, highlighting conserved and species-specific mechanisms in somitogenesis and neural development. We synthesize data from recent CRISPR screens, chromatin immunoprecipitation sequencing (ChIP-seq), and single-cell transcriptomic studies to delineate the ZEB2 regulon across developmental contexts. Quantitative comparisons of target genes, binding sites, and functional pathways are presented alongside detailed experimental methodologies and essential research tools. This analysis frames ZEB2 function within the broader thesis of its non-redundant roles in mammalian development and its implications for neurodevelopmental disorders such as Mowat-Wilson syndrome.
ZEB2 (also known as SIP1/ZFHX1B) is a DNA-binding transcription factor containing two zinc finger clusters and a homeodomain-like sequence that recognizes E-box-like motifs (CACCT sequences) in target genes [10] [47]. During embryonic development, ZEB2 exhibits spatiotemporally regulated expression in the neural tube, neural crest cells, hippocampus, and cerebral cortex [35]. Its non-redundant functions are evidenced by severe developmental defects in knockout models and human haploinsufficiency syndromes. Mouse Zeb2 knockout embryos die around embryonic day (E) 9.5 with profound neural plate and neural crest defects [5], while heterozygous mutations in humans cause Mowat-Wilson syndrome (MOWS), characterized by intellectual disability, epilepsy, Hirschsprung disease, and distinctive facies [10] [65].
The positioning of ZEB2 within developmental GRNs enables it to coordinate multiple processes including epithelial-to-mesenchymal transition (EMT), neural differentiation, and somitogenesis. This review systematically compares ZEB2-dependent networks across species and experimental systems, with particular emphasis on its role in the segmentation clock governing somitogenesis - the process of sequential somite formation from presomitic mesoderm that establishes the vertebrate body plan [66].
Comparative Analysis of ZEB2 Target Networks Across Biological Contexts
ZEB2 Regulon in Mouse Embryonic Stem Cells and Neural Differentiation
In mouse embryonic stem cells (mESCs), ZEB2 is undetectable in the naïve state but undergoes strong upregulation during neural differentiation, where it is essential for exit from primed pluripotency and progression to neuroprogenitor cells (NPCs) [10]. A comprehensive ChIP-seq analysis in ESC-derived NPCs identified 2,432 ZEB2 binding sites, with 2,294 sites mapping to 1,952 protein-encoding genes [10]. This study revealed ZEB2 autoregulation through a promoter-proximal binding site, with deletion of this site demonstrating that Zeb2 autoregulation is necessary for appropriate Zeb2-dependent effects in ESC-to-NPC differentiation.
Table 1: Key ZEB2 Target Genes in mESC Neural Differentiation
| Target Gene | Function | Regulation by ZEB2 | Experimental Evidence |
|---|---|---|---|
| Nanog | Pluripotency factor | Repression | ChIP-seq, RNA-seq [10] |
| Sox2 | Pluripotency factor | Repression | ChIP-seq, RNA-seq [10] |
| Cdh1 (E-cadherin) | Cell adhesion | Repression | ChIP-seq, functional validation [10] [5] |
| Id1 | Differentiation inhibitor | Repression | ChIP-seq, RNA-seq [10] |
| Smad7 | TGF-β signaling inhibitor | Repression | ChIP-seq, RNA-seq [10] |
| Ntf3 | Neurotrophin | Activation | ChIP-seq, RNA-seq [10] |
| Sox6 | Neural differentiation | Activation | ChIP-seq, RNA-seq [10] |
| Zeb2 (autoregulation) | Self-regulation | Both positive and negative | Promoter binding site deletion [10] |
ZEB2 orchestrates the transition from pluripotency to committed neural fates through direct repression of core pluripotency factors (Nanog, Sox2) and activation of neural differentiation genes (Ntf3, Sox6) [10]. Additionally, ZEB2 represses Id1 and Smad7, modulating BMP/TGF-β signaling responses during neural induction [5]. The direct repression of Cdh1 (E-cadherin) facilitates epithelial-to-mesenchymal transition, enabling neural crest migration and neuroepithelial remodeling [47].
ZEB2 in Human Pluripotent Stem Cell Models and Anterior Neural Tube Closure
Recent advances in human pluripotent stem cell (hPSC)-derived organoids have enabled systematic dissection of ZEB2 function in human neurulation. An arrayed CRISPR interference (CRISPRi) screen of 77 transcription factors in anterior neural tube organoids revealed ZEB2 as a key regulator of neural tube closure [67] [68]. Single-cell RNA sequencing of perturbed organoids positioned ZEB2 within a network of transcription factors including ZIC2, SOX11, and ZNF521 that jointly govern neural tube morphogenesis in the anterior forebrain region [67].
In this human organoid system, ZEB2 expression marked the neural ectoderm population, which also expressed forebrain-associated transcription factors including LHX2, HESX1, PAX6, SIX3, ZEB1, and ZEB2 itself [67] [68]. The neural ectoderm displayed a medial-to-lateral SOX2 gradient, with highest expression in neural ectoderm, intermediate in neural plate border, and lowest in surface ectoderm [68]. This human organoid model successfully recapitulated the three ectodermal cell types observed in vivo, with matching gene expression patterns to 4-week human embryos [68].
Table 2: ZEB2-Dependent GRN Components in Human Neural Tube Organoids
| Gene | Role in Neural Tube Closure | Expression Pattern | Functional Relationship with ZEB2 |
|---|---|---|---|
| ZEB2 | Neural tube morphogenesis | Neural ectoderm | Core regulator [67] |
| ZIC2 | Required for closure | Neural ectoderm | Co-regulated program [67] |
| SOX11 | Required for closure | Neural ectoderm | Co-regulated program [67] |
| ZNF521 | Prevents ectopic closure | Neural ectoderm | Opposing role to ZEB2 [67] |
| SOX2 | Neural progenitor identity | Gradient: high medial, low lateral | Co-expressed with ZEB2 [68] |
| PAX6 | Forebrain specification | Neural ectoderm | Co-expressed with ZEB2 [68] |
| SIX3 | Forebrain specification | Neural ectoderm | Co-expressed with ZEB2 [68] |
ZEB2 in Somitogenesis: Mouse and Human Conservation
Somitogenesis represents a crucial developmental process where ZEB2 functions have been conserved across species. A multilayered mass spectrometry-based proteomics study of gastruloid differentiation identified ZEB2 as playing a key role in both mouse and human somitogenesis [12]. The segmentation clock governing somitogenesis involves oscillatory gene expression in the presomitic mesoderm, with HES7 as a core oscillator and ZEB2 functioning downstream to regulate boundary formation [66].
In the context of somitogenesis, ZEB2 interacts with multiple signaling pathways. Computational modeling of the presomitic mesoderm GRN positions ZEB2 as integrating inputs from the Notch, FGF, and WNT pathways to regulate the transition from mesodermal progenitors to segmented somites [66]. This modeling predicts that the Hes7 oscillator shows interference with Hes1 when their intrinsic frequencies differ, with ZEB2 potentially modulating this interaction [66].
Recent single-cell time-lapse analysis of mouse prenatal development from gastrula to birth has provided unprecedented resolution of ZEB2 expression dynamics during posterior embryo development and somitogenesis [22]. This comprehensive dataset profiling 12.4 million nuclei from 83 embryos reveals ZEB2 involvement in neuromesodermal progenitor (NMP) differentiation and notochord development during the trunk-to-tail transition [22].
Experimental Methodologies for ZEB2 GRN Analysis
Chromatin Immunoprecipitation Sequencing (ChIP-seq)
Protocol for ZEB2 ChIP-seq in Mouse ESC-Derived NPCs [10]:
- Cell Line Generation: Edit one Zeb2 allele in mESCs by inserting a Flag-V5 epitope tag just before the Zeb2 stop codon, in-frame with the last exon.
- Neural Differentiation: Differentiate Zeb2-V5 mESCs toward neural lineage using embryoid body formation. On day 0, plate 4×10^6 cells in non-adherent dishes in EB medium. On day 4, add 5μM retinoic acid. Harvest aggregates at day 8 for NPC analysis.
- Cross-linking and Cell Lysis: Fix cells with 1% formaldehyde for 10min at room temperature. Quench with 125mM glycine.
- Chromatin Preparation: Lyse cells and sonicate chromatin to 200-500bp fragments.
- Immunoprecipitation: Incubate chromatin with V5 antibody-conjugated beads overnight at 4°C.
- Library Preparation: Reverse cross-links, purify DNA, and prepare sequencing libraries.
- Bioinformatic Analysis: Map sequences to reference genome, call peaks, and annotate binding sites.
This approach identified 2,432 high-confidence ZEB2 binding sites in NPCs, providing the foundation for ZEB2 regulon analysis [10].
Arrayed CRISPRi Screening in Human Organoids
Protocol for Arrayed CRISPRi Screening in hPSC-Derived Anterior Neural Tube Organoids [67] [68]:
- Organoid Generation: Seed hPSCs onto micropatterned glass coverslips with 250μm diameter ECM micropatterns. Embed in Matrigel to form epithelial cysts with apicobasal polarity.
- Neural Induction: Inhibit BMP, TGFβ, and WNT signaling for 2 days, then add recombinant human BMP4 for 4 days with continued TGFβ and WNT inhibition.
- Lentiviral Library Delivery: Develop high-titer lentivirus production method for efficient delivery to stem cells in parallelized small volumes.
- CRISPRi Implementation: Target 77 transcription factors with individual guide RNAs in arrayed format.
- Phenotypic Screening: Assess neural tube closure morphology via apical NCAD staining on day 4.
- Single-Cell RNA Sequencing: Profile perturbed organoids to identify co-regulated gene programs.
This platform enabled identification of ZIC2, SOX11, and ZNF521 as essential regulators of neural tube closure operating in conjunction with ZEB2 [67].
Single-Cell RNA Sequencing Analysis
Protocol for scRNA-seq in Developing Embryos [22]:
- Embryo Collection and Staging: Collect mouse embryos from E8 to P0, stage by somite number and limb bud morphology.
- Nuclei Isolation: Flash-freeze embryos, pulverize, and isolate nuclei.
- Combinatorial Indexing: Perform sci-RNA-seq3 with optimized single-nucleus combinatorial indexing.
- Sequencing: Generate 160 billion reads across 15 sci-RNA-seq experiments.
- Data Processing: Demultiplex, trim, map, and filter data to obtain 11.4 million high-quality nuclear transcriptomes.
- Cell Type Annotation: Use Scanpy for clustering and annotate 190 cell types based on marker genes.
This massive dataset enabled reconstruction of developmental trajectories and identification of ZEB2 expression dynamics across cell types and stages [22].
ZEB2 Signaling Pathways and Regulatory Circuits
ZEB2 in TGF-β/SMAD Signaling
ZEB2 physically interacts with SMAD proteins, intracellular effectors of TGF-β signaling, forming a core component of its mechanistic function [35] [47]. Upon TGF-β stimulation, SMAD2 and SMAD3 are phosphorylated and form complexes with SMAD4. These complexes then interact with ZEB2, which recruits co-repressor complexes (CtBP, NuRD) to target genes [47]. This ZEB2-SMAD complex represses epithelial genes like E-cadherin while activating mesenchymal genes, facilitating EMT during neural crest development and somitogenesis [47].
Figure 1: ZEB2 in TGF-β/SMAD Signaling Pathway. ZEB2 interacts with phosphorylated SMAD complexes to regulate target gene expression.
ZEB2 in the Segmentation Clock Network
During somitogenesis, ZEB2 operates within a complex gene regulatory network that integrates oscillatory signaling from the Notch, Wnt, and FGF pathways [66]. The core oscillator involves HES7, which directly represses its own expression and regulates ZEB2 activity. ZEB2 functions downstream to translate the oscillatory clock into spatial boundaries through regulation of Mesp2 and Ephrin signaling.
Figure 2: ZEB2 in the Segmentation Clock Network. ZEB2 integrates oscillatory signals to regulate boundary formation during somitogenesis.
ZEB2 Autoregulatory Circuitry
ZEB2 participates in an autoregulatory feedback loop critical for maintaining its expression levels during development [10]. This circuit involves direct binding of ZEB2 to its own promoter region, creating a self-reinforcing loop that stabilizes cell fate decisions. Additionally, ZEB2 is regulated by upstream factors including DLX1/2 in the subpallium and AP-1 in response to TNFα signaling [35].
Figure 3: ZEB2 Autoregulatory Circuit. ZEB2 maintains its expression through direct promoter binding while regulating downstream differentiation genes.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for ZEB2 GRN Analysis
| Reagent/Resource | Function/Application | Key Features | Reference |
|---|---|---|---|
| ZEB2-V5 mESCs | ChIP-seq; neural differentiation | Flag-V5 epitope tag at endogenous Zeb2 locus | [10] |
| Arrayed CRISPRi Library | Functional screening | 77 TF targets; high-titer lentiviral delivery | [67] [68] |
| Anterior Neural Tube Organoids | Human neurulation modeling | Micropatterned; reproducible morphogenesis | [67] [68] |
| sci-RNA-seq3 Protocol | Single-cell transcriptomics | Combinatorial indexing; 11.4M nuclei dataset | [22] |
| Zeb2flox/flox Mice | Conditional knockout | Tissue-specific deletion; MOWS modeling | [5] |
| H1 and H9 hESC Lines | Human neural differentiation | MGE-like progenitors; interneuron differentiation | [35] |
This comparative analysis reveals both conserved and species-specific features of ZEB2-dependent gene regulatory networks. Across mouse and human systems, ZEB2 consistently operates as a transcriptional repressor that promotes exit from pluripotent states and facilitates neural differentiation through direct repression of core pluripotency factors (Nanog, Sox2) and epithelial genes (Cdh1). In somitogenesis, ZEB2 functions within the evolutionarily conserved segmentation clock network but shows species-specific regulatory connections evidenced by distinct enhancer utilization between mice and humans [35].
The positioning of ZEB2 at the interface of multiple signaling pathways (TGF-β/SMAD, Notch, FGF, Wnt) enables it to integrate diverse developmental cues and translate them into precise transcriptional programs governing cell fate decisions. Mutations disrupting these networks cause Mowat-Wilson syndrome, highlighting the functional importance of precise ZEB2 dosage and spatiotemporal regulation during human development [65].
Future studies leveraging emerging technologies like single-cell multi-omics and CRISPR-based screening in advanced organoid models will further refine our understanding of ZEB2-dependent GRNs, potentially revealing therapeutic opportunities for modulating ZEB2 function in developmental disorders and tissue regeneration.
The zinc finger E-box-binding homeobox 2 (ZEB2) transcription factor exemplifies functional duality in biological systems. While critically important for normal embryonic development, stem cell differentiation, and tissue homeostasis, ZEB2 plays complex and often contradictory roles in cancer pathogenesis. This review systematically analyzes ZEB2's contrasting functions, examining its master regulatory role in developmental processes such as somitogenesis, neurogenesis, and myogenic differentiation alongside its context-dependent oncogenic and tumor-suppressive activities across various cancers. We integrate structural insights, molecular interaction networks, and experimental evidence to provide a comprehensive comparison of ZEB2's dichotomous nature, with implications for targeted therapeutic interventions.
ZEB2 (also known as SIP1 or Zfhx1b) is a member of the Zfh1 family of two-handed zinc finger/homeodomain proteins located on chromosome 2q22.3 [7]. As a DNA-binding transcriptional regulator, ZEB2 recognizes specific E-box sequences (CACCTG) in promoter regions of target genes through two clusters of zinc finger domains located at its N- and C-termini [69]. The protein's central homeodomain facilitates protein-protein interactions rather than DNA binding [69].
Structural and Functional Domains: ZEB2 contains several critical domains that determine its functional capabilities:
- NZF and CZF clusters: Display 88% and 93% amino acid sequence similarity with ZEB1, respectively, enabling binding to similar DNA regions [69]
- NuRD complex interacting motif (NIM): Facilitates interaction with nucleosome remodeling and histone deacetylation complex [69]
- SMAD binding domains (SBD): Play a key role in regulating SMAD protein activity [69]
- CtBP-interacting domain (CID): Determines transcriptional repressor capability through interaction with histone deacetylase HDAC1 [69]
Unlike ZEB1, ZEB2 contains a unique 51-amino acid segment with a critical tandem repeat (QXVX)2 essential for binding all activated SMAD proteins [69]. This structural difference accounts for the antagonistic regulatory effects ZEB1 and ZEB2 exert on SMAD protein activity, with ZEB2 primarily functioning as a repressor while ZEB1 can recruit coactivators p300 and P/CAF to enhance TGFβ-dependent gene transcription [69].
Table 1: Key Structural Domains of ZEB2 and Their Functions
| Domain | Location | Function |
|---|---|---|
| N-terminal zinc finger cluster (NZF) | N-terminus | DNA binding to E-box sequences |
| C-terminal zinc finger cluster (CZF) | C-terminus | DNA binding to E-box sequences |
| Homeodomain (HD) | Central region | Protein-protein interactions |
| SMAD binding domain (SBD) | N-terminal region | Regulation of SMAD protein activity |
| CtBP-interacting domain (CID) | Multiple regions | Transcriptional repression via HDAC1 |
| NuRD complex interacting motif (NIM) | Central region | Interaction with chromatin remodeling complex |
ZEB2 in Development and Stem Cell Biology
Neural Development and Differentiation
ZEB2 plays indispensable roles in neural development, as demonstrated by multiple stem cell differentiation models. In mouse embryonic stem cells (mESCs), Zeb2 knockout causes cells to stall in an early epiblast-like state, impairing both neural and mesendodermal differentiation [5]. During neural differentiation, Zeb2 mRNA and protein upregulation is essential for the exit from primed pluripotency and progression to neuroprogenitor cells (NPCs) [10].
Mechanistic Insights: Zeb2 regulates the pluripotency network by controlling the expression of key genes including Nanog, Sox2, and DNA methylation regulators [5]. Zeb2-deficient ESCs show elevated Tet1 levels, resulting in aberrant DNA methylation patterns that prevent irreversible commitment to differentiation [5]. Knockdown of Tet1 in Zeb2 knockout cells partially rescues their impaired differentiation capacity, establishing a direct link between Zeb2 and epigenetic regulation [5].
Chromatin immunoprecipitation sequencing (ChIP-seq) in ESC-derived NPCs has identified 2,432 Zeb2 DNA-binding sites, with 2,294 mapping to 1,952 protein-encoding genes [10]. Notably, a major binding site maps promoter-proximal to Zeb2 itself, demonstrating critical autoregulation necessary for appropriate Zeb2-dependent effects in ESC-to-NPC differentiation [10].
Myogenic Differentiation
Beyond neural development, Zeb2 positively regulates skeletal muscle differentiation. During myogenic induction of pluripotent stem cells, Zeb2 overexpression upregulates key myogenic markers (Pax3, Pax7, MyoD, Myogenin) and myomiRs (miR-1, miR-133b, miR-206, miR-208) [3]. Zeb2 also enhances the expression of late myogenic markers like myosin heavy chain (MyHC) and sarcomeric α-actinin [3].
Functional Validation: In C2C12 myoblast cells, Zeb2 overexpression upregulates Myf5, MyoD, and Myogenin, confirming its pro-differentiation role [3]. The DNA-binding capability of Zeb2 is essential for this function, as a zinc finger mutant (ZnfZeb2) fails to similarly enhance myogenic differentiation [3].
Implications for Somitogenesis Research
The fundamental principles of ZEB2 function established in stem cell models directly inform its role in somitogenesis. During embryonic development, ZEB2 coordinates with signaling pathways (TGFβ/BMP, Wnt, Notch) to regulate mesodermal patterning and somite formation [3]. The protein's ability to fine-tune transcriptional responses to TGFβ/Nodal-Activin and BMP signaling positions it as a critical modulator of somite boundary formation and differentiation [3].
Oncogenic Functions of ZEB2
Epithelial-Mesenchymal Transition and Metastasis
ZEB2 is widely recognized as a master regulator of epithelial-mesenchymal transition (EMT) in carcinoma progression. Through repression of epithelial markers like E-cadherin (CDH1) and induction of mesenchymal markers (vimentin), ZEB2 enhances cell migration, invasion, and metastatic dissemination [69] [70].
Experimental Evidence in Glioblastoma: In both pediatric and adult glioblastoma cells, ZEB2 inhibition upregulates E-cadherin expression, downregulates vimentin, and significantly reduces invasion and migration capabilities [70]. The functional consequences of ZEB2 suppression, however, differ between pediatric and adult glioblastoma cells, suggesting distinct underlying molecular mechanisms in these tumor types [70].
Table 2: ZEB2 Functional Effects Across Different Cancer Types
| Cancer Type | ZEB2 Function | Experimental Evidence | Molecular Targets |
|---|---|---|---|
| Glioblastoma (adult) | Pro-invasive | Reduced migration/invasion after siRNA knockdown | E-cadherin ↑, Vimentin ↓ |
| Glioblastoma (pediatric) | Pro-invasive & pro-proliferative | Reduced migration, invasion, AND proliferation after siRNA knockdown | E-cadherin ↑, Vimentin ↓, Cell cycle arrest |
| Non-small cell lung cancer | Immunomodulatory | Drives CD8+ T-cell differentiation toward cytotoxic effector phenotype | T-bet/ZEB2 axis |
| Laryngeal squamous cell carcinoma | Oncogenic | Silencing inhibits viability, migration, invasion; induces cell cycle arrest and apoptosis | EMT markers |
| Gastric cancer | Oncogenic | Promotes proliferation, migration, invasion, EMT via Wnt/β-catenin pathway | ZEB2-AS1 lncRNA, β-catenin |
ZEB2 in Tumor Microenvironment and Immunomodulation
Recent research has revealed ZEB2's critical functions within the tumor microenvironment, particularly in immune cell regulation.
CD8+ T-cell Differentiation: In non-small cell lung cancer (NSCLC), ZEB2 drives CD8+ T-cell differentiation along the cytotoxic effector trajectory [40]. Single-cell RNA sequencing analysis of CD8+ cells from treatment-naïve NSCLC patients identified ZEB2 as a master regulon promoting the differentiation of tumor-resident effector CD8+ T-cells (Teff) over exhausted (Texh) or dysfunctional (TLdys) populations [40]. The T-bet/ZEB2 axis stimulates lung tumor-reactive Teff cell differentiation and displays immunotherapeutic effects independent of immune checkpoint blockade therapy [40].
Tumor-Associated Macrophage Programming: ZEB2 functions as a master molecular switch controlling the pro-tumor program in tumor-associated macrophages (TAMs) [71]. Single-cell CRISPR screens in primary TAM models demonstrated that ZEB2 induces immunosuppression programs while suppressing antigen presentation and type-I interferon inflammation [71]. Targeting ZEB2 reprograms TAMs, enhances anti-tumor T-cell responses, and enables robust tumor control [71].
Contrasting ZEB2 Functions: Experimental Methodologies
Genetic Manipulation Approaches
Knockout Models: Zeb2 knockout mouse ESCs are generated through cre-loxP mediated recombination or similar gene editing technologies [5]. These are maintained in 2i + LIF medium (containing PD0325901, CHIR99021, and leukemia inhibitory factor) to preserve ground-state pluripotency [5].
Knockdown Strategies: ZEB2 inhibition is typically achieved using small interfering RNAs (siRNAs) targeting specific regions of ZEB2 mRNA [70]. Standard protocols involve transfection with 25-50 nM siRNA using Lipofectamine RNAiMAX reagent, with efficiency measured at mRNA level by quantitative RT-PCR 48-72 hours post-transfection [70].
Overexpression Systems: For gain-of-function studies, researchers employ R26_Zeb2 mESCs generated via RMCE technology to insert N-terminally Flag epitope-tagged wild-type Zeb2 cDNA into the ROSA26 locus [3]. This enables controlled Zeb2 expression at physiological levels.
Differentiation Protocols
Neural Differentiation: The standard protocol involves plating ESCs in bacterial petri dishes to form embryoid bodies (EBs) in EB medium, followed by sequential exposure to retinoic acid (500 nM) and N2B27 medium over 8-15 days [5]. The resulting neural progenitors can be assessed for marker expression and functional properties.
Myogenic Differentiation: Skeletal muscle differentiation is induced through transient transfection with MyoD expression constructs, with differentiation markers analyzed at specific timepoints (typically up to 22 days) [3]. Assessment includes qRT-PCR for myogenic factors, immunofluorescence for myosin heavy chain, and Western blot for sarcomeric α-actinin.
Functional Assays
Invasion and Migration: Transwell migration assays (using 8 μm pore size membranes) and wound healing assays quantitatively measure cellular invasion and migration capabilities following ZEB2 manipulation [70].
Proliferation and Viability: Cell Counting Kit-8 (CCK-8) and BrdU proliferation ELISA assays determine cell viability and proliferation rates [70]. Flow cytometry with propidium iodide staining assesses cell cycle distribution and apoptosis.
Molecular Analyses: Western blotting for E-cadherin, vimentin, and other EMT markers; qRT-PCR for lineage-specific genes; and chromatin immunoprecipitation (ChIP) for DNA-binding site identification provide mechanistic insights [70] [10].
Signaling Pathways and Molecular Networks
ZEB2 operates within complex signaling networks, with context-dependent outcomes determined by specific interacting partners and cellular environments.
Diagram 1: ZEB2 Signaling Network. ZEB2 integrates TGFβ/BMP signaling through SMAD interactions, functioning as either a repressor or activator depending on co-factor recruitment (NuRD/CtBP vs. p300/PCAF). This dual capacity enables context-specific regulation of target genes involved in differentiation and EMT.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for ZEB2 Investigation
| Reagent/Category | Specific Examples | Application | Function |
|---|---|---|---|
| Cell Lines | Mouse ESCs (CGR8), Human ESCs, U87/U373 (adult GBM), KNS42 (pediatric GBM) | In vitro modeling | Provide context-specific systems for ZEB2 functional studies |
| Genetic Tools | ZEB2-specific siRNAs, Cre-loxP systems, Flag-V5 epitope-tagged Zeb2 | Genetic manipulation | Enable controlled knockdown, knockout, or overexpression of ZEB2 |
| Differentiation Media | 2i+LIF medium, N2B27, retinoic acid (500 nM) | Stem cell differentiation | Support directed differentiation into neural or other lineages |
| Antibodies | Anti-ZEB2 (ProSci), Anti-E-cadherin (Abcam), Anti-vimentin (Cell Signaling) | Protein detection | Enable Western blot, immunohistochemistry, and functional characterization |
| Assay Kits | Cell Counting Kit-8 (CCK-8), BrdU proliferation ELISA, Transwell invasion assays | Functional analysis | Quantify proliferation, viability, invasion, and migration capabilities |
| Sequencing Tools | ChIP-seq grade reagents, RNA-seq kits | Molecular profiling | Identify genome-wide binding sites and transcriptomic changes |
The dualistic nature of ZEB2 in development and cancer underscores the critical importance of context in considering its therapeutic targeting. In developmental contexts, ZEB2 deficiency causes severe defects, as evidenced by Mowat-Wilson syndrome characterized by intellectual disability, epilepsy, Hirschsprung disease, and other anomalies [10] [3]. By contrast, in numerous cancers, ZEB2 overexpression drives malignant progression through EMT induction, invasion, and metastasis [69] [70].
However, this paradigm is further complicated by ZEB2's recently discovered immunomodulatory functions. In CD8+ T-cells, ZEB2 promotes anti-tumor immunity by driving cytotoxic effector differentiation [40], while in tumor-associated macrophages, it orchestrates pro-tumor programs [71]. These opposing immune functions highlight the cellular context-dependency of ZEB2 activity and suggest that therapeutic strategies must be precisely tailored to specific tumor compartments.
Future research should focus on developing compartment-specific ZEB2 modulators that can inhibit its oncogenic functions in carcinoma cells and pro-tumor macrophages while preserving or enhancing its differentiation-promoting and immunostimulatory roles in cytotoxic T-cells. The experimental methodologies and reagents outlined herein provide the essential toolkit for these next-generation investigations, moving the field toward sophisticated therapeutic approaches that respect the contextual duality of this multifunctional transcription factor.
Integration of Proteomic and Transcriptomic Data for Pathway Validation
Within the field of developmental biology, the integration of proteomic and transcriptomic data has emerged as a powerful methodology for validating molecular pathways controlling critical processes such as somitogenesis. This multi-omics approach enables researchers to move beyond correlation to causation by identifying coherent signals across molecular layers, providing stronger evidence for pathway involvement in complex biological systems. The transcription factor ZEB2 has been identified through such integrated analyses as a critical regulator in mouse and human somitogenesis, offering a compelling case study for the power of these methodologies [5] [12]. This guide objectively compares experimental approaches and computational frameworks for multi-omics data integration, with a specific focus on validating ZEB2's role in somitogenesis, providing researchers with practical methodologies for pathway validation in developmental contexts.
Comparative Analysis of Multi-Omics Integration Methods
Table 1: Comparison of Multi-Omics Data Integration Methods
| Method Name | Core Approach | Directional Integration | Key Advantages | ZEB2 Research Applicability |
|---|---|---|---|---|
| DPM (Directional P-value Merging) | P-value fusion with directional constraints | Yes | Prioritizes genes with consistent directionality; reduces false positives | High - Ideal for testing directional hypotheses in ZEB2 perturbations |
| ActivePathways | Gene prioritization through data fusion | No (standard version) | Identifies pathways with multi-omics support; intuitive visualization | Medium - Useful for exploratory analysis of ZEB2 networks |
| Empirical Brown's Method | P-value merging with covariance adjustment | No | Accounts for gene-to-gene covariation in omics data | Medium - General purpose significance testing |
| Stouffer's Method | Weighted Z-score integration | No (with directional extension available) | Simple implementation; handles weighted integration | Low - Limited directional capability without modification |
| scDCC | Deep learning for single-cell clustering | N/A | Top-performing for both transcriptomic and proteomic clustering | High - For single-cell analyses of ZEB2 knockout models |
Performance Metrics in Biological Contexts
Recent benchmarking studies evaluating 28 computational algorithms on 10 paired transcriptomic and proteomic datasets revealed that methods like scDCC, scAIDE, and FlowSOM demonstrate top performance across both omics modalities, with scAIDE ranking first for proteomic data and scDCC for transcriptomic data [72]. The Directional P-value Merging (DPM) method has shown particular utility for pathway validation in developmental contexts, as it incorporates user-defined directional constraints to prioritize genes whose changes across omics datasets align with biological expectations (e.g., mRNA-protein pairs that coherently increase or decrease) while penalizing those with inconsistent directions [73].
For ZEB2 research, the DPM framework enables testing specific hypotheses about downstream targets by defining expected directional relationships. For instance, if ZEB2 acts as a transcriptional repressor, its knockout should show inverse relationships between mRNA expression of direct targets and subsequent protein abundance, which DPM can systematically evaluate across the entire transcriptome and proteome [5] [73].
Experimental Design for ZEB2 Pathway Validation
Model Systems and Perturbation Strategies
Table 2: Experimental Approaches for ZEB2 Functional Validation
| Experimental Component | Mouse Model Approach | Human Stem Cell Approach | Comparative Advantages |
|---|---|---|---|
| Genetic perturbation | Conventional KO; Conditional KO | CRISPRi; CRISPR knockout in hPSCs | Mouse: Full developmental context; Human: Direct relevance |
| Tissue analysis | Embryonic dissection at E8.5-E12.5 | hPSC-derived gastruloids and organoids | Mouse: Native tissue environment; Human: Scalable for screening |
| Multi-omics profiling | FACS sorting → RNA-seq + LC-MS/MS | scRNA-seq + Mass cytometry | Mouse: Cell-type specific in vivo; Human: Reduced heterogeneity |
| Functional validation | Lineage tracing; Morphological analysis | Immunofluorescence; Morphometric scoring | Mouse: Physiological relevance; Human: Genetic manipulability |
Integrated Workflow for Pathway Validation
The following diagram illustrates a comprehensive experimental workflow for validating ZEB2 pathways using integrated proteomic and transcriptomic data:
Case Study: Validating ZEB2 Function in Somitogenesis
Key Molecular Pathways Regulated by ZEB2
Integrated transcriptomic and proteomic analyses of ZEB2 knockout models have revealed its essential role in regulating several critical pathways during somitogenesis:
Table 3: ZEB2-Regulated Pathways Identified Through Multi-Omics Integration
| Pathway Category | Specific Pathway | Direction of Change in KO | Functional Role in Somitogenesis | Multi-Omics Support |
|---|---|---|---|---|
| Pluripotency Network | Nanog, Oct4 | Upregulated | Prevents exit from pluripotent state | Transcriptomic & Proteomic |
| EMT Regulation | E-cadherin, N-cadherin | Deregulated | Impairs epithelial-to-mesenchymal transition | Transcriptomic & Proteomic |
| DNA Methylation | Tet1, Dnmt family | Deregulated | Disrupts epigenetic programming | Primarily Transcriptomic |
| Neural Differentiation | Neural specification genes | Downregulated | Impairs neural differentiation programs | Transcriptomic & Proteomic |
| Mesendodermal Formation | Mesendodermal markers | Variable | Alters mesendoderm specification | Transcriptomic & Proteomic |
ZEB2 Regulatory Network in Somitogenesis
The following diagram illustrates the core regulatory network through which ZEB2 controls somitogenesis, as revealed through integrated multi-omics studies:
Detailed Experimental Protocols
Cell Sorting and Multi-Omics Profiling
For validating ZEB2 pathways in developmental contexts, the following protocol adapted from human lung cell analyses provides a robust framework [74]:
Fluorescence Activated Cell Sorting (FACS) Protocol:
- Dissociate embryonic tissue or gastruloids to single-cell suspension using enzymatic digestion (Trypsin-EDTA or collagenase)
- Stain cells with fluorophore-conjugated antibodies against surface markers (CD45, CD326, CD31, CD144 for major lineages)
- Sort purified cell populations using FACS Aria II or similar instrument with strict gating controls
- Split sorted cells for parallel transcriptomic and proteomic analysis
Transcriptomic Profiling:
- Extract RNA using column-based purification kits with DNase treatment
- Assess RNA quality (RIN > 8.0 recommended)
- Prepare libraries using SMARTer or similar kit for full-length transcript coverage
- Sequence on Illumina platform (minimum 30M reads per sample)
- Map reads to reference genome using STAR or HiSAT2
- Quantify expression as counts per million (CPM) for downstream analysis
Proteomic Sample Processing:
- Extract proteins using modified Folch method (chloroform:methanol)
- Reduce proteins with DTT (5 mM, 30 min at 60°C) and alkylate with iodoacetamide (400 mM, 1 hr at 37°C)
- Digest with trypsin overnight at 37°C (1:50 enzyme:protein ratio)
- Desalt peptides using C18 SPE cartridges
- Analyze via LC-MS/MS using label-free quantification or TMT multiplexing
- Identify and quantify proteins using MaxQuant software with log2 transformation and median normalization
Data Integration and Pathway Analysis
Directional P-value Merging (DPM) Implementation:
- Process upstream omics datasets into matrices of gene P-values and directional changes
- Define constraints vector (CV) based on biological hypotheses (e.g., [+1, +1] for coherent mRNA-protein pairs)
- Compute directionally weighted score for each gene: XDPM = -2(-|Σ{i=1}^j ln(Pi)oiei| + Σ{i=j+1}^k ln(P_i))
- Calculate merged P-value using cumulative χ2 distribution adjusted for covariation
- Perform pathway enrichment analysis using ranked hypergeometric algorithm
- Visualize results as enrichment maps highlighting directional evidence [73]
Validation by Parallel Reaction Monitoring (PRM):
- Select candidate proteins based on integrated analysis
- Synthesize stable isotope-labeled standard peptides
- Digested sample peptides are spiked with heavy labeled standards
- Analyze using nanoLC-MS/MS with targeted MS2 acquisition
- Quantify by comparing light/heavy peptide peak areas
- Validate protein abundance changes across conditions [75]
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Research Reagent Solutions for ZEB2 Multi-Omics Studies
| Reagent Category | Specific Products | Application in ZEB2 Research | Technical Considerations |
|---|---|---|---|
| Cell Sorting Antibodies | CD45-, CD326-, CD31+, CD144+ | Isolation of embryonic cell populations | Validate cross-reactivity for model species |
| CRISPR Tools | Cas9, gRNAs targeting ZEB2 | Genetic perturbation in model systems | Optimize delivery efficiency and off-target control |
| Mass Spectrometry Standards | TMT 10-plex, iRT peptides | Multiplexed proteomic quantification | Consider dynamic range and labeling efficiency |
| Library Prep Kits | SMARTer Seq v4, NEBNext Ultra | Transcriptomic library construction | Assess compatibility with low-input samples |
| Validation Antibodies | Anti-ZEB2, Anti-ACTA2, Anti-FOXF1 | Immunofluorescence validation | Confirm specificity for embryonic tissues |
| Bioinformatics Tools | ActivePathways R package, MaxQuant | Multi-omics data integration | Ensure version compatibility and computational resources |
The integration of proteomic and transcriptomic data provides a powerful framework for validating molecular pathways in complex developmental processes such as somitogenesis. The case of ZEB2 illustrates how multi-omics approaches can move beyond correlation to establish causal relationships between transcription factor perturbation and phenotypic outcomes. The methodologies and experimental designs presented here offer researchers a comprehensive toolkit for applying these approaches to their own systems of interest, with particular relevance for studying developmental transcription factors where protein-level validation of transcriptomic findings remains essential. As single-cell multi-omics technologies continue to advance, these integrated approaches will become increasingly central to unraveling the complex regulatory networks governing embryogenesis and tissue patterning.
The zinc finger E-box-binding homeobox 2 (ZEB2) protein, initially discovered in 1999 as a SMAD-interacting transcription factor, has emerged as a critical molecular switch governing cell fate decisions from embryogenesis to adult tissue homeostasis [1] [8]. Also known as SIP1 (SMAD Interacting Protein 1), ZEB2 functions as a transcriptional regulator that coordinates complex gene expression programs through its DNA-binding capabilities and protein-protein interactions [1]. While heterozygous mutations in ZEB2 cause Mowat-Wilson syndrome (MOWS)—a rare neurodevelopmental disorder—research has progressively unveiled ZEB2's fundamental roles in pathological contexts, particularly cancer and immune regulation [1] [71] [40]. This article synthesizes current understanding of ZEB2's dualistic functions, comparing its roles in developmental disorders and cancer therapeutics, with specific emphasis on experimental methodologies enabling these discoveries.
Table 1: ZEB2 Functional Roles Across Biological Contexts
| Biological Context | Primary Role of ZEB2 | Downstream Consequences | Therapeutic Implications |
|---|---|---|---|
| Neural Development | Timing of neuro-/gliogenesis [1] | Regulates cortical layer formation; cell non-autonomous control of paracrine signaling [1] | Explains neurodevelopmental defects in Mowat-Wilson syndrome |
| Stem Cell Differentiation | Exit from pluripotency [5] | Prevents stalling in epiblast-like state; enables neural and mesendodermal commitment [5] | Informs regenerative medicine strategies |
| Cancer Immunity | Master regulator of TAM programming [71] | Promotes pro-tumor TAM functions; suppresses antigen presentation [71] | Potential target for myeloid-based immunotherapies |
| CD8+ T-cell Differentiation | Drives cytotoxic effector trajectory [40] | Enhances Teff cell differentiation; improves anti-tumor response [40] | Could enhance efficacy of T-cell therapies |
ZEB2 in Development: Insights from Mowat-Wilson Syndrome
Developmental Pathogenesis and Genetic Landscape
Mowat-Wilson syndrome (MOWS; OMIM #235730) represents the most definitive clinical manifestation of ZEB2 haploinsufficiency, occurring in approximately 1:50,000 to 1:70,000 live births [1] [8]. This monogenic disorder exhibits multi-system involvement with variable penetrance, including severe intellectual disability, distinctive facial dimorphism, Hirschsprung disease (occurring in ~50% of patients), seizures, and structural anomalies affecting the corpus callosum, hippocampus, heart, kidneys, and eyes [1]. Genotype-phenotype analyses of approximately 350 documented patients reveal that severe cases typically involve complete gene deletions (19%), nonsense mutations (34%), or frameshift mutations (40%) that trigger nonsense-mediated mRNA decay, resulting in functional null alleles [1] [8]. Notably, rare missense mutations (1.5% of cases) and C-terminal truncations correlate with milder phenotypes, suggesting preserved functionality of specific protein domains [1].
ZEB2 Functions in Neural and Neural Crest Development
Conditional knockout mouse models have been instrumental in deciphering ZEB2's cell-autonomous and non-autonomous functions during embryogenesis. In the developing forebrain, ZEB2 represents the first transcription factor demonstrated to control the timing of neurogenic-to-gliogenic transition in a cell non-autonomous manner [1]. Specifically, Zeb2 deletion in cortical neurons disrupts the neuron-to-progenitor feedback signaling that normally maintains radial glial cells in a proliferative state, resulting in premature differentiation and disrupted cortical lamination [1]. Furthermore, Zeb2 deficiency in neural crest cell lineages explains the enteric nervous system defects (Hirschsprung disease) and craniofacial abnormalities characteristic of MOWS, through disrupted migration, differentiation, and survival of these multipotent progenitors [1] [8].
ZEB2 in Cancer and Immunity: Therapeutic Targeting Opportunities
ZEB2 as a Master Regulator of Tumor-Associated Macrophages
Recent single-cell RNA sequencing and CRISPR-based functional studies have identified ZEB2 as a molecular switch controlling tumor-associated macrophage (TAM) polarization states [71]. In a comprehensive analysis of 832,107 single cells from 123 patients with breast, lung, and colon cancers, ZEB2 emerged as the predominant regulator of pro-tumor TAM functions by inducing immunosuppressive gene programs while simultaneously repressing antigen presentation and type-I interferon inflammatory responses [71]. Mechanistically, ZEB2 perturbation in TAMs resulted in functional reprogramming toward anti-tumor phenotypes, enhanced T-cell-mediated immunity, and robust tumor control in experimental models, presenting ZEB2 inhibition as a promising strategy for myeloid-directed cancer immunotherapy [71].
ZEB2 in CD8+ T-cell Differentiation and Anti-Tumor Immunity
Paradoxically, while ZEB2 promotes pro-tumor functions in macrophages, it exhibits anti-tumor activity in the context of CD8+ T-cell biology [40]. MetaVIPER-based analysis of single-cell RNA sequencing data from treatment-naïve NSCLC patients revealed that ZEB2 drives CD8+ T-cell differentiation along the cytotoxic effector (Teff) trajectory, as opposed to exhausted (Texh) or dysfunctional (TLdys) states [40]. The T-bet/ZEB2 axis was shown to stimulate lung tumor-reactive Teff cell differentiation in vitro and mediate the therapeutic efficacy of IL-2/IL-12 combination immunotherapy in murine lung cancer models [40]. This context-dependent duality underscores the complexity of ZEB2 targeting in cancer therapy and highlights the need for cell-type-specific intervention strategies.
Table 2: Experimental Models for ZEB2 Functional Analysis
| Experimental System | Key Findings | Methodological Approaches |
|---|---|---|
| Zeb2-V5 epitope-tagged mESCs [10] | Mapped 2432 Zeb2 DNA-binding sites in neuroprogenitor cells; identified Zeb2 autoregulation [10] | Chromatin immunoprecipitation sequencing (ChIP-seq); neural differentiation protocols |
| Zeb2 knockout mESCs [5] | Zeb2 essential for exit from epiblast state; links pluripotency network with DNA methylation [5] | RNA-sequencing; reduced representation bisulfite sequencing (RRBS); embryoid body differentiation |
| R26_Zeb2 overexpression mESCs [32] | Zeb2 enhances skeletal muscle differentiation; upregulates myogenic markers and myomiRs [32] | Myogenic induction with MyoD transfection; single-cell RNA-sequencing; immunofluorescence |
| Conditional KO mice [1] | Cell-type specific functions in forebrain development, neural crest migration, and organogenesis [1] | Tissue-specific Cre recombinase systems; transcriptional profiling; histological analysis |
Experimental Approaches: Methodologies for ZEB2 Functional Analysis
Mapping ZEB2 Genomic Binding Landscapes
Chromatin immunoprecipitation followed by sequencing (ChIP-seq) has been pivotal for identifying direct ZEB2 target genes. A recent study established a Zeb2-V5 epitope-tagged mouse embryonic stem cell (mESC) model, enabling mapping of 2432 Zeb2 DNA-binding sites during neural differentiation into neuroprogenitor cells (NPCs) without antibody-related limitations [10]. Critical findings included the identification of a promoter-proximal Zeb2 binding site that facilitates positive autoregulation, essential for appropriate Zeb2-dependent neural differentiation [10]. The experimental workflow comprised:
- Gene Editing: Insertion of Flag-V5 epitope tag in-frame before the stop codon of one Zeb2 allele in mESCs using CRISPR-Cas9.
- Neural Differentiation: Conversion of Zeb2-V5 mESCs into neuroprogenitor cells via embryoid body formation in DMEM with 10% FBS, followed by retinoic acid induction (5µM) from day 4, and plating on poly-DL-ornithine/laminin-coated surfaces at day 8.
- Chromatin Immunoprecipitation: Crosslinking, nuclear extraction, chromatin shearing, and immunoprecipitation with V5 antibody at differentiation day 8-10.
- Sequencing and Analysis: Library preparation, sequencing, peak calling, and integration with transcriptome data from Zeb2 perturbations [10].
Functional Perturbation in Stem Cell Differentiation
Zeb2 knockout mESC models have revealed its critical role in exit from naïve pluripotency. Zeb2-deficient ESCs fail to silence the pluripotency network (including Oct4 and Nanog), stall at an epiblast-like state, and exhibit impaired neural and mesendodermal differentiation capacity [5]. The protocol for this analysis involves:
- ESC Culture: Maintenance in 2i/LIF medium (1µM PD0325901, 3µM CHIR99021, 1000U/mL LIF) to preserve ground state pluripotency.
- Neural Differentiation: Formation of embryoid bodies in KO-DMEM with 15% FBS for 4 days, then transition to N2B27 medium with 500nM retinoic acid (days 4-8), followed by N2B27 alone (days 8-15).
- Molecular Phenotyping: Temporal RNA-sequencing and reduced representation bisulfite sequencing (RRBS) to assess transcriptome and methylome dynamics [5].
This approach demonstrated that Zeb2 KO cells correctly acquire DNA methylation marks early in differentiation but fail to maintain them, associated with elevated Tet1 levels. Subsequent Tet1 knockdown partially rescued differentiation impairment, establishing a ZEB2-TET1 regulatory axis in cell fate commitment [5].
Diagram 1: Dualistic roles of ZEB2 in development and cancer. ZEB2 regulates normal neural and neural crest development, with deficiency causing Mowat-Wilson syndrome. In cancer, ZEB2 exhibits context-dependent functions, promoting pro-tumor TAM programming while enhancing anti-tumor CD8+ T-cell differentiation.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for ZEB2 Investigation
| Reagent / Tool | Function / Application | Example Use Case |
|---|---|---|
| Zeb2-floxed mice (Zeb2flox/flox) [5] [1] | Enables cell-type specific knockout via Cre recombinase | Conditional deletion in neural crest, forebrain, hematopoietic lineages |
| R26-Zeb2 cDNA mice [32] | Conditional overexpression from Rosa26 safe harbor locus | Gain-of-function studies in myogenesis, neurogenesis |
| Zeb2-V5 epitope-tagged mESCs [10] | Enables ChIP-seq with standard V5 antibody | Genome-wide mapping of Zeb2 binding sites in neural progenitors |
| 2i/LIF medium [5] | Maintains naïve pluripotent state in mESCs | Ground state ESC culture before differentiation assays |
| Anti-ZEB2 antibodies | Detection, localization, and immunoprecipitation | Western blot, immunofluorescence; limited by cross-reactivity with ZEB1 |
| ChIP-grade V5 antibody [10] | Chromatin immunoprecipitation of tagged Zeb2 | Identification of direct Zeb2 target genes |
Comparative Analysis: ZEB2 in Development versus Cancer
The functional duality of ZEB2 across developmental and cancer contexts presents both challenges and opportunities for therapeutic targeting. In development, ZEB2 acts as a differentiation-promoting factor, driving lineage commitment through coordinated repression of pluripotency networks and modulation of key signaling pathways (TGF-β/BMP, Wnt, Notch) [5] [1]. By contrast, in cancer, ZEB2 frequently exhibits oncogenic properties, particularly through its regulation of epithelial-mesenchymal transition (EMT) and immune cell functions [71] [76] [77]. However, this paradigm is complicated by ZEB2's anti-tumor activity in CD8+ T-cells, where it promotes desirable cytotoxic effector differentiation [40].
This contextual duality necessitates cell-type-specific therapeutic approaches. While ZEB2 inhibition may reprogram pro-tumor macrophages and counteract EMT in carcinoma cells, ZEB2 enhancement could potentially improve T-cell-mediated anti-tumor immunity [71] [40]. The successful targeting of ZEB2 will require sophisticated delivery systems that account for these opposing functions across different cell populations within the tumor microenvironment.
Diagram 2: ZEB2 regulatory networks in development versus cancer. ZEB2 represses pluripotency factors while activating differentiation programs during development. In cancer, ZEB2 exhibits context-dependent regulation, activating EMT and suppressing anti-tumor immunity in TAMs while promoting desirable Teff differentiation in CD8+ T-cells.
The investigation of ZEB2 has traversed from clinical genetics of a rare developmental disorder to cutting-edge cancer immunotherapy research, revealing unexpected complexities in its functional repertoire. The experimental data compiled herein demonstrates that ZEB2 operates as a contextual master regulator rather than a uniformly oncogenic or developmental factor. Future therapeutic strategies must account for this duality, particularly as immunotherapeutic approaches advance. The methodologies and reagents summarized provide a roadmap for continued investigation of ZEB2's multifaceted roles and the development of targeted interventions that can selectively modulate its activity in specific cell types. As single-cell technologies and CRISPR-based screening methods continue to evolve, our understanding of ZEB2's therapeutic implications will undoubtedly expand, potentially offering new avenues for treating both developmental disorders and malignant diseases.
Conclusion
The critical role of ZEB2 in mouse and human somitogenesis is now firmly established through integrated approaches combining advanced model systems like gastruloids with multi-omics technologies. Recent research confirms ZEB2 as a master regulator steering the complex process of embryonic segmentation, with its dysfunction having profound implications for human developmental disorders such as Mowat-Wilson Syndrome. The methodological advances discussed, particularly degron-based perturbations combined with single-cell analyses, provide powerful tools for continued dissection of ZEB2's mechanisms. Future research should focus on elucidating the complete ZEB2-dependent regulatory network in somitogenesis, developing more refined human model systems, and exploring the therapeutic potential of modulating ZEB2 activity in both developmental disorders and cancers where its pathways are co-opted. This knowledge creates new avenues for diagnostic and therapeutic innovation in developmental biology and oncology.
References
- 1. ZEB2, the Mowat-Wilson Syndrome Transcription Factor [https://pmc.ncbi.nlm.nih.gov/articles/PMC8304685/]
- 2. Zeb2: A multifunctional regulator of nervous system ... [https://pubmed.ncbi.nlm.nih.gov/26193487/]
- 3. Zeb2 Regulates Myogenic Differentiation in Pluripotent ... [https://www.mdpi.com/1422-0067/21/7/2525]
- 4. ZEB2 [https://en.wikipedia.org/wiki/ZEB2]
- 5. Zeb2 Regulates Cell Fate at the Exit from Epiblast State in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5396376/]
- 6. GSE229025 - GEO Accession viewer - NIH [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE229025]
- 7. ZEB2 | Cancer Genetics Web [http://www.cancerindex.org/geneweb/ZEB2.htm]
- 8. ZEB2, the Mowat-Wilson Syndrome Transcription Factor [https://www.mdpi.com/2073-4425/12/7/1037]
- 9. Mowat-Wilson syndrome factor ZEB2 controls early formation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10679662/]
- 10. Zeb2 DNA-Binding Sites in Neuroprogenitor Cells Reveal ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10048071/]
- 11. Deciphering lineage specification during early ... [https://pubmed.ncbi.nlm.nih.gov/38754429/]
- 12. Deciphering lineage specification during early ... [https://www.sciencedirect.com/science/article/pii/S1934590924001449]
- 13. Generation of a ZEB2 deficient human iPSC line ... [https://www.sciencedirect.com/science/article/pii/S1873506124002198]
- 14. The EMT regulator Zeb2/Sip1 is essential for murine ... [https://pubmed.ncbi.nlm.nih.gov/21355089/]
- 15. Pluripotency circuit members mediate germ layer fate ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5603300/]
- 16. The exit from naive pluripotency: a platform for the study of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12493175/]
- 17. In vivo Human Somitogenesis Guides Somite Development ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5327729/]
- 18. A transcriptional and regulatory map of mouse somite maturation [https://pmc.ncbi.nlm.nih.gov/articles/PMC10563765/]
- 19. ZEB2 Gene Pathogenic Variants Across Protein-Coding ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11818587/]
- 20. Identification of the DNA methylation signature of Mowat ... [https://www.nature.com/articles/s41431-024-01548-4]
- 21. Mowat–Wilson Syndrome: Case Report and Review of ... [https://www.mdpi.com/1422-0067/25/5/2838]
- 22. A single-cell time-lapse of mouse prenatal development ... [https://www.nature.com/articles/s41586-024-07069-w]
- 23. Alteration of Hair Melanin in Patients With Mowat-Wilson ... [https://pubmed.ncbi.nlm.nih.gov/40415661/]
- 24. Gastruloids — a minimalistic model to study complex ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10754324/]
- 25. GSE123187 - GEO Accession viewer - NIH [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE123187]
- 26. A spatiotemporal atlas of mouse gastrulation and early ... [https://www.sciencedirect.com/science/article/pii/S2211124725008186]
- 27. An Optimized Protocol for the Extended Culture of ... [https://pubmed.ncbi.nlm.nih.gov/40973928/]
- 28. Large-scale deep multi-layer analysis of Alzheimer's ... [https://www.nature.com/articles/s41593-021-00999-y]
- 29. Proteomics-based multi-omics identifies the roadmap of ... [https://www.sciencedirect.com/science/article/abs/pii/S1534580725005362]
- 30. Multi-layered proteomic analyses decode compositional ... [https://www.nature.com/articles/s41467-020-17387-y]
- 31. Integrated multi-dimensional comparison of proteomic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12483590/]
- 32. Zeb2 Regulates Myogenic Differentiation in Pluripotent Stem ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7177401/]
- 33. Comparative analysis and directed protein evolution yield an ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11601833/]
- 34. Systematic comparison and base-editing-mediated ... [https://www.nature.com/articles/s41467-025-61848-1]
- 35. Functional characterization of the ZEB2 regulatory landscape [https://pmc.ncbi.nlm.nih.gov/articles/PMC6466108/]
- 36. Unveiling Lineage Specification in Mouse Gastruloids [https://www.sonnenlab.org/2024/news-from-the-sonnen-lab/new-publication-unveiling-lineage-specification-in-mouse-gastruloids/]
- 37. Comparison of visualization tools for single-cell RNAseq data [https://pmc.ncbi.nlm.nih.gov/articles/PMC7391988/]
- 38. Comparison of Single Cell Data Visualization Tools | Part 1 [https://www.elucidata.io/blog/comparison-of-single-cell-data-visualization-tools]
- 39. Best single-cell RNA-sequencing data analysis tools in 2024 [https://www.biomage.net/blog/best-single-cell-rna-sequencing-data-analysis-tools-in-2024]
- 40. The transcription factor ZEB2 mediates the antitumor ... [https://www.nature.com/articles/s41419-025-08112-y]
- 41. The transcription factor ZEB2 mediates the antitumor ... [https://pubmed.ncbi.nlm.nih.gov/41203628/]
- 42. The transcription factor ZEB2 drives formation of age- ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7616037/]
- 43. novel E-box interactors revealed by proximity labeling - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11610934/]
- 44. High-density P300 enhancers control cell state transitions - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4636788/]
- 45. STATs Shape the Active Enhancer Landscape of T Cell ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3509201/]
- 46. Enhancer–promoter interactions become more instructive ... [https://www.nature.com/articles/s41588-024-01678-x]
- 47. ZEB2: a multifunctional regulator of neural injury repair [https://www.sciencedirect.com/science/article/abs/pii/S1567576925012561]
- 48. The EMT transcription factor Zeb2 controls adult murine ... [https://www.sciencedirect.com/science/article/pii/S0006497120337551]
- 49. Multiple Gene Transfer and All-In-One Conditional ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8995969/]
- 50. Rapid generation of conditional knockout mice using the ... [https://molecularbrain.biomedcentral.com/articles/10.1186/s13041-021-00859-7]
- 51. A Convenient Cas9-based Conditional Knockout Strategy ... [https://www.nature.com/articles/s41598-017-00654-2]
- 52. A concise guide to choosing suitable gene expression ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10643540/]
- 53. Effective Gene Expression Prediction and Optimization ... [https://pubmed.ncbi.nlm.nih.gov/39783932/]
- 54. Rapid and site-specific deep phosphoproteome profiling by ... [https://www.nature.com/articles/s41467-020-14609-1]
- 55. Optimizing differential expression analysis for proteomics ... [https://www.nature.com/articles/s41467-024-47899-w]
- 56. Top 6 Proteomics Databases and How to Access Them [https://bigomics.ch/blog/guide-to-top-proteomics-databases-and-how-to-access-them/]
- 57. Retinoic acid induces human gastruloids with posterior ... [https://www.nature.com/articles/s41556-024-01487-8]
- 58. Stem cell culture conditions affect in vitro differentiation ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0317309]
- 59. Towards an integrated view and understanding of ... [https://www.sciencedirect.com/science/article/pii/S266729012500035X]
- 60. Development of large-scale gastruloid array to identify ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12158465/]
- 61. A quantitative pipeline for whole-mount deep imaging and ... [https://elifesciences.org/reviewed-preprints/107154]
- 62. Stem cell-based modeling and single-cell multiomics ... [https://www.sciencedirect.com/science/article/pii/S2211124723002875]
- 63. Zeb2 MGI Mouse Gene Detail - MGI:1344407 - zinc finger E ... [https://www.informatics.jax.org/marker/MGI:1344407]
- 64. Speed of life: tuning the ticktock of the segmentation clock [https://pmc.ncbi.nlm.nih.gov/articles/PMC12404190/]
- 65. ZEB2 gene: MedlinePlus Genetics [https://medlineplus.gov/genetics/gene/zeb2/]
- 66. From Dynamic Expression Patterns to Boundary Formation in ... [https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1002586]
- 67. Arrayed single-gene perturbations identify drivers of ... [https://elifesciences.org/reviewed-preprints/108224]
- 68. Arrayed single-gene perturbations identify drivers of human ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12330715/]
- 69. Dualistic role of ZEB1 and ZEB2 in tumor progression - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11927373/]
- 70. The Role of ZEB2 Expression in Pediatric and Adult ... [https://ar.iiarjournals.org/content/41/1/175]
- 71. ZEB2 is a master switch controlling the tumor-associated ... [https://www.sciencedirect.com/science/article/abs/pii/S1535610825001229]
- 72. Comparative benchmarking of single-cell clustering ... [https://genomebiology.biomedcentral.com/articles/10.1186/s13059-025-03719-y]
- 73. Directional integration and pathway enrichment analysis ... [https://www.nature.com/articles/s41467-024-49986-4]
- 74. Integration of transcriptomic and proteomic data identifies ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6766718/]
- 75. Integration Analysis of Transcriptome and Proteome ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2022.836913/full]
- 76. The molecular mechanisms and therapeutic strategies of EMT ... [https://jhoonline.biomedcentral.com/articles/10.1186/s13045-022-01347-8]
- 77. Dualistic role of ZEB1 and ZEB2 in tumor progression [https://biologydirect.biomedcentral.com/articles/10.1186/s13062-025-00604-3]