Mastering DNA Repair Pathways in Zebrafish: A Comprehensive Guide to NHEJ and HDR for Precision Genome Editing
This comprehensive review explores the mechanisms and applications of double-strand break repair pathways—Non-Homologous End Joining (NHEJ) and Homology-Directed Repair (HDR)—in the zebrafish model.
Mastering DNA Repair Pathways in Zebrafish: A Comprehensive Guide to NHEJ and HDR for Precision Genome Editing
Abstract
This comprehensive review explores the mechanisms and applications of double-strand break repair pathways—Non-Homologous End Joining (NHEJ) and Homology-Directed Repair (HDR)—in the zebrafish model. Tailored for researchers and drug development professionals, we examine the fundamental biology distinguishing these pathways, practical methodologies for implementing CRISPR/Cas9-mediated editing, strategies for optimizing repair outcomes, and rigorous validation approaches. The article synthesizes current best practices from recent studies, including chemical enhancement of HDR efficiency and standardization protocols, providing an essential resource for advancing precision genetic modeling and therapeutic discovery in biomedical research.
The Cellular Machinery: Understanding NHEJ and HDR Mechanisms in Zebrafish
Fundamental Principles of Double-Strand Break Repair
DNA double-strand breaks (DSBs) represent one of the most critical forms of DNA damage, posing an immediate threat to genomic integrity through potential chromosome rearrangements and disruption of gene function [1]. The fundamental principles governing DSB repair are essential knowledge for researchers utilizing model organisms like zebrafish in biomedical research and drug development. In mammalian cells, two major pathways predominate in DSB repair: non-homologous end joining (NHEJ) and homologous recombination (HR), with homology-directed repair (HDR) representing a precise subset of homologous recombination [1] [2]. The cellular choice between these pathways is not random but is tightly regulated by cell cycle phase, chromatin context, and the specific nature of the DNA break itself [1]. Understanding these principles provides the foundation for developing precise genome-editing tools and therapeutic strategies aimed at manipulating DNA repair for research and clinical applications.
Core DSB Repair Pathways
Non-Homologous End Joining (NHEJ)
Classical Non-Homologous End Joining (cNHEJ) operates as a rapid, high-capacity pathway that functions throughout the cell cycle, making it the default DSB repair mechanism in mammalian cells [1] [3]. This pathway initiates with the binding of the Ku70-Ku80 heterodimer to DSB ends, which nucleates the recruitment of other essential cNHEJ factors including DNA-dependent protein kinase catalytic subunit (DNA-PKcs), DNA ligase IV (LIG4), and associated scaffolding factors XRCC4, XRCC4-like factor (XLF), and paralogue of XRCC4 and XLF (PAXX) [1] [3]. The cNHEJ mechanism involves a two-stage synapsis process where Ku70-Ku80 and DNA-PKcs first establish long-range synapsis, followed by close end alignment requiring XLF, non-catalytic functions of XRCC4-LIG4, and DNA-PKcs kinase activity [1]. End processing by nucleases like Artemis and specialized DNA polymerases ensures compatibility of ligated ends [1]. A significant characteristic of cNHEJ is its ability to join DNA ends with minimal reference to DNA sequence, though it can accommodate very limited base-pairing (up to ~4 base pairs of "microhomology") between processed DNA ends [1]. While this makes cNHEJ efficient throughout the cell cycle, it also renders it potentially error-prone, often resulting in small insertions or deletions (indels) at the repair junction [3].
Homologous Recombination and Homology-Directed Repair
Homologous Recombination (HR) represents a more precise DSB repair pathway that is largely restricted to the S and G2 phases of the cell cycle when an undamaged sister chromatid is available as a repair template [1] [2] [3]. The critical step committing a DSB to HR is 5'-to-3' resection of DNA ends to form 3' single-stranded DNA (ssDNA) overhangs [1] [3]. This process initiates with the MRE11-RAD50-NBS1 (MRN) complex, which recruits CtBP-interacting protein (CtIP) to begin resection [1]. Subsequently, Exonuclease 1 (EXO1) and the BLM-DNA2 complex perform long-range resection, generating extensive 3' ssDNA tails [1] [3]. The resulting ssDNA is rapidly bound by replication protein A (RPA), which must later be replaced by the RAD51 recombinase with assistance from recombination mediators including BRCA1, BRCA2, and PALB2 [1] [3]. The RAD51-nucleoprotein filament then mediates homology search and strand invasion into the homologous DNA template, generating a displacement loop (D-loop) [1]. Homology-Directed Repair (HDR) specifically refers to the process where this homologous recombination machinery copies information from a provided DNA template to repair the break precisely [3]. Several subpathways exist beyond D-loop formation, including double-strand break repair (DSBR), synthesis-dependent strand annealing (SDSA), and break-induced replication (BIR), with SDSA being the most preferred in somatic cells as it yields non-crossover products [2].
Table 1: Key Characteristics of Major DSB Repair Pathways
| Feature | Non-Homologous End Joining (NHEJ) | Homologous Recombination (HR) |
|---|---|---|
| Core Function | Direct ligation of broken ends | Templated repair using homologous sequence |
| Cell Cycle Phase | Throughout cell cycle | Primarily S and G2 phases |
| Template Required | No | Yes (sister chromatid or donor DNA) |
| Key Initiating Factor | Ku70-Ku80 heterodimer | MRN complex with CtIP |
| Resection Dependent | No | Yes (5'-to-3' resection) |
| Fidelity | Error-prone (small indels) | High-fidelity |
| Primary Regulatory Kinase | DNA-PKcs | ATM |
| Essential Mediators | DNA-PKcs, XRCC4-LIG4 complex, Artemis | BRCA1, BRCA2, RAD51, PALB2 |
Pathway Choice and Regulatory Mechanisms
The decision between NHEJ and HR pathways represents a critical juncture in DSB repair with significant implications for genomic integrity. Mammalian cells preferentially employ NHEJ over HDR through several biological mechanisms: NHEJ is active throughout the cell cycle except mitosis, while HDR is restricted to S and G2 phases; NHEJ operates more rapidly than HDR; and NHEJ actively represses HDR through a series of mechanisms [3]. A key determinant of pathway choice is the initiation of DNA end resection, which commits breaks to the HR pathway while preventing NHEJ [1] [3]. The MRN complex serves as a central player in this decision point, functioning as a scaffold for ATM activation while also initiating resection in conjunction with CtIP [1]. BRCA1 promotes end resection and later stages of HR, working in complex with its heterodimeric partner BARD1 and interacting with CtIP and MRN [1]. The Ku70-Ku80 complex not only initiates NHEJ but also protects DNA ends from resection, thereby antagonizing HR [1]. Additionally, 53BP1 promotes NHEJ by protecting DNA ends from resection and counteracting BRCA1 function [3]. Recent research has revealed that the regulatory "rules" governing stalled replication fork repair differ substantially from those operating at conventional two-ended DSBs, suggesting contextual modulation of pathway choice [1].
Experimental Applications in Zebrafish Research
Zebrafish (Danio rerio) have emerged as a pivotal model organism for DSB repair research and genome engineering applications due to their genetic similarity to humans, transparent embryos, rapid development, and high fecundity [4] [5]. The application of DSB repair principles in zebrafish research has enabled sophisticated genome editing approaches that leverage both NHEJ and HDR pathways.
CRISPR-Cas9 and DSB Repair-Mediated Editing
Traditional CRISPR-Cas9 editing in zebrafish introduces targeted DSBs that are subsequently repaired by endogenous cellular machinery, primarily resulting in NHEJ-mediated indels that can disrupt gene function [4]. While HDR-mediated knock-in approaches using exogenous donor DNA templates enable precise genome modifications, this process occurs less efficiently than NHEJ in zebrafish, mirroring the challenge observed in mammalian systems [6]. To address this limitation, researchers have developed strategies to modulate repair pathway choice, including cell cycle synchronization and inhibition of NHEJ factors to favor HDR outcomes [3].
Advanced Genome Editing Technologies
Base editing technology has revolutionized precise genome engineering in zebrafish by enabling direct chemical conversion of one DNA base into another without inducing DSBs, thereby bypassing the competitive repair pathway choice altogether [4] [5]. Cytosine base editors (CBEs) facilitate C•G to T•A conversions through fusion of catalytically impaired Cas9 with cytidine deaminase enzymes, while adenine base editors (ABEs) promote A•T to G•C conversions using engineered adenine deaminases [4] [5]. The development of zebrafish-codon-optimized editors like AncBE4max has enhanced editing efficiency approximately threefold compared to earlier systems [4]. More recently, prime editing systems have been employed in zebrafish, utilizing Cas9-reverse transcriptase fusion proteins programmed with prime editing guide RNAs (pegRNAs) to directly copy edited sequences into target genomic loci without requiring DSBs [6]. Comparative studies in zebrafish demonstrate that PE2 (nickase-based) editors are more effective for single-nucleotide substitutions, while PEn (nuclease-based) editors show superior efficiency for inserting short DNA fragments up to 30 bp [6].
Table 2: Genome Editing Approaches Leveraging DSB Repair Principles in Zebrafish
| Editing Technology | Mechanism | Key Components | Efficiency in Zebrafish | Primary Applications |
|---|---|---|---|---|
| CRISPR-Cas9 (NHEJ) | DSB induction with error-prone repair | Cas9 nuclease, sgRNA | High (varies by target) | Gene knockout, random mutagenesis |
| HDR-Mediated Knock-in | DSB induction with templated repair | Cas9 nuclease, sgRNA, donor DNA template | Low (<10% typically) | Precise sequence insertion, gene correction |
| Cytosine Base Editing (CBE) | Direct base conversion without DSB | dCas9 or nCas9, cytidine deaminase, UGI | 9-28% (BE3); ~90% (AncBE4max) [4] | Point mutation introduction, disease modeling |
| Adenine Base Editing (ABE) | Direct base conversion without DSB | dCas9 or nCas9, adenine deaminase | Similar range to CBE | Point mutation introduction, disease modeling |
| Prime Editing (PE2) | Reverse transcription without DSB | Cas9 nickase, reverse transcriptase, pegRNA | 8.4% for nucleotide substitution [6] | Single-nucleotide variants, small edits |
| Prime Editing (PEn) | DSB induction with homology-assisted repair | Cas9 nuclease, reverse transcriptase, pegRNA | 4.4% for substitution; higher for insertions [6] | Short DNA fragment insertion (up to 30 bp) |
HDR Enhancement Strategies and Associated Risks
Recent research has explored pharmacological inhibition of NHEJ factors to enhance HDR efficiency in genome editing. Inhibition of DNA-PKcs using compounds like AZD7648 has shown potential to increase HDR rates by redirecting repair toward homologous recombination [7]. However, a 2025 study revealed that despite increasing apparent HDR efficiency, AZD7648 treatment during genome editing causes frequent kilobase-scale and megabase-scale deletions, chromosome arm loss, and translocations that evade detection by standard short-read sequencing methods [7]. In RPE-1 p53-null cells, AZD7648 increased kilobase-scale deletion frequency by 2.0-fold to 35.7-fold depending on the locus, reaching 43.3% of reads at the GAPDH target site [7]. Similarly, in primary human CD34+ hematopoietic stem and progenitor cells (HSPCs), AZD7648 increased large deletion frequency by 1.2-fold to 4.3-fold across three target loci [7]. These findings highlight the critical importance of comprehensive genotyping when deploying HDR-enhancing strategies and suggest that AZD7648 converts small-scale NHEJ outcomes into larger genetic alterations [7].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagent Solutions for DSB Repair Studies in Zebrafish
| Reagent Category | Specific Examples | Function in DSB Repair Research |
|---|---|---|
| CRISPR-Cas9 Systems | Wild-type SpCas9, HiFi Cas9 | Induces targeted DSBs to engage endogenous repair pathways |
| Base Editors | BE3, BE4max, AncBE4max, Target-AID, ABE | Enables precise nucleotide conversion without DSB induction |
| Prime Editors | PE2, PEn | Programs precise edits without donor templates via reverse transcription |
| HDR Enhancement Reagents | AZD7648 (DNA-PKcs inhibitor) | Shifts repair balance toward HDR by inhibiting NHEJ [7] |
| Repair Pathway Reporters | FIRE (Fluorescent Insertional Repair) reporter | Quantifies HDR vs. NHEJ efficiency in live cells [7] |
| Zebrafish-Specific Delivery Tools | Codon-optimized editors, hei-tag nuclear localization | Enhances efficiency in zebrafish models through improved nuclear import [4] |
| Analytical Tools | Long-read sequencing (Oxford Nanopore), ddPCR, scRNA-seq | Detects large-scale structural variations from editing [7] |
Detailed Experimental Protocol: Prime Editing in Zebrafish
The following protocol outlines the methodology for precise genome editing in zebrafish using prime editing technology, based on established research [6]:
Reagent Preparation
- Prime Editor Selection: Choose between nickase-based PE2 for single-nucleotide substitutions or nuclease-based PEn for short DNA fragment insertions (up to 30 bp).
- Guide RNA Design: Design prime editing guide RNAs (pegRNAs) containing:
- Target-specific spacer sequence (typically 20 nt)
- Reverse transcriptase (RT) template encoding desired edit
- Primer binding site (PBS, ~13 nt)
- Homology arm extension for PEn/pegRNA experiments (~13 nt)
- Optional Refolding Procedure: To prevent misfolding between spacer and PBS/RT template sequences, implement a refolding protocol involving denaturation at 65°C for 5 minutes followed by slow cooling to room temperature.
Microinjection Setup
- Sample Preparation: Combine Prime Editor mRNA (100-200 ng/μL) with synthesized pegRNA (50-100 ng/μL) in nuclease-free injection buffer.
- Embryo Collection: Harvest zebrafish embryos within 30 minutes post-fertilization at the one-cell stage.
- Microinjection: Inject 1-2 nL of the ribonucleoprotein (RNP) complex into the cell cytoplasm or yolk near the cell interface.
- Incubation Conditions: Maintain injected embryos at 32°C for optimal prime editor activity [6].
Genotype Analysis
- DNA Extraction: At 96 hours post-fertilization (hpf), extract genomic DNA from pooled embryos (n=10) or individual specimens.
- Target Amplification: Perform PCR amplification of the target genomic region using primers flanking the edit site.
- Editing Assessment:
- T7 Endonuclease I (T7E1) Assay: Detect sequence modifications through mismatch cleavage of heteroduplex PCR products.
- Amplicon Sequencing: Clone PCR products and sequence individual clones or perform next-generation sequencing to characterize editing precision and efficiency.
- Efficiency Calculation: Determine precise editing efficiency as the ratio of correctly edited sequences to total analyzed sequences.
The fundamental principles of double-strand break repair revolve around the competitive interplay between the rapid but error-prone NHEJ pathway and the precise but context-restricted HR/HDR pathway. In zebrafish research, understanding these mechanisms has enabled the development of increasingly sophisticated genome editing technologies that either exploit endogenous repair pathways or bypass them entirely. While base editing and prime editing represent significant advances for precise genome manipulation, traditional CRISPR-Cas9 approaches coupled with HDR enhancement strategies continue to evolve, albeit with newly recognized risks of large-scale genomic alterations. The continued refinement of these tools, guided by fundamental principles of DSB repair, promises to enhance both basic research and therapeutic applications in zebrafish models and beyond.
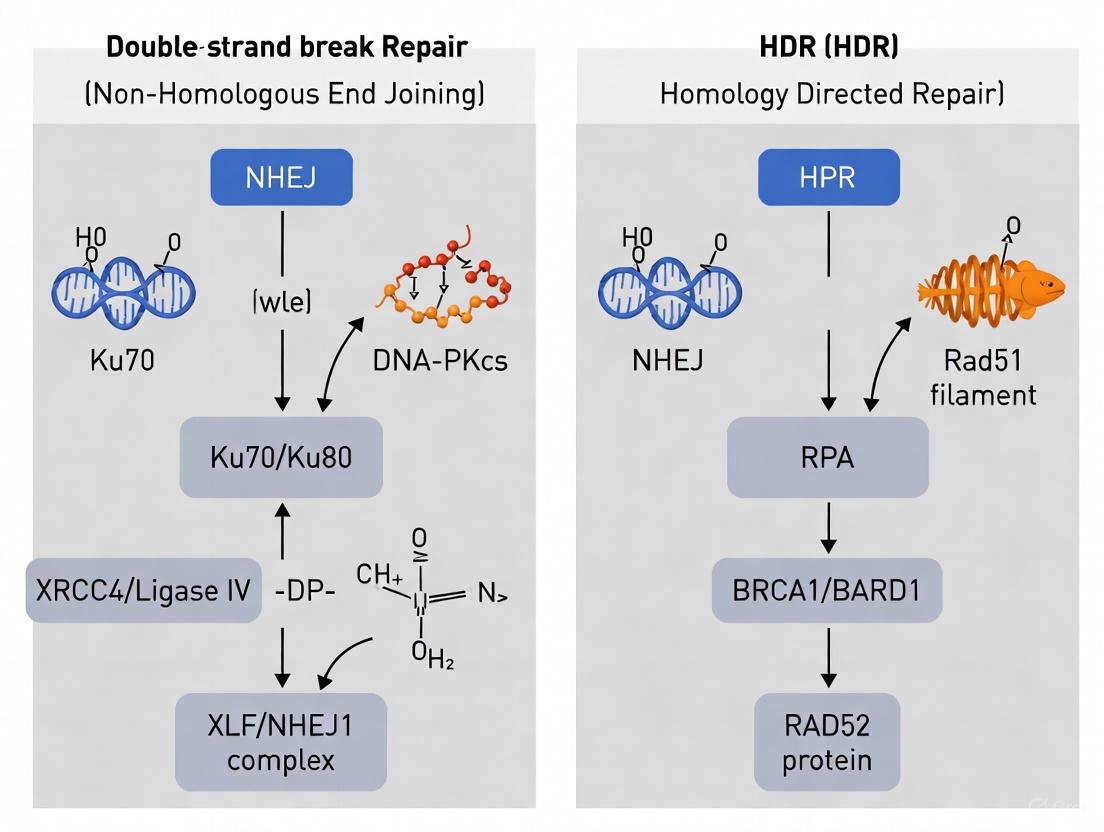
Non-homologous end joining (NHEJ) serves as the primary and most rapid cellular defense mechanism for repairing DNA double-strand breaks (DSBs) across all cell cycle stages, with particular dominance during G0 and G1 phases when sister chromatids are unavailable as repair templates [8]. This pathway functions as a constant genomic guardian, directly ligating broken DNA ends without requiring a homologous template [9] [10]. While this template-independent nature enables rapid repair, it also renders NHEJ inherently error-prone, often resulting in small insertions or deletions (indels) at the repair junction [9] [8]. In the context of zebrafish research, understanding the intricate balance between NHEJ and homology-directed repair (HDR) is fundamental for designing effective gene editing strategies, as NHEJ often competes with precise HDR-based editing approaches [11] [10].
Molecular Mechanism of NHEJ
The NHEJ pathway operates through a coordinated sequence of protein recruitment and catalytic activities that recognize, process, and ligate broken DNA ends. The process can be delineated into three core stages, visualized in the following diagram and detailed in subsequent sections.
End Binding and Tethering
NHEJ initiation occurs when the Ku70/Ku80 heterodimer recognizes and tightly binds to exposed DNA ends [9] [8]. This basket-shaped complex slides onto the DNA end and translicates inward, forming a stable ring that protects ends from degradation and prevents premature separation [9]. In vertebrates, Ku recruits the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), which becomes activated upon DNA binding and phosphorylates various substrates to coordinate subsequent repair steps [8] [12]. Simultaneously, the Mre11-Rad50-Xrs2 (MRX in yeast) or Mre11-Rad50-Nbs1 (MRN in mammals) complex promotes bridging of the DNA ends, maintaining them in proximity for repair [9].
End Processing
Before ligation can occur, damaged or incompatible DNA ends often require processing to create ligatable termini [9]. The Artemis nuclease plays crucial roles in opening DNA hairpins generated during V(D)J recombination and trimming damaged nucleotides during general NHEJ [9] [8]. For gap filling, the X-family DNA polymerases Pol λ and Pol μ (Pol4 in yeast) perform template-independent synthesis to add missing nucleotides, with Pol μ being particularly important for gap filling at 3' overhangs where the primer terminus is less stable [9] [8].
Ligation
The final and definitive step in NHEJ involves ligation of processed DNA ends by the specialized DNA ligase IV complex [9]. This complex consists of the catalytic subunit DNA ligase IV and its essential cofactor XRCC4, which stabilizes the ligase and enhances its activity [9] [12]. XLF (also known as Cernunnos) interacts with this complex and likely promotes re-adenylation of DNA ligase IV after ligation, recharging the enzyme for multiple catalytic cycles [9]. The rejoining of DNA ends by this complex restores chromosomal integrity, albeit potentially with small sequence alterations [8].
Quantitative Analysis of NHEJ Activity
Cell Cycle Regulation and Efficiency
NHEJ operates throughout the cell cycle but demonstrates variable activity and dominance compared to homologous recombination (HR), as quantitatively demonstrated in normal human fibroblasts:
Table 1: NHEJ and HR Efficiency Across Cell Cycle Phases in Human Fibroblasts [13]
| Cell Cycle Phase | NHEJ Activity | HR Activity | Relative Pathway Dominance |
|---|---|---|---|
| G1 | Active | Nearly absent | NHEJ exclusively dominant |
| S | Increases 1.5-3x vs. G1 | Most active | Both active, HR peaks |
| G2/M | Highest activity | Declines from peak | NHEJ elevated, HR declining |
This cell cycle regulation stems from mechanistic constraints: NHEJ can function without a sister chromatid, making it essential in G1, while HR requires a homologous template primarily available during and after DNA replication [13] [12]. The critical regulatory step involves 5' end resection, which commits breaks to HR and inhibits NHEJ; this resection is controlled by cyclin-dependent kinases that are inactive in G1 [9].
NHEJ in Zebrafish Gene Editing
In zebrafish research, NHEJ represents both a challenge and opportunity for genome engineering. When creating specific mutations via HDR, NHEJ competes with precise editing, often resulting in unintended indels. Analysis of successful HDR experiments in zebrafish reveals optimal conditions for suppressing NHEJ while promoting HDR:
Table 2: Experimentally Determined Optimal Conditions for HDR in Zebrafish [11]
| Parameter | Optimal Condition | Rationale |
|---|---|---|
| sgRNA Cutting Efficiency | >60% | Ensures sufficient DSB induction to engage repair mechanisms |
| DSB-Target Proximity | Within 20 nucleotides | Facilitates homologous template access to break site |
| Homology Arm Symmetry | Symmetric or asymmetric | Both can be effective with proper design |
| PAM Site Modification | Essential | Prevents re-cleavage of successfully edited loci |
| Microinjection Stage | 1-2 cell stage | Enables incorporation into germline |
| Template Topology | Single-stranded or double-stranded DNA | Both effective with proper design considerations |
Experimental Approaches for NHEJ Inhibition in Research
Chemical Inhibition Strategies
The development of NHEJ inhibitors provides powerful tools for dissecting pathway functions and potentially enhancing cancer therapies. Recent research has investigated SCR130, a specific ligase IV inhibitor, for its potential to radiosensitize cancer cells:
Methodology Overview:
- Cell Lines: Multiple HNSCC cell lines (Cal33, CLS-354, Detroit 562, HSC4, RPMI 2650, UD-SCC-2, UM-SCC-47) and healthy fibroblasts (SBLF9, 01-GI-SBL) [12]
- Inhibitor Treatment: 30 µM SCR130 (diluted in DMSO) [12]
- Irradiation Protocol: Single dose of 2 Gy ionizing radiation using ISOVOLT Titan X-ray generator [12]
- Assessment Endpoints: Cell death (flow cytometry), clonogenicity (colony formation), DNA damage (γH2AX foci), cell cycle distribution (propidium iodide staining), gene expression (qPCR) [12]
Key Findings: SCR130 treatment combined with IR showed limited radiosensitizing effects that were highly cell line-specific. However, it consistently increased G0/G1 phase arrest concomitant with gained p21 expression, suggesting anti-proliferative effects rather than direct cell death induction [12].
Genetic and Molecular Approaches
Beyond chemical inhibition, genetic approaches provide alternative strategies for NHEJ manipulation:
- NHEJ Gene Disruption: CRISPR-mediated knockout of core NHEJ factors (Ku70/80, DNA-PKcs, Ligase IV, XRCC4) [14]
- NHEJ Inhibition in HDR Workflows: Using NHEJ inhibitors to enhance HDR efficiency in precision genome editing [11]
- Variant Analysis: Computational assessment of missense variants in NHEJ components to understand molecular consequences on protein stability, interactions, and function [14]
The Scientist's Toolkit: Essential Reagents for NHEJ Research
Table 3: Key Research Reagents for NHEJ Investigation
| Reagent / Tool | Primary Function | Research Application |
|---|---|---|
| SCR130 | Selective DNA ligase IV inhibitor | Probing NHEJ function; potential radiosensitizer in cancer cells [12] |
| Ku Antibodies | Immunodetection of Ku70/Ku80 heterodimer | Verifying protein expression and cellular localization [14] [8] |
| NHEJ Reporter Cassettes | GFP-based systems with engineered I-SceI endonuclease sites | Quantifying NHEJ efficiency in different cell types and conditions [13] |
| DNA-PKcs Inhibitors (e.g., Peposertib) | Block DNA-PKcs kinase activity | Clinical investigation of NHEJ inhibition combined with radiotherapy [12] |
| Pol λ/μ Antibodies | Detect X-family polymerases | Assessing polymerase recruitment to break sites [9] [8] |
| XRCC4/LIG4 Variant Libraries | Collections of clinically identified mutations | Studying molecular drivers of NHEJ-related diseases [14] |
Implications for Zebrafish Research and Therapeutic Development
In zebrafish models, the interplay between NHEJ and HDR has profound implications for disease modeling and functional genomics. The error-prone nature of NHEJ is frequently exploited to generate gene knockouts through frameshift mutations, while HDR enables precise genetic modifications [11] [10]. The competition between these pathways necessitates strategic intervention; inhibiting NHEJ through chemical or genetic approaches can significantly enhance HDR efficiency in zebrafish embryo microinjections [11].
Beyond basic research, understanding NHEJ has direct therapeutic applications. Defects in NHEJ components are linked to human disorders including severe combined immunodeficiency (SCID), microcephaly, growth delay, and cancer predisposition [9] [14]. Conversely, cancer cells often exhibit increased reliance on NHEJ due to HR deficiencies, creating therapeutic opportunities for NHEJ inhibitors in combination with DNA-damaging agents [12]. As zebrafish continue to emerge as valuable models for human disease and drug discovery, precisely manipulating DNA repair pathway choices remains essential for advancing both basic science and therapeutic development.
In the landscape of CRISPR-based genome editing, the controlled repair of CRISPR-induced double-strand breaks (DSBs) is paramount. While non-homologous end joining (NHEJ) offers efficient but error-prone repair, homology-directed repair (HDR) provides a precise, template-dependent pathway for accurate genome modification [10] [15]. In zebrafish research, a premier model for vertebrate biology and human disease modeling, mastering HDR is particularly valuable. Zebrafish share approximately 70% of human disease-related genes, making them an essential tool for functional validation [6] [16]. However, HDR-mediated precise genome editing occurs less efficiently than random mutagenesis, presenting a significant challenge for researchers [6]. This in-depth technical guide explores the mechanisms, optimization strategies, and experimental protocols for enhancing HDR efficiency in zebrafish, framed within the broader context of DSB repair pathways.
Core Mechanisms of DNA Repair Pathways
The Cellular Repair Landscape
When a DSB occurs, multiple competing repair pathways are activated. The choice between these pathways is a critical decision point that researchers can influence to achieve desired editing outcomes.
The diagram above illustrates the fundamental decision point after a DSB. NHEJ is the dominant, error-prone pathway that ligates broken ends without a template, often resulting in small insertions or deletions (indels) ideal for gene knockout studies [10] [15]. In contrast, HDR uses a homologous DNA template to precisely repair the break, enabling accurate sequence integration [10]. Alternative pathways like microhomology-mediated end joining (MMEJ) and single-strand annealing (SSA) also contribute to repair outcomes and can be targeted to improve HDR efficiency [17].
The Critical Role of Pathway Competition
A primary challenge in precise genome editing is the innate cellular preference for NHEJ over HDR. NHEJ is active throughout the cell cycle and is faster, while HDR is restricted primarily to the S and G2 phases when homologous templates are available [15]. This competition significantly limits HDR efficiency, with studies in zebrafish showing that even under optimized conditions, imprecise integration can account for nearly half of all integration events despite NHEJ inhibition [17]. Consequently, shifting the repair equilibrium toward HDR is a central focus of optimization efforts.
Quantitative Analysis of HDR Efficiency and Optimization
Template Design and Composition
The design and type of repair template significantly influence HDR efficiency. The table below summarizes key findings from quantitative studies in zebrafish.
Table 1: Impact of Template Design on HDR Efficiency in Zebrafish
| Template Type | Experimental Efficiency | Key Advantages | Reported Germline Transmission |
|---|---|---|---|
| Long ssDNA (zLOST) | ~98% phenotypic rescue [18] | High efficiency, precise modification | Up to 31.8% [18] |
| Chemically Modified Templates | Outperformed plasmid-released templates [19] | Reduced degradation/concatemerization | >20% at multiple loci [19] |
| ssODN with Optimized Arms | 1-4% error-free repair rate [20] | Suitable for point mutations | Sufficient for F1 transmission [20] |
| Plasmid DNA (circular) | Variable, often lower efficiency [18] [19] | Convenient for larger inserts | Highly variable |
Chemical and Genetic Reprogramming of Repair Pathways
Strategic inhibition of competing repair pathways can substantially enhance HDR efficiency. Research has identified several small molecules that modulate key pathway components.
Table 2: Small Molecule Modulators to Enhance HDR Efficiency in Zebrafish
| Small Molecule | Target Pathway | Effect on HDR | Reported Efficacy in Zebrafish |
|---|---|---|---|
| NU7441 | NHEJ (DNA-PK inhibitor) | Dramatic HDR enhancement [16] | Up to 13.4-fold increase [16] |
| ART558 | MMEJ (POLQ inhibitor) | Reduces large deletions [17] | Increases perfect HDR frequency [17] |
| D-I03 | SSA (Rad52 inhibitor) | Reduces asymmetric HDR [17] | Decreases imprecise donor integration [17] |
| RS-1 | HDR (RAD51 activator) | Modest HDR stimulation [16] | Statistically significant but modest increase [16] |
Nuclease Selection and Cutting Characteristics
The choice of CRISPR nuclease and the proximity of the cut site to the intended edit are critical parameters.
- Cas9 vs. Cas12a: While Cas9 is the most widely used nuclease, Cas12a recognizes a different PAM sequence (TTTN) and creates a 5-nt 5' overhang rather than a blunt end. This distinct cutting profile is thought to potentially contribute to higher HDR rates at some loci [19].
- Cut-to-Edit Distance: A consistent finding across studies is that HDR efficiency is highly dependent on the distance between the DSB and the target sequence for modification. Designs should place the cut site as close as possible to the intended edit, ideally within 20 nucleotides [11] [19].
Advanced HDR Techniques and Experimental Protocols
The zLOST Protocol for Efficient Knock-In
The zebrafish Long Single-Stranded DNA Template (zLOST) method represents a significant advancement for precise mutation introduction [18].
Step-by-Step Workflow:
- Template Generation: Produce a long single-stranded DNA (lssDNA) donor (e.g., 299-512 nt) containing the desired modification flanked by homology arms.
- Microinjection Setup: Co-inject into one-cell stage zebrafish embryos:
- zCas9 mRNA or recombinant Cas9 protein
- Target-specific gRNA
- zLOST donor template
- Embryo Incubation: Incubate injected embryos at standard laboratory temperatures (e.g., 28.5°C) until analysis.
- Efficiency Assessment: For visible phenotypes (e.g., tyr rescue), score phenotypic rescue. For non-visible edits, use PCR-based methods and sequencing.
- Germline Transmission: Raise injected embryos (F0) to adulthood and outcross to wild-type fish. Screen F1 progeny for precise integration.
Key Advantages:
- Demonstrated precise repair at the tyr locus in nearly 98% of injected embryos [18]
- Achieved germline transmission rates up to 31.8% [18]
- Successfully applied to model human disease mutations (e.g., twist2 E78Q and rpl18 L51S) [18]
Chemical Reprogramming Protocol
This protocol utilizes small molecule inhibitors to shift the repair equilibrium toward HDR [16].
Step-by-Step Workflow:
Critical Optimization Notes:
- Treatment Timing: Inhibitor treatment should begin immediately post-injection and continue for 24 hours, covering the window when HDR typically occurs after Cas9 delivery [17] [16].
- Quantitative Assessment: Use single-cell resolution analysis (e.g., counting individual fluorescent cells) rather than qualitative presence/absence assessment, as the latter can mask the effects of HDR stimulation [16].
Prime Editing as an Alternative Strategy
Prime editing offers a template-dependent editing approach that does not require a DSB or donor DNA template [6].
Implementation Considerations:
- PE2 vs. PEn: The nickase-based PE2 editor is more effective for single-nucleotide substitutions, while the nuclease-based PEn editor shows higher efficiency for inserting short DNA fragments (up to 30 bp) [6].
- Applications: Particularly valuable for introducing stop codons or short peptide tags without requiring a donor DNA template [6].
The Scientist's Toolkit: Essential Reagents and Materials
Table 3: Key Research Reagent Solutions for HDR in Zebrafish
| Reagent Category | Specific Examples | Function/Purpose |
|---|---|---|
| CRISPR Nucleases | Cas9 mRNA/protein, Cas12a (Cpf1) | Induces targeted double-strand breaks at genomic loci of interest [17] [19] |
| Repair Templates | zLOST (lssDNA), ssODN, dsDNA with chemical modifications | Serves as homologous donor for precise HDR-mediated repair [18] [19] |
| Pathway Inhibitors | NU7441 (NHEJi), ART558 (MMEJi), D-I03 (SSAi) | Shifts repair equilibrium toward HDR by blocking competing pathways [17] [16] |
| HDR Enhancers | RS-1 (RAD51 activator) | Stimulates the HDR pathway directly [16] |
| Validation Tools | Long-read sequencing (PacBio), T7E1 assay, flow cytometry | Confirms precise editing outcomes and quantifies efficiency [17] [18] [19] |
HDR remains the gold standard for precise genome editing in zebrafish, yet its efficiency is constrained by cellular pathway competition. Through optimized template design (e.g., zLOST, chemically modified donors), strategic pathway modulation (NHEJ/MMEJ/SSA inhibition), and careful nuclease selection, researchers can significantly enhance HDR outcomes. The integration of advanced techniques like prime editing and deep-learning-assisted template design [21] promises further improvements. As these methodologies continue to evolve, HDR will become increasingly robust, enabling more sophisticated genetic modeling of human diseases in zebrafish and accelerating drug discovery pipelines.
Zebrafish (Danio rerio) have emerged as a preeminent vertebrate model for elucidating the complexities of DNA repair mechanisms. Their genetic architecture shares a remarkable 71% of protein-coding genes and 82% of human disease-associated genes, providing a highly relevant system for translational research [22]. This whitepaper details the foundational attributes—including external development, optical transparency, and genetic tractability—that position zebrafish as an ideal organism for dissecting double-strand break (DSB) repair pathways such as Non-Homologous End Joining (NHEJ) and Homology-Directed Repair (HDR). We further present advanced CRISPR-based functional genomics tools, quantitative experimental protocols, and key reagent solutions that leverage the zebrafish system to accelerate discovery in genome maintenance and therapeutic development.
The study of DNA repair mechanisms is critical for understanding genome integrity, disease etiology, and cancer biology. Zebrafish offer a unique combination of vertebrate biology and experimental practicality that is unparalleled for in vivo investigation of DNA repair pathways. Several intrinsic characteristics solidify their status as a powerful model system. Their high fecundity and rapid ex utero development facilitate the generation of large cohorts for high-throughput genetic and chemical screens [22]. The optical transparency of embryos and availability of pigment mutants enables real-time, high-resolution imaging of cellular processes, including the recruitment of repair factors to damage sites in living organisms.
Furthermore, the zebrafish genome has been fully sequenced, and a rich repository of genetic tools is available. A key advantage is the ease of genetic manipulation; CRISPR-Cas technologies enable highly efficient gene knockout and precise genome editing [23]. The establishment of mutant lines for DNA repair genes, such as those involved in the Fanconi anemia pathway, has revealed that these genes are not only crucial for genome maintenance but also impact fundamental biological processes like sex determination and differentiation [22]. This ability to model complex human disease phenotypes in a tractable vertebrate system makes zebrafish an indispensable asset for functional genomics and preclinical research.
The DNA Repair Landscape in Zebrafish
Double-strand breaks (DSBs) are among the most deleterious DNA lesions, and their accurate repair is essential for cell viability. Zebrafish possess the full repertoire of conserved DSB repair pathways, each with distinct mechanisms and outcomes.
Key Repair Pathways
- Non-Homologous End Joining (NHEJ): This is the dominant, error-prone pathway that ligates broken DNA ends without a template, often resulting in small insertions or deletions (indels). It is a rapid process active throughout the cell cycle and is commonly exploited to generate gene knockouts [15] [10] [23].
- Microhomology-Mediated End Joining (MMEJ): Also an error-prone pathway, MMEJ relies on short sequence microhomologies (2-20 bp) flanking the break to guide repair, typically resulting in deletions of the intervening sequence. A key factor in this pathway is DNA polymerase theta (Polθ/POLQ), which is highly conserved between zebrafish and humans [24] [25].
- Homology-Directed Repair (HDR): This is a high-fidelity pathway that uses a homologous DNA template (such as a sister chromatid or an exogenously supplied donor) for precise repair. HDR is the preferred mechanism for introducing specific point mutations or inserting sequences like fluorescent protein tags, though its efficiency is lower than that of NHEJ [15] [10] [16].
Table 1: Characteristics of Major DNA Double-Strand Break Repair Pathways in Zebrafish
| Pathway | Template Required | Fidelity | Key Zebrafish Factors | Primary Application in Research |
|---|---|---|---|---|
| Non-Homologous End Joining (NHEJ) | No | Error-prone | DNA Ligase 4 (Lig4) | Gene knockout studies via indel generation [15] [23] |
| Microhomology-Mediated End Joining (MMEJ) | No (uses microhomology) | Error-prone | DNA Polymerase Theta (Polθ), nuclear DNA Ligase 3 (nLig3) [24] | Studying repair-associated mutagenesis; modeling genomic instability |
| Homology-Directed Repair (HDR) | Yes | High-fidelity | Rad51, BRCA2 [22] | Precise gene knock-in, point mutation modeling, and endogenous tagging [26] |
Pathway Interplay and Expression Dynamics
The choice of repair pathway is not static but is dynamically regulated during development. Research has shown that the expression of genes related to cNHEJ and MMEJ is dynamic during zebrafish embryonic development and often increases in specific tissues [24]. Studies mutating key pathway components reveal a complex interplay:
- Loss of Polθ (MMEJ) sensitizes embryos to ionizing radiation and alters the mutation spectrum from Cas9-induced DSBs [24].
- Loss of Lig4 (cNHEJ) leads to significant larval growth defects but does not profoundly affect embryo survival under non-stressed conditions, suggesting other pathways can compensate [24].
This context-dependent requirement of repair pathways underscores the importance of using an in vivo model like zebrafish to understand their regulation in a developing, multicellular organism.
Advanced Functional Genomics Tools
The CRISPR-Cas revolution has been fully embraced in zebrafish research, enabling sophisticated functional genomics at an unprecedented scale and precision.
High-Throughput Mutagenesis and Screening
The scalability of CRISPR in zebrafish allows for systematic, high-throughput interrogation of gene function. Pioneering studies have successfully targeted hundreds of genes to identify those essential for specific biological processes. Examples include:
- A screen of 254 genes to identify regulators of hair cell and tissue regeneration [23].
- A screen of over 300 genes for their role in retinal regeneration or degeneration [23].
- Targeted mutation of zebrafish orthologs of 132 human schizophrenia-associated genes and 40 childhood epilepsy-associated genes [23].
These large-scale efforts demonstrate the power of zebrafish for directly linking human genetic variants to physiological outcomes.
Precision Genome Editing with Base Editors
Beyond inducing DSBs, base editors have revolutionized functional genomics by enabling precise single-nucleotide modifications without creating double-strand breaks [5]. Both cytosine base editors (CBEs) and adenine base editors (ABEs) have been widely applied in zebrafish.
- CBEs catalyze C:G to T:A conversions and have been used to model diseases like oculocutaneous albinism [5].
- ABEs catalyze A:T to G:C conversions, expanding the scope of possible edits [5].
Recent developments, such as "near PAM-less" editors (e.g., CBE4max-SpRY), have further expanded the targeting scope, allowing access to virtually all genomic sequences with efficiencies as high as 87% at some loci [5]. This level of precision is invaluable for modeling human genetic diseases caused by point mutations.
Table 2: Evolution of Key Base Editing Tools in Zebrafish
| Editor System | Key Features and Improvements | Demonstrated Application in Zebrafish |
|---|---|---|
| BE3 | First CBE system used in zebrafish [5] | Microinjection of mRNA or RNP complexes; efficiency of 9.25–28.57% [5] |
| Target-AID | Unique editing window targeting −19 to −16 nucleotides upstream of PAM [5] | Complementary targeting range to other base editors [5] |
| AncBE4max | Codon-optimized for zebrafish; ~3x higher efficiency than BE3 [5] | Inducing oncogenic mutations in tumor suppressor genes (e.g., tp53) for cancer modeling [5] |
| CBE4max-SpRY | "Near PAM-less" cytidine base editor [5] | Bypasses traditional NGG PAM requirement; achieves editing efficiencies up to 87% [5] |
Experimental Protocols for DNA Repair Studies
This section provides detailed methodologies for investigating and manipulating DNA repair pathways in zebrafish.
Visual Quantification of HDR Efficiency
An in vivo visual reporter assay allows for the quantitative analysis of HDR events at single-cell resolution in live zebrafish embryos [16].
Workflow Description: The diagram illustrates a transgenic zebrafish embryo assay used to quantify Homology-Directed Repair (HDR). The process starts with a transgenic embryo expressing eBFP2 in fast-muscle fibers. A donor DNA template containing the tdTomato gene flanked by homology arms is designed. The embryo is co-injected with Cas9 protein, eBFP2-targeting sgRNA, and the donor template. When Cas9 creates a double-strand break in the eBFP2 gene, the donor template can be used for HDR, leading to the replacement of eBFP2 with tdTomato. Successful HDR is quantified by counting the resulting red fluorescent muscle fibers in the live embryo.
Protocol Steps:
- Reporter Strain: Use a stable transgenic zebrafish line expressing eBFP2 (blue fluorescent protein) specifically in fast-muscle fibers under the control of the
acta1promoter [16]. - Donor Template Construction: Design a donor DNA fragment containing the
tdTomato(red fluorescent protein) gene flanked by homology arms (e.g., 303 bp left arm, 1022 bp right arm) that are homologous to sequences in theeBFP2transgene. The donor should include the sgRNA target site within the homology arm to promote HDR [16]. - Microinjection: Co-inject one-cell stage embryos with:
- Cas9 protein complexed with sgRNA targeting
eBFP2. - The linearized
tdTomatodonor DNA template.
- Cas9 protein complexed with sgRNA targeting
- Chemical Enhancement (Optional): To shift repair toward HDR, include the NHEJ inhibitor NU7441 (50 µM), which has been shown to enhance HDR efficiency up to 13.4-fold in this assay [16].
- Quantification: At 72 hours post-fertilization (hpf), image the trunk musculature. Count the number of fast-muscle fibers that have successfully switched fluorescence from blue (eBFP2) to red (tdTomato) using fluorescence microscopy. This provides a direct, quantitative readout of somatic HDR events [16].
Analyzing DSB Repair Pathway Outcomes
To dissect the contributions of different repair pathways to the mutation spectrum, one can sequence the outcomes of CRISPR-induced breaks in wild-type and DNA repair-deficient mutants.
Workflow Description: The diagram outlines an experimental pipeline to analyze DNA double-strand break (DSB) repair outcomes. The process begins by introducing DSBs into zebrafish embryos via microinjection of Cas9 and guide RNAs (gRNAs). Genomic DNA is then extracted from the embryos. The target loci are amplified from the DNA using polymerase chain reaction (PCR). These amplicons are then subjected to long-read sequencing (e.g., PacBio). The resulting sequencing data is analyzed using a computational genotyping framework (e.g., knock-knock) to classify each read into specific repair outcomes, such as perfect HDR, indels from NHEJ, or deletions characteristic of MMEJ. The frequency of these outcomes can be compared between wild-type embryos and those deficient in specific repair pathways (e.g., polq MMEJ mutants).
Protocol Steps:
- DSB Induction: Microinject wild-type and mutant (e.g.,
polq,lig3,lig4) zebrafish embryos with Cas9 ribonucleoprotein (RNP) complexes targeting specific genomic loci [24] [17]. - DNA Extraction and Amplification: At the desired stage, extract genomic DNA from pools of embryos. Perform PCR to amplify the genomic regions flanking the Cas9 target site(s) [17].
- Deep Sequencing: Subject the PCR amplicons to long-read amplicon sequencing (e.g., PacBio) to capture the full spectrum of repair events, including large deletions [17].
- Computational Genotyping: Analyze the sequencing data using a framework like
knock-knockto classify each sequencing read into specific categories:- Wild-type sequence
- Perfect HDR (if a donor was supplied)
- Small indels (characteristic of NHEJ)
- Deletions using microhomology (characteristic of MMEJ)
- Other complex rearrangements [17].
- Comparative Analysis: Compare the distribution of repair outcomes between genotypes. For example,
polq(MMEJ) mutants show a reduction in large deletions and complex indels, revealing the specific mutagenic signature of the MMEJ pathway [24] [17].
The Scientist's Toolkit: Essential Research Reagents
Successful DNA repair studies in zebrafish rely on a suite of well-defined reagents and tools.
Table 3: Essential Research Reagents for Zebrafish DNA Repair Studies
| Reagent / Tool | Function and Specification | Application Example |
|---|---|---|
| Cas9 Nuclease (protein or mRNA) | Induces site-specific double-strand breaks guided by sgRNA [15] [23] | Gene knockout via NHEJ; creating DSBs for HDR and MMEJ studies [23] |
| Base Editor Systems (e.g., AncBE4max) | Fuses catalytically impaired Cas9 to a deaminase enzyme for precise single-base changes without DSBs [5] | Modeling human genetic diseases caused by point mutations (e.g., in oncogenes or tumor suppressors) [5] |
| Homology-Directed Repair Donor Template | Provides the correct DNA sequence for precise repair; can be single-stranded oligos or double-stranded DNA with homology arms [26] | Introducing specific point mutations or inserting protein tags (e.g., GFP) into endogenous genes [16] [26] |
| NHEJ Inhibitors (e.g., NU7441) | Chemical inhibitor of DNA-PK, a key kinase in the NHEJ pathway [16] | Enhancing HDR efficiency by suppressing the competing error-prone NHEJ pathway; shown to increase HDR up to 13.4-fold in zebrafish [16] |
| MMEJ/SSA Inhibitors (e.g., ART558, D-I03) | ART558 inhibits Polθ (MMEJ); D-I03 inhibits Rad52 (SSA) [17] | Reducing specific imprecise repair patterns in knock-in experiments; improving the accuracy of genomic integrations [17] |
| DNA Repair-Deficient Mutant Lines (e.g., polq, lig4, unga) | Zebrafish strains with loss-of-function mutations in specific DNA repair genes [22] [24] | Studying the in vivo function of a specific repair gene and its interaction with other pathways; analyzing mutation spectra [24] |
Zebrafish provide an unmatched combination of physiological relevance and experimental power for the study of DNA repair mechanisms. Their high genetic homology to humans, coupled with advanced, scalable CRISPR-Cas tools, enables the direct functional validation of variants identified in human patients. The ability to quantitatively monitor and manipulate the interplay between NHEJ, HDR, and MMEJ pathways in a living, developing vertebrate offers insights that are simply not attainable in cell culture systems. As CRISPR technologies continue to evolve toward ever-greater precision and scope, the zebrafish model will undoubtedly remain at the forefront of functional genomics, disease modeling, and the development of novel therapeutic strategies aimed at safeguarding genomic integrity.
Double-strand breaks (DSBs) in DNA are critical lesions that, if not properly repaired, can lead to genomic instability, carcinogenesis, and cell death [15] [27]. In vertebrate cells, including zebrafish, two principal pathways compete to repair DSBs: the error-prone non-homologous end joining (NHEJ) and the high-fidelity homology-directed repair (HDR) [15] [27]. The cellular decision-making process that determines pathway choice is a crucial biological phenomenon with profound implications for genome editing, disease modeling, and therapeutic development. The zebrafish (Danio rerio) has emerged as a powerful vertebrate model for elucidating these mechanisms due to its genetic tractability, optical transparency, and conservation of DNA repair pathways with humans [28] [29] [27]. This review synthesizes current understanding of the factors governing NHEJ/HDR pathway choice, with specific focus on insights gained from zebrafish models.
DNA Repair Pathways: Mechanisms and Key Players
Non-Homologous End Joining (NHEJ)
NHEJ is an error-prone DNA repair pathway that functions throughout the cell cycle by directly ligating broken DNA ends without requiring a homologous template [15] [10]. This pathway is faster and more efficient than HDR but often results in small insertions or deletions (indels) at the repair site [15]. The classic NHEJ pathway involves the Ku70/Ku80 heterodimer recognizing and binding to DSB ends, followed by recruitment of DNA-PKcs, Artemis, XLF, XRCC4, and DNA Ligase IV to process and ligate the ends [30]. In zebrafish, NHEJ is the dominant DSB repair pathway and is particularly efficient for generating gene knockouts [15] [16].
Homology-Directed Repair (HDR)
HDR is a precise repair mechanism that utilizes homologous sequences (typically a sister chromatid or exogenously supplied donor template) as a blueprint for accurate DSB repair [15] [10]. This pathway is restricted to the late S and G2 phases of the cell cycle when homologous templates are available [15]. The core HDR mechanism in zebrafish involves resection of DNA ends to create 3' single-stranded overhangs, followed by RAD51 filament formation with the assistance of BRCA2, and strand invasion into the homologous template [27]. HDR is essential for precise genome editing applications, including knock-ins and specific point mutations [10] [18].
Table 1: Key Characteristics of NHEJ and HDR Pathways in Zebrafish
| Feature | NHEJ | HDR |
|---|---|---|
| Template Requirement | No homologous template needed | Requires homologous template (sister chromatid or donor DNA) |
| Cell Cycle Phase | Active throughout cell cycle | Primarily late S and G2 phases |
| Fidelity | Error-prone (often creates indels) | High-fidelity (precise repair) |
| Efficiency in Zebrafish | High (dominant pathway) | Low (typically <10% without intervention) |
| Key Proteins | Ku70/Ku80, DNA-PKcs, Ligase IV | BRCA2, RAD51, RAD52 |
| Primary Applications | Gene knockouts, gene disruption | Precise knock-ins, point mutations, tag insertions |
Alternative Repair Pathways
Beyond classical NHEJ and HDR, zebrafish possess additional DSB repair pathways including microhomology-mediated end joining (MMEJ) and single-strand annealing (SSA) [30]. MMEJ relies on 2-20 nucleotide microhomologous sequences flanking the broken junction and frequently results in deletions [30]. SSA utilizes Rad52-dependent annealing of longer homologous sequences and can lead to significant sequence deletions between repeats [30]. These alternative pathways contribute to the complex landscape of DSB repair outcomes in CRISPR-mediated genome editing.
Factors Governing Pathway Choice
The decision between NHEJ and HDR pathways is influenced by a complex interplay of cellular, molecular, and experimental factors. Research in zebrafish has been instrumental in elucidating these determinants.
Cell Cycle Phase
The cell cycle represents a fundamental determinant of pathway choice, with HDR restricted to late S and G2 phases when sister chromatids are available as repair templates [15] [27]. Evidence from zebrafish embryos demonstrates that HDR-capable cells are those in late S-/G2-phase, as visualized through geminin positivity in intestinal cells [27]. This cell cycle dependency fundamentally limits HDR efficiency, as only a subset of cells are competent for homologous recombination at any given time.
DNA End Resection
The initial processing of DSB ends represents a critical branch point in repair pathway choice [11]. Limited resection promotes NHEJ by preserving DNA ends for direct ligation, while extensive 5'→3' resection creates 3' single-stranded overhangs that favor HDR [11]. In zebrafish, the balance between resection factors and NHEJ machinery determines the cellular commitment to either pathway, with proteins like CtIP and MRE11 promoting resection and HDR.
Expression and Activity of Repair Proteins
The relative abundance and activity of key repair proteins significantly influence pathway choice. Zebrafish studies have demonstrated that BRCA2 deficiency essentially abolishes RAD51 foci formation following irradiation, indicating complete abrogation of HDR [27]. Similarly, heterozygous deficiency of Brca2 results in significantly reduced RAD51 foci, suggesting haploinsufficiency that may predispose to tumorigenesis [27]. Competition between Ku70/80 (NHEJ) and BRCA2/RAD51 (HDR) for binding to DSB ends constitutes a crucial mechanistic point of pathway regulation.
Developmental Stage
DNA repair pathway activity and regulation vary significantly during zebrafish embryonic development [31]. Early embryos possess maternally deposited DNA repair transcripts but have compromised DNA damage recognition and checkpoint activation until the mid-blastula transition (MBT) [31]. The delayed activation of proper DNA damage response mechanisms may represent an adaptation to ensure rapid embryonic cell divisions, even under genotoxic stress.
Template Availability and Nature
The availability, design, and delivery method of homologous templates significantly impact HDR efficiency in zebrafish. Research demonstrates that long single-stranded DNA (lssDNA) templates (zLOST method) achieve dramatically higher HDR efficiency (up to 98.5% phenotypic rescue at the tyr locus) compared to double-stranded or short single-stranded templates [18]. Optimal homology arm length and symmetry further enhance HDR rates [11] [18].
Table 2: Experimental Factors Influencing HDR Efficiency in Zebrafish
| Factor | Optimal Condition | Effect on HDR Efficiency |
|---|---|---|
| Template Type | Long ssDNA (zLOST) | Up to 98.5% rescue at tyr locus [18] |
| Template Length | 299-512 nt | Significant improvement over shorter templates [18] |
| Homology Arm Symmetry | Symmetric arms | Moderate improvement (≤3%) [18] |
| sgRNA Efficiency | >60% cutting efficiency | Critical for successful HDR [11] |
| PAM Site Alteration | Modified in repair template | Prevents re-cutting of repaired targets [11] |
| DSB-Target Proximity | Within 20 nucleotides | Standard for efficient HDR [11] |
Experimental Modulation of Pathway Choice
Chemical Reprogramming
Small molecule inhibitors provide a powerful approach to shift the repair equilibrium toward HDR in zebrafish. NU7441, a DNA-PK inhibitor that blocks NHEJ, enhances HDR efficiency up to 13.4-fold in zebrafish embryos [16]. Similarly, RS-1 (RAD51 stimulator) shows a modest but significant increase in HDR, while SCR7 (Lig4 inhibitor) demonstrates minimal effects in zebrafish despite efficacy in other models [16]. These findings highlight the species-specific nature of chemical modulation and the importance of empirical validation in zebrafish.
Diagram 1: DNA Repair Pathway Decision and Chemical Modulation. The diagram illustrates key steps in NHEJ (red) and HDR (green) pathways, highlighting points of chemical intervention with NU7441 (NHEJ inhibitor) and RS-1 (HDR enhancer).
Multi-Pathway Suppression Strategies
Recent evidence suggests that simultaneous suppression of multiple repair pathways can further enhance precise editing outcomes. While NHEJ inhibition alone significantly increases perfect HDR events, imprecise integration still accounts for nearly half of all integration events [30]. Additional suppression of MMEJ (via POLQ inhibition) or SSA (via Rad52 inhibition) reduces nucleotide deletions around the cut site and decreases asymmetric HDR events, thereby further improving knock-in accuracy [30]. This multi-pathway approach represents a promising strategy for achieving ultra-precise genome editing in zebrafish.
Zebrafish-Specific Methodological Advances
The zLOST Method
The zebrafish long single-stranded DNA template (zLOST) method represents a significant advancement in HDR-mediated genome editing in zebrafish [18]. This approach utilizes long single-stranded DNA donors (299-512 nt) containing symmetrical homology arms and achieves remarkable efficiency - restoring pigmentation in close to 98% of albino tyr25del/25del embryos [18]. The method demonstrates precise HDR-dependent repair and achieves germline transmission rates of up to 31.8% [18].
Quantitative HDR Assessment Methods
Zebrafish researchers have developed sophisticated assays for quantifying HDR efficiency. A visual reporter assay using fast-muscle fiber conversion from eBFP2 to tdTomato expression enables quantitative in vivo analysis of HDR events at single-cell resolution [16]. Similarly, the RAD51 foci assay in embryonic intestinal tissue provides accurate quantification of HR activity under various experimental conditions [27]. These quantitative approaches provide robust assessment tools for evaluating HDR enhancement strategies.
Diagram 2: Experimental Workflow for HDR Assessment in Zebrafish. The diagram outlines key components for microinjection (left) and methods for evaluating HDR efficiency (right) in zebrafish embryos.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Studying DNA Repair Pathways in Zebrafish
| Reagent/Chemical | Function/Application | Specific Use in Zebrafish Research |
|---|---|---|
| NU7441 | DNA-PK inhibitor, blocks NHEJ | Enhances HDR efficiency up to 13.4-fold in zebrafish embryos [16] |
| RS-1 | RAD51 stimulator, enhances HDR | Modest but significant increase in HDR efficiency [16] |
| ART558 | POLQ inhibitor, suppresses MMEJ | Reduces large deletions and complex indels in knock-in experiments [30] |
| D-I03 | Rad52 inhibitor, suppresses SSA | Decreases asymmetric HDR and imprecise donor integration [30] |
| Long ssDNA Templates | Donor template for HDR | zLOST method achieves up to 98.5% phenotypic rescue at tyr locus [18] |
| Anti-RAD51 Antibody | Immunostaining for HR quantification | Enables Rad51 foci assay in proliferative intestinal tissue [27] |
| BrdU | S-phase marker | Identifies proliferating cells for HR capability assessment [27] |
| γH2AX Antibody | DSB marker | Confirms DSB induction after irradiation [27] |
The cellular decision-making process governing NHEJ versus HDR pathway choice represents a complex biological phenomenon influenced by cell cycle phase, DNA end resection, repair protein expression, developmental stage, and template availability. Zebrafish has proven to be an invaluable model for elucidating these mechanisms, offering insights that bridge fundamental biology and applied genome editing. Methodological advances such as the zLOST platform, combined with chemical reprogramming strategies and sophisticated quantitative assays, have dramatically improved precise genome editing outcomes in zebrafish. These developments not only enhance our fundamental understanding of DNA repair pathway choice but also establish zebrafish as a powerful platform for modeling human diseases and advancing therapeutic development. Future research directions include refining multi-pathway suppression strategies, developing temporal control over repair pathway choice, and applying these insights to improve precision in therapeutic genome editing.
Key Protein Players and Their Roles in Each Repair Pathway
DNA double-strand breaks (DSBs) represent one of the most deleterious forms of DNA damage, posing a serious threat to genomic stability. If left unrepaired or misrepaired, DSBs can lead to cell death, chromosomal aberrations, and oncogenic transformations [32]. Eukaryotic cells have evolved two primary mechanisms to repair DSBs: non-homologous end joining (NHEJ) and homology-directed repair (HDR). The zebrafish (Danio rerio) has emerged as a powerful vertebrate model for studying these DNA repair pathways due to its high genetic conservation with humans, optical transparency during embryonic development, and genetic tractability [24] [16]. This review provides a comprehensive technical guide to the key protein players in NHEJ and HDR pathways, framed within the context of zebrafish research, to facilitate advanced studies in functional genomics and drug development.
Non-Homologous End Joining (NHEJ) Pathway
NHEJ is the predominant DSB repair pathway in vertebrate cells, responsible for repairing up to ~80% of all DSBs [32]. It is characterized by the direct ligation of broken DNA ends without a homologous template, making it active throughout all phases of the cell cycle, particularly in G0 and G1 [8]. While this pathway is fast and efficient, it is inherently error-prone, often resulting in small insertions or deletions (indels) at the repair junction [15]. In zebrafish, NHEJ is the dominant pathway for repairing DSBs induced by CRISPR-Cas9, making it crucial for gene knockout studies [24] [15].
Core NHEJ Protein Players and Their Functions
The NHEJ pathway employs a sophisticated array of proteins that recognize, process, and ligate broken DNA ends. The table below summarizes the key protein players and their specific roles in the NHEJ pathway.
Table 1: Key Protein Players in the NHEJ Pathway
| Protein Complex/Enzyme | Proposed Role(s) in NHEJ |
|---|---|
| Ku70/Ku80 Heterodimer | Initial DSB sensor and interaction hub; forms a ring-shaped structure that encircles DNA ends; recruits downstream NHEJ factors [32] [33]. |
| DNA-PKcs | Serine/threonine protein kinase activated by DNA binding; phosphorylates NHEJ substrates; involved in end synapsis and acts as a molecular "gate" regulating access to DNA ends [32] [33]. |
| XRCC4 | Forms a constitutive complex with DNA Ligase IV; interacts with XLF to promote synapsis [32] [33]. |
| DNA Ligase IV (LIG4) | Catalyzes the final ligation step; can tolerate certain terminal mismatches and damaged bases [32] [33]. |
| XLF (XRCC4-like factor) | Interacts with XRCC4 to stabilize synapsis; functions redundantly with PAXX [32]. |
| PAXX (Paralog of XRCC4 and XLF) | Promotes synapsis; provides redundant functions with XLF [32]. |
| Artemis | Endonuclease activated by DNA-PKcs; processes DNA ends by opening hairpin structures (critical for V(D)J recombination) and trimming damaged nucleotides [32] [33]. |
| Pol μ and Pol λ | X-family DNA polymerases that perform template-independent synthesis; fill gaps during end processing [32] [33] [8]. |
Specialized NHEJ Sub-pathways
NHEJ can proceed through distinct sub-pathways depending on the nature of the DNA ends [33]:
- Blunt-end ligation-dependent sub-pathway: For clean breaks, the Ku-XRCC4-DNA Ligase IV complex directly catalyzes ligation.
- Nuclease-dependent sub-pathway: For ends with damaged nucleotides or overhangs, the Artemis:DNA-PKcs complex is recruited to process the ends before ligation.
- Polymerase-dependent sub-pathway: For ends with missing nucleotides, Pol μ and Pol λ are recruited to fill in gaps, sometimes utilizing terminal microhomology.
Diagram: The Core Mechanism of Non-Homologous End Joining (NHEJ)
Homology-Directed Repair (HDR) Pathway
HDR is a precise, template-dependent repair pathway that utilizes a homologous DNA sequence—typically a sister chromatid or an exogenously provided donor template—to accurately repair the break [15] [34]. This pathway is predominantly active during the S and G2 phases of the cell cycle, when a sister chromatid is available [32]. In zebrafish CRISPR research, HDR is the preferred mechanism for introducing precise point mutations, inserting fluorescent protein tags, or creating other specific genomic modifications, though it is generally less efficient than NHEJ [11] [16].
Core HDR Protein Players and Their Functions
HDR involves a more complex sequence of events than NHEJ, requiring a coordinated interplay between numerous proteins responsible for end resection, strand invasion, and synthesis.
Table 2: Key Protein Players in the HDR Pathway
| Protein Complex/Enzyme | Proposed Role(s) in HDR |
|---|---|
| MRN Complex (MRE11-RAD50-NBS1) | Initiates DNA end resection; generates 3' single-stranded DNA (ssDNA) overhangs [24]. |
| CtIP | Promotes extensive end resection alongside the MRN complex [11]. |
| RPA | Binds to and stabilizes ssDNA overhangs after resection, preventing secondary structure formation [24]. |
| RAD51 | The central recombinase; forms a nucleoprotein filament on ssDNA and catalyzes strand invasion into the homologous donor template [34]. |
| BRCA2 | Mediator protein that facilitates the loading of RAD51 onto ssDNA [32]. |
| DNA Polymerases (δ/ε) | Perform DNA synthesis using the homologous template to copy genetic information across the break site. |
| DNA Ligase I | Seals the nicks in the DNA backbone after synthesis is complete, finalizing the repair. |
The HDR Process in Zebrafish
The HDR pathway can be broken down into several key stages, as illustrated in the workflow below. This process is leveraged in zebrafish genome editing by co-injecting a donor DNA template alongside CRISPR-Cas9 components.
Diagram: Experimental Workflow for HDR-Based Genome Editing in Zebrafish
Experimental Protocols for Zebrafish Research
Enhancing HDR Efficiency in Zebrafish Embryos
A significant challenge in zebrafish precision genome editing is the low efficiency of HDR compared to the competing NHEJ pathway. Research has identified several chemical and technical strategies to shift this balance. A seminal study established a quantitative in vivo reporter assay in zebrafish muscle fibers to screen for small-molecule HDR enhancers [16]. Key findings from this and other studies are summarized below.
Table 3: Strategies to Modulate DNA Repair Pathway Choice in Zebrafish
| Method | Example Agent/Target | Effect on Repair Pathways | Reported Outcome in Zebrafish |
|---|---|---|---|
| NHEJ Inhibition | NU7441 (DNA-PKcs inhibitor) [16] | Suppresses c-NHEJ | 13.4-fold enhancement of HDR efficiency; most effective treatment identified [16]. |
| NHEJ Inhibition | SCR7 (Ligase IV inhibitor) | Suppresses c-NHEJ | No significant effect on HDR efficiency in zebrafish [16]. |
| HDR Activation | RS-1 (RAD51 stimulator) [16] | Enhances RAD51 activity | Modest but significant increase in HDR efficiency [16]. |
| Template Design | Asymmetric repair templates [11] | Optimizes donor usability | A standard for improving HDR success rates; cut site should be within 20 nt of the target [11]. |
| Template Topology | Single-stranded oligodeoxynucleotides (ssODNs) vs. double-stranded DNA (dsDNA) [11] | Influences repair template accessibility | Varies by study; both are successfully used with optimized protocols [11]. |
The Scientist's Toolkit: Essential Reagents for Zebrafish DNA Repair Studies
This table compiles key reagents and their applications for studying or manipulating DNA repair pathways in zebrafish models.
Table 4: Research Reagent Solutions for Zebrafish DNA Repair Studies
| Research Reagent | Function/Application | Example Use in Zebrafish |
|---|---|---|
| CRISPR-Cas9 System | Induces targeted DSBs at genomic loci of interest. | Foundation for both NHEJ-mediated knockout and HDR-mediated knock-in studies [33] [35]. |
| NU7441 | Small-molecule inhibitor of DNA-PKcs; inhibits c-NHEJ. | Co-injected with CRISPR components to enhance HDR efficiency up to 13.4-fold [16]. |
| RS-1 | Small-molecule enhancer of RAD51 activity; stimulates HDR. | Co-injected to modestly improve HDR-mediated repair [16]. |
| High-Efficiency sgRNA | Guides Cas9 to the specific target locus. | Essential prerequisite; >60% cutting efficiency recommended for HDR experiments [11] [35]. |
| Homology-Donor Template | Provides the homologous sequence for precise repair (ssODN or dsDNA). | Designed with homology arms and altered PAM site to prevent re-cutting [11] [16]. |
| Protein Disulfide Isomerase (PDI) Modulators | Targets a novel redox-dependent regulator of NHEJ. | Protective against DNA damage in whole zebrafish; potential therapeutic target [36]. |
The intricate interplay of protein players in NHEJ and HDR pathways underpins the maintenance of genomic integrity in zebrafish and other vertebrates. NHEJ, driven by the rapid action of Ku, DNA-PKcs, and the Ligase IV/XRCC4/XLF complex, offers a swift but error-prone repair solution. In contrast, HDR, orchestrated by the MRN complex, RAD51, and associated mediators, provides a template-dependent mechanism for high-fidelity repair, albeit with lower intrinsic efficiency in zebrafish. The continued refinement of chemical modulation strategies, such as using NU7441 to inhibit NHEJ, and technical optimizations in reagent design are critical for advancing precise genome editing in this model organism. A deep mechanistic understanding of these pathways and their key protein constituents is indispensable for developing novel therapeutic interventions for human diseases, including cancer and neurodegenerative disorders, leveraging the unique experimental strengths of the zebrafish system.
From Theory to Practice: Implementing CRISPR/Cas9-Mediated NHEJ and HDR in Zebrafish
The zebrafish (Danio rerio) has emerged as a premier vertebrate model for functional genomics and disease modeling, largely due to its genetic similarity to humans, external development, and optical transparency during early stages [23] [5]. The advent of CRISPR/Cas9 technology has revolutionized genetic research in this model organism, enabling precise genome modifications that were previously challenging or impossible. CRISPR/Cas9 functions as a programmable gene-editing tool that utilizes a guide RNA (gRNA) to direct the Cas9 nuclease to a specific DNA sequence, where it creates a double-strand break (DSB) [37]. This break activates the cell's endogenous DNA repair mechanisms, primarily non-homologous end joining (NHEJ) and homology-directed repair (HDR), which researchers can harness to achieve different genetic outcomes [10].
Understanding these repair pathways is fundamental to designing effective CRISPR experiments. NHEJ is an error-prone pathway that directly ligates broken DNA ends, often resulting in small insertions or deletions (indels) that disrupt gene function—making it ideal for gene knockout studies [38] [10]. In contrast, HDR uses a homologous DNA template to repair the break accurately, allowing for precise genetic modifications such as point mutations, gene insertions, or reporter knock-ins [38] [18]. The choice between these pathways depends on the experimental goals, and successful genome editing requires careful design of both gRNAs and repair templates to maximize efficiency while minimizing off-target effects [37].
Guide RNA (gRNA) Design and Optimization
Principles of gRNA Design
The guide RNA is the targeting component of the CRISPR/Cas9 system, responsible for directing the Cas9 nuclease to specific genomic loci. Effective gRNA design must balance on-target efficiency with minimal off-target effects [38] [37]. The gRNA consists of a ~20 nucleotide spacer sequence that complementary base pairs with the target DNA, immediately adjacent to a protospacer adjacent motif (PAM) sequence (5'-NGG-3' for standard Streptococcus pyogenes Cas9) [37]. The target sequence should be unique within the genome to prevent off-target editing at similar sites.
Several factors influence gRNA efficiency. The nucleotide composition near the PAM-distal region affects Cas9 binding stability, with guanine-rich sequences often exhibiting higher efficiency [37]. The GC content of the gRNA should ideally range between 40-60%, as extremely low or high GC content can impair binding or promote non-specific interactions [38]. Additionally, the accessibility of the target chromatin region influences editing efficiency, with open chromatin regions typically being more accessible than tightly packed heterochromatin [38].
Bioinformatics Tools for gRNA Design
Several bioinformatics tools have been developed to facilitate optimal gRNA design and off-target prediction. These tools analyze potential gRNA sequences against reference genomes to identify unique targets with minimal off-site activity [37]. Commonly used platforms include:
- CHOPCHOP: A web tool that evaluates gRNA efficiency, scores potential off-target sites, and provides primer design for validation [37].
- Cas-OFFinder: Identifies potential off-target sites by allowing mismatches and bulges in the gRNA-DNA pairing [37].
- CRISPResso: Provides quantitative analysis of CRISPR editing outcomes from sequencing data, helping validate gRNA efficiency and specificity [37].
These tools typically generate efficiency scores for each potential gRNA target, enabling researchers to select the most promising candidates for their experiments. It is recommended to design and test multiple gRNAs for each target gene to ensure at least one produces efficient editing [38].
Repair Template Design for Precise Editing
Homology-Directed Repair (HDR) Templates
HDR enables precise genome modification by using an exogenous DNA template containing the desired edit flanked by homology arms complementary to the target locus [38] [18]. The design of this repair template significantly impacts HDR efficiency.
Table 1: Comparison of HDR Repair Template Types in Zebrafish
| Template Type | Description | Optimal Length | Advantages | Limitations | Reported Efficiency |
|---|---|---|---|---|---|
| ssODN (Single-Stranded Oligodeoxynucleotide) [39] [18] | Short, single-stranded DNA | 60-180 nt | Easy to synthesize; reduced random integration | Lower efficiency for large inserts; error-prone (1-4% error-free HDR) [39] | 2-8% total HDR (1-4% perfect HDR) [39] |
| lssDNA (Long Single-Stranded DNA) [18] | Long, single-stranded DNA | ~300-500 nt | Higher efficiency than ssODNs; suitable for small inserts | Limited commercial availability | Up to 98.5% phenotypic rescue (tyr locus) [18] |
| dsDNA (Double-Stranded DNA) [18] | Double-stranded DNA (plasmid or PCR product) | >500 bp with homology arms | Suitable for large insertions (e.g., reporters, tags) | Low efficiency; requires larger homology arms | ≤3% (high variability) [18] |
Key considerations for HDR template design include:
- Homology Arm Length: For ssODNs, symmetric arms of 30-60 nucleotides each are commonly used, with longer arms (e.g., 90 nt) potentially increasing HDR rates [39]. For dsDNA templates, arms of 500-1000 bp are typically required.
- Strand Complementarity: Templates complementary to the non-target strand (the strand not binding the gRNA) may slightly improve HDR efficiency [39].
- Modification Protection: Introducing silent mutations in the PAM sequence or the gRNA binding site within the repair template can prevent re-cleavage of successfully edited alleles [38].
Microhomology-Mediated End Joining (MMEJ) Templates
MMEJ is an alternative repair pathway that utilizes microhomology regions (5-25 bp) flanking the DSB [21]. Recent advances leverage predictable MMEJ outcomes for precise integration. The Pythia design tool uses deep learning to predict optimal microhomology sequences, improving frame retention and reducing deletions at integration sites [21]. This approach is particularly valuable in post-mitotic cells where HDR efficiency is low.
Design strategies include:
- Tandem Repeat Arms: Adding 3-6 bp microhomology sequences as tandem repeats at the edges of the transgene cassette safeguards against DNA trimming during integration [21].
- Sequence Context: The nucleotide composition at position -4 relative to the PAM influences integration efficiency, with a guanine (G) base enhancing the process [21].
Advanced Genome Editing Technologies
Base Editing
Base editors enable direct, irreversible conversion of a single DNA base without requiring a DSB or donor template, thereby minimizing indel formation [5]. These systems fuse a catalytically impaired Cas9 (nCas9) to a deaminase enzyme.
Table 2: Base Editing Systems for Precision Genome Editing
| Editor Type | Mechanism | Editing Window | Key Applications in Zebrafish | Considerations |
|---|---|---|---|---|
| Cytosine Base Editors (CBEs) [5] | C•G to T•A conversion | ~5 nt window (positions 4-8 upstream of PAM) | Modeling point mutations; introducing stop codons | Bystander edits possible within window |
| Adenine Base Editors (ABEs) [5] | A•T to G•C conversion | ~5 nt window (positions 4-8 upstream of PAM) | Correcting pathogenic G•C to A•T mutations | High fidelity with minimal off-target effects |
| Near PAM-less CBE (CBE4max-SpRY) [5] | C•G to T•A conversion with relaxed PAM requirement | Broad editing window | Targeting previously inaccessible genomic sites | Expands targetable sites almost genome-wide |
Base editors have achieved efficiencies ranging from 9% to over 87% in zebrafish, depending on the target locus and specific editor used [5]. Recent developments include codon-optimized versions (e.g., AncBE4max) that show approximately threefold higher efficiency than early BE3 systems [5].
Prime Editing
Prime editors represent a versatile "search-and-replace" editing technology that can mediate all 12 possible base-to-base conversions, as well as small insertions and deletions, without requiring DSBs [6]. The system utilizes a Cas9 nickase fused to a reverse transcriptase and a prime editing guide RNA (pegRNA) that specifies both the target site and the desired edit.
In zebrafish, two prime editor architectures have been compared:
- PE2 (Nickase-based): More effective for single-nucleotide substitutions, with higher precision scores (40.8% vs. 11.4% for PEn) and lower indel rates [6].
- PEn (Nuclease-based): More efficient for inserting short DNA fragments (up to 30 bp), facilitating applications such as stop codon integration or small tag insertion [6].
Prime editing efficiency can be enhanced by optimizing the pegRNA design, including refining the primer binding site and reverse transcriptase template sequences [6].
Experimental Protocols for Zebrafish Genome Editing
Microinjection Setup for CRISPR/Cas9 Delivery
The standard method for delivering CRISPR components into zebrafish embryos is microinjection at the one-cell stage to ensure widespread distribution of editing components [38] [18].
Reagent Preparation:
- gRNA Production: Synthesize gRNAs via in vitro transcription using T7 RNA polymerase or purchase chemically modified synthetic gRNAs for enhanced stability [5].
- Cas9 Source: Use Cas9 protein (as ribonucleoprotein complexes), Cas9 mRNA, or express Cas9 from an injected plasmid. RNP complexes often show reduced off-target effects and faster activity [5].
- Repair Template: Prepare the appropriate template (ssODN, lssDNA, dsDNA) in purified form. For HDR experiments, co-inject the repair template with Cas9 and gRNA.
- Injection Mix: Combine components in nuclease-free water: 100-300 ng/μL Cas9 protein or 150-300 ng/μL mRNA, 50-100 ng/μL gRNA, and 50-100 ng/μL repair template when applicable [18].
Injection Procedure:
- Calibrate the injection needle to deliver 1-2 nL per embryo.
- Inject directly into the cell cytoplasm or yolk of one-cell stage embryos.
- Maintain injected embryos at 28-32°C for development. For prime editing, incubation at 32°C has been shown to improve efficiency [6].
Validation and Screening Methods
Confirming successful genome editing requires a combination of molecular and phenotypic assays:
Initial Screening (T7 Endonuclease I Assay):
- Extract genomic DNA from pooled embryos (24-48 hpf).
- PCR amplify the target region.
- Digest PCR products with T7E1 enzyme, which cleaves heteroduplex DNA formed by wild-type and mutant alleles.
- Analyze fragments on agarose gel; cleavage products indicate mutagenesis [6].
Amplicon Sequencing (Next-Generation Sequencing):
Germline Transmission Screening:
- Raise injected embryos (G0) to adulthood.
- Outcross G0 founders to wild-type fish.
- Screen F1 progeny for the desired mutation using PCR, restriction fragment length polymorphism (if a site was introduced or disrupted), or sequencing.
- Establish stable lines from identified F1 carriers [38].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for CRISPR/Cas9 Genome Editing in Zebrafish
| Reagent / Tool | Function | Application Notes |
|---|---|---|
| Cas9 Nuclease [37] | Creates DSB at target DNA site | Use as mRNA, protein (RNP), or plasmid; RNP reduces off-target effects |
| Guide RNA (gRNA) [37] | Targets Cas9 to specific genomic locus | Chemically modified sgRNAs enhance stability and efficiency |
| ssODN Repair Template [39] [18] | Donor for precise HDR-mediated editing | 60-180 nt; design with homology arms and protected PAM site |
| lssDNA Repair Template (zLOST) [18] | Long donor for efficient HDR | 300-500 nt; significantly higher efficiency than ssODNs for small inserts |
| Base Editor mRNA [5] | Enables single-nucleotide conversion | ABE or CBE mRNA for precise point mutations without DSBs |
| Prime Editor System [6] | Enables search-and-replace editing | PE2 mRNA + pegRNA for substitutions; PEn for small insertions |
| T7 Endonuclease I [6] | Detects mutagenesis events | Used for initial efficiency screening of injected populations |
| Pythia Design Tool [21] | Predicts optimal microhomology for MMEJ | Web-based tool for designing precise integration templates |
Workflow and Pathway Diagrams
Step-by-Step Protocol for Microinjection at Early Developmental Stages
Microinjection is a foundational technique in zebrafish research, enabling the direct delivery of genetic materials into embryos and oocytes for functional genomic studies. Within the context of double-strand break (DSB) repair research, this method is particularly crucial for investigating the balance between non-homologous end joining (NHEJ) and homology-directed repair (HDR) pathways [4] [40]. The early developmental stages of zebrafish offer a unique window for manipulating and observing these fundamental cellular processes, making standardized microinjection protocols essential for generating reproducible data in genome editing experiments [41] [42].
This technical guide provides a comprehensive protocol for microinjection at early zebrafish developmental stages, with specific applications for DSB repair studies. We detail equipment requirements, step-by-step procedures, and analytical methods to enable researchers to effectively utilize this technique for probing the mechanisms of DNA repair in a vertebrate model system.
Equipment and Reagent Setup
Essential Equipment
- Micromanipulator and Microinjector: Systems such as the Sutter Instrument XenoWorks Digital Microinjector provide precise control over injection parameters [41]. Both manual and semi-automatic modes are available, with studies indicating that semi-automatic mode may improve cell survival rates (86% vs. 73% with smaller needle diameters) [43].
- Needle Puller: A programmable puller (e.g., Sutter Instrument P-97) is essential for creating consistent glass micropipettes with appropriate tip diameters [41] [43].
- Stereomicroscope: High-magnification (80×–100×) capability is necessary for visualizing early-stage oocytes (40–100 μm) during injection [41].
- Glass Capillaries: Borosilicate glass capillaries with filament (e.g., World Precision Instruments TW100F-4) are recommended for optimal needle pulling [41].
Research Reagent Solutions
Table 1: Essential Materials for Zebrafish Microinjection
| Item | Function | Specifications/Alternatives |
|---|---|---|
| Juvenile zebrafish (5-6 wpf, SL 10-15 mm) | Source of early-stage oocytes | Juvenile ovaries mainly contain early stage oocytes [41] |
| Leibovitz's L-15 Medium | Oocyte maintenance during dissection | Supplement with 1× Antibiotic-Antimycotic and 1× Glutamax [41] |
| Phenol Red (0.5%) | Injection volume indicator | Added to injection solution to visualize successful delivery [41] [44] |
| Agarose (0.8% in PBS) | Oocyte embedding for stabilization | Kept molten at 42°C before use [41] |
| Morpholinos or mRNA | Gene knockdown or overexpression | Typical morpholino concentrations: 200-500 μM; mRNA concentrations variable [42] |
| CRISPR-Cas9 components | Genome editing induction | Cas9 mRNA with sgRNAs; RNP complexes for direct editing [4] [44] |
Microinjection Procedure
Preparation of Injection Materials
For DSB repair studies, prepare CRISPR-Cas9 components as either mRNA or ribonucleoprotein (RNP) complexes. Base editors (e.g., BE3, AncBE4max) should be codon-optimized for zebrafish to enhance editing efficiency [4]. Add phenol red (0.5%) to the injection solution at a 1:5-1:10 ratio to visualize delivery [41] [42].
Pull injection needles using parameters that produce a taper suitable for early-stage oocytes. For larger tip diameters (Type I), use heat values at Ramp-10, pull at 30, and velocity at 30. For smaller tips (Type II) that improve cell survival, increase heat to Ramp+10 while maintaining pull and velocity at 30 [43].
Oocyte Collection and Preparation
- Oocyte Extraction: Euthanize juvenile zebrafish (∼5–6 weeks post fertilization, standard length 10–15 mm) in ice-cold water. The juvenile ovary predominantly contains early-stage oocytes, making it ideal for these studies [41].
- Dissection: Place the euthanized fish on a plastic Petri dish and remove the head and tail using a razor blade. Open the body cavity along the ventral midline using forceps and spring scissors to expose the internal organs [41].
- Ovary Isolation: Transfer the trunk to a glass Petri dish with 1× PBS and carefully remove the digestive system. The ovary will typically be attached to the swim bladder – gently separate it and clean off connective tissue, veins, and fat droplets [41].
- Oocyte Selection: Place the ovary in a glass Petri dish with L-15 medium. Under a microscope, remove late-stage oocytes (opaque Stage III oocytes or those with grainy texture) as their yolk can compromise survival of surrounding early-stage oocytes when broken. Injection works best with small clumps containing only Stage I oocytes, as the connective tissue improves oocyte survival [41].
- Embedding: Transfer selected oocyte clumps to a drop of L-15 media in a glass-bottom 10 mm microwell dish. Quickly remove excess media and secure oocytes in a dome of molten 0.8% agarose added drop by drop. This embedding process stabilizes oocytes for microinjection [41].
Microinjection Execution
- Needle Preparation: Backload a pulled needle with 3 μL of injection material using a microloader pipette. Remove air bubbles by shaking the bolus toward the needle tip. Insert the needle into the microinjector and break the tip at a point that creates an opening narrow enough to pierce the yolk but sufficient to deliver a consistent volume [41] [42].
- Volume Calibration: Calibrate injection volume by injecting into mineral oil on a micrometer. A bead diameter of 0.1 mm corresponds to approximately 500 pL, a typical volume for early-stage oocytes. Adjust injection pressure and time to achieve consistent bead sizes [42].
- Injection Process: Lower the needle toward the embedded oocytes at a 45° angle. Pierce the oocyte membrane in one smooth stroke and expel the injection material using predetermined pressure and time settings. Avoid injecting air bubbles or excessive stretching of the oocyte, as this can be lethal [41] [42].
- Post-Injection Care: After injection, gently transfer oocytes to a clean dish with fresh L-15 medium. Maintain at 28°C and periodically replace the medium to reduce infection risk [41].
Application to DSB Repair Research
DNA Repair Pathway Analysis
Microinjection enables precise delivery of CRISPR-Cas9 components to study DSB repair mechanisms in early developmental stages. Research indicates that DSB repair pathway choice follows a developmental progression: microhomology-mediated end joining (MMEJ) or insertion events predominate during early rapid mitotic cell cycles, followed by a switch to distinct subsets of NHEJ alleles, and finally to HDR-based gene conversion in later stages [40].
Base editors delivered via microinjection provide particularly valuable tools for DSB repair studies. These include cytosine base editors (CBEs) for C:G to T:A conversions and adenine base editors (ABEs) for A:T to G:C changes, which can introduce precise single-nucleotide modifications without inducing double-strand breaks [4]. Recent advancements such as the "near PAM-less" cytidine base editor (CBE4max-SpRY) can achieve editing efficiencies up to 87% in zebrafish [4].
Quantitative Analysis of Repair Outcomes
Table 2: Microinjection Parameters and Cell Viability
| Parameter | Manual Mode | Semi-Automatic Mode |
|---|---|---|
| Typical Cell Survival | 43-73% | 58-86% |
| Injection Rate | 100-200 cells/30min | 200-300 cells/30min |
| Needle Diameter Impact | Smaller diameter improves survival | Smaller diameter improves survival |
| Key Advantage | Higher injection efficiency | Better cell viability and consistency |
To analyze DSB repair outcomes, implement bioinformatic pipelines such as the Integrated Classification Pipeline (ICP) which categorizes mutations into distinct repair signatures: PAM-End Proximal Protected Repair (PEPPR), MMEJ, pure deletion (DELET), and insertion (INSRT) classes [40]. This enables single-allele resolution analysis of repair pathway utilization.
For HDR-specific studies, screen for chemical enhancers using high-throughput approaches. The LacZ colorimetric assay provides a quantifiable readout for HDR efficiency when combined with cell viability assays [45].
Troubleshooting and Optimization
Low Survival Rates: Optimize needle diameter, as reducing tip size significantly improves oocyte survival (from 43% to 73% in manual mode) [43]. Ensure minimal injection volume (500 pL or less for early-stage oocytes) and avoid excessive membrane disruption [41] [42].
Variable Editing Efficiency: For base editing applications, utilize nuclear localization signal (NLS) tags such as the "hei-tag" to improve nuclear import and editing efficiency [4]. Codon-optimize editors for zebrafish and consider RNP delivery for more immediate activity.
Inconsistent Injection Volume: Regularly calibrate volume delivery using the droplet method in oil. Maintain consistent injection pressure (e.g., 21.4 kPa) and time (e.g., 100 ms) parameters throughout the experiment [43] [46].
Mastering microinjection at early developmental stages provides researchers with a powerful tool for interrogating DSB repair mechanisms in zebrafish. This detailed protocol emphasizes the critical parameters that influence both technical success and experimental outcomes, particularly in the context of NHEJ and HDR pathway analysis. Through careful attention to oocyte preparation, injection parameters, and downstream analysis, researchers can generate robust data on DNA repair dynamics during early vertebrate development.
The generation of robust and reproducible knockout models is a cornerstone of functional genomics, enabling researchers to elucidate gene function in development, physiology, and disease. The error-prone nature of the non-homologous end joining (NHEJ) pathway, once a challenge for precision editing, has been effectively harnessed to create gene disruptions with high efficiency. This technical guide details a streamlined NHEJ workflow for creating knockout models in vertebrate systems, with a particular focus on the zebrafish, a model organism prized for its genetic tractability and physiological similarity to humans. We provide a comprehensive overview of the molecular principles, detailed protocols for CRISPR-Cas9-mediated mutagenesis, quantitative analysis of editing outcomes, and advanced strategies to optimize the efficiency of generating null alleles, framing these methodologies within the broader context of double-strand break repair research.
In response to CRISPR-Cas9-induced double-strand breaks (DSBs), eukaryotic cells activate a hierarchically regulated network of repair pathways. The choice between these pathways—primarily non-homologous end joining (NHEJ) and homology-directed repair (HDR)—has profound implications for genome editing outcomes [40] [47].
- NHEJ is often described as the cell's "first responder" to DSBs. This pathway operates throughout the cell cycle and functions by directly ligating broken DNA ends with minimal processing. The core mechanism involves the Ku70-Ku80 heterodimer recognizing and binding to broken DNA ends, effectively preventing extensive resection and recruiting subsequent factors like DNA-PKcs, Artemis, and the XRCC4-DNA ligase IV complex for final ligation [47] [48]. While this process can be accurate, the persistent re-cleavage of the target site by Cas9 and the inherent nature of the repair process often result in small insertions or deletions (indels). When these indels occur within a protein-coding exon and shift the reading frame, they lead to premature stop codons and effective gene knockouts [23] [47].
- HDR provides a high-fidelity alternative by using a homologous DNA template (such as a sister chromatid or an exogenously supplied donor) to precisely repair the break. However, HDR is restricted to the late S and G2 phases of the cell cycle and is generally far less efficient than NHEJ in most contexts, making it less suitable for simple gene disruption [47].
- Alternative Pathways: Other repair pathways, including microhomology-mediated end joining (MMEJ) and single-strand annealing (SSA), also contribute to DSB repair. These pathways typically result in larger deletions and can be activated under specific conditions, such as during early embryonic development where MMEJ has been observed to predominate [40] [17].
The competition between these pathways is influenced by factors such as cell cycle stage, the extent of end resection, and the availability of repair templates. For the specific goal of generating knockout models, NHEJ is the most exploited pathway due to its high activity and predictable outcome of generating disruptive indels [23] [47]. The following diagram illustrates the critical decision points that determine how a cell repairs a Cas9-induced DSB, leading to these distinct outcomes.
A Practical Workflow for NHEJ-Mediated Knockout in Zebrafish
The following section provides a detailed, step-by-step protocol for generating knockout zebrafish lines using CRISPR-Cas9, a system that has revolutionized targeted mutagenesis in this model organism [49] [50].
sgRNA Design and Synthesis
The process begins with the careful design and production of single guide RNAs (sgRNAs).
- Design Principles: sgRNAs are typically 17-20 base pairs in length and must be located immediately upstream of a protospacer adjacent motif (PAM) sequence (5'-NGG-3' for standard S. pyogenes Cas9) [49]. The target site should be within an early exon of the gene of interest to maximize the likelihood of a disruptive frameshift. Tools like CHOPCHOP or CRISPRscan are recommended for selecting sgRNAs with high predicted efficiency and minimal off-target effects [49].
- Synthesis via In Vitro Transcription (IVT): A common and cost-effective method is to produce sgRNAs using IVT.
- Template Preparation: A DNA template is generated via PCR using a gene-specific forward primer (containing the T7 promoter sequence followed by the target-specific crRNA sequence) and a common reverse primer (encoding the tracrRNA scaffold) [49].
- IVT Reaction: The purified PCR product is used as a template in a T7 IVT reaction.
- Purification: The synthesized sgRNA is column-purified, quantified, and stored in aliquots at -80°C [49].
Microinjection into Zebrafish Embryos
To ensure heritable mutations, the CRISPR-Cas9 components are delivered into one-cell stage embryos.
- Ribonucleoprotein (RNP) Complex Assembly: For high efficiency and reduced off-target effects, pre-assemble sgRNA with purified Cas9 protein to form RNP complexes. This approach leads to more immediate editing upon delivery [49] [50] [51].
- Microinjection: Using a fine glass needle and a microinjector, inject approximately 1 nL of the RNP mixture directly into the cytoplasm or cell yolk of one-cell stage zebrafish embryos [49] [50]. A typical injection mixture might contain 300 ng/μL of sgRNA and 500 ng/μL of Cas9 protein.
- Embryo Rearing: After injection, incubate embryos at 28.5°C in embryo medium (E3). Raise the injected embryos (referred to as crispants or F0 founders) to adulthood. These animals will be mosaic for induced mutations [49].
Germline Transmission and Line Establishment
The mosaic F0 fish are outcrossed with wild-type fish to screen for those that can transmit mutations to the next generation.
- Genotyping F1 Progeny: At 3-5 days post-fertilization, collect fin clips or a portion of the tail from F1 progeny for DNA extraction. Screen for mutations using methods such as:
- Heteroduplex Mobility Assay (HMA): A rapid, gel-based method to detect the presence of indels [50].
- Restriction Fragment Length Polymorphism (RFLP): If the edit disrupts a restriction site.
- Sanger Sequencing or Next-Generation Sequencing (NGS): For definitive confirmation of the exact sequence changes [49] [50].
- Establish Stable Lines: Identify F1 offspring carrying frameshift mutations and raise them to establish stable heterozygous lines. Intercrossing these heterozygotes will yield homozygous knockout mutants for phenotypic analysis [49].
The entire workflow, from design to validation, is summarized in the following diagram.
Quantitative Analysis of NHEJ Outcomes
Understanding the efficiency and mutational profile of NHEJ is critical for interpreting experimental results. The following tables summarize key quantitative data from recent studies in zebrafish and other models.
Table 1: Efficiency of CRISPR-Cas9 NHEJ Workflow in Zebrafish
| Metric | Typical Efficiency | Context and Notes |
|---|---|---|
| Somatic Mutation Rate (F0) | Very High (>70% for many loci) | Efficiency can vary based on sgRNA and delivery method. High mosaicism is typical in F0 crispants [23]. |
| Germline Transmission Rate | ~28% (Average) | Reported average from a screen of 162 loci; individual rates can vary widely [23]. |
| Biallelic Disruption in F0 | Common | Enables phenotypic screening in the injected generation, though with mosaic presentation [23] [49]. |
| HDR vs. NHEJ Ratio | Heavily favors NHEJ | NHEJ is the dominant repair pathway, especially in G0 injected embryos, making it ideal for knockout generation [6] [47]. |
Table 2: Spectrum of NHEJ-Generated Mutations in Vertebrate Models
| Mutation Type | Frequency | Characteristics and Functional Impact |
|---|---|---|
| Small Deletions (<50 bp) | Most Common (~60-80% of indels) | Often result in frameshifts, leading to premature stop codons and effective gene knockouts [40] [17]. |
| Small Insertions | Less Common | Typically templated from nearby genomic sequences or result from micro-insertions, also causing frameshifts [40]. |
| Large Deletions | Variable | More frequent when using dual sgRNAs or associated with MMEJ/SSA repair; can delete entire exons [49] [17]. |
| Complex Indels | Less Frequent | Combinations of insertions and deletions; still highly likely to be disruptive [40]. |
Advanced sequencing studies using specialized classifiers like the Integrated Classification Pipeline (ICP) have revealed that NHEJ outcomes are not random but can be highly reproducible and even dependent on developmental stage, with a switch from MMEJ to more classic NHEJ signatures observed during development [40].
Successful execution of the NHEJ knockout workflow relies on a core set of reagents and materials. The following table details these essential components.
Table 3: Essential Research Reagents for Zebrafish CRISPR Knockouts
| Reagent / Tool | Function / Description | Examples / Notes |
|---|---|---|
| Cas9 Protein | Bacterial-derived nuclease that creates DSBs at target sites. | Commercially available (e.g., NEB). Using protein (RNP) allows for immediate activity and can reduce off-target effects [49] [50]. |
| sgRNAs | Synthetic guide RNA that directs Cas9 to the specific genomic locus. | Designed using tools like CHOPCHOP [49]. Can be synthesized via IVT or purchased from companies like IDT or Synthego [49]. |
| Microinjection Equipment | Apparatus for delivering RNP complexes into embryos. | Includes micropipette puller, microinjector, micromanipulator, and injection molds [49]. |
| 5' Modified Primers | For producing knock-in donors with enhanced efficiency. | Primers with 5'AmC6 modifications protect dsDNA donors from degradation and multimerization, boosting HDR/MMEJ knock-in efficiency over 5-fold in some cases [51]. |
| NHEJ Inhibitors | Small molecules to suppress the NHEJ pathway. | Alt-R HDR Enhancer V2. Used to bias repair toward HDR for knock-ins, but not typically used for NHEJ knockout workflows [17] [47]. |
| POLQ/Rad52 Inhibitors | Small molecules to suppress alternative repair pathways. | ART558 (POLQ/MMEJ inhibitor) and D-I03 (Rad52/SSA inhibitor). Their use can reduce specific imprecise editing patterns and may enhance precise HDR when combined with NHEJ inhibition [17]. |
Advanced Strategies and Future Directions
While the basic NHEJ workflow is highly effective, several advanced strategies can further optimize knockout generation and application.
- Leveraging Alternative Repair Pathways: Understanding the interplay of repair pathways allows for more sophisticated editing strategies. For instance, the MMEJ pathway, which is highly active during early, rapid mitotic cycles, can be harnessed with specific donor designs to generate predictable larger deletions, potentially excising entire functional domains [40] [51].
- Multiplexing for Robust Knockouts: To ensure complete loss of gene function and circumvent issues like in-frame deletions or truncated proteins with residual function, researchers can co-inject multiple sgRNAs targeting different exons of the same gene. This strategy generates large deletions and provides a more reliable route to null alleles [49].
- Base and Prime Editing for Precision: Although NHEJ is ideal for gene disruption, newer technologies offer alternative routes to precise mutations. Base editors enable direct, single-nucleotide conversions without inducing DSBs, while prime editors allow for targeted insertions, deletions, and all 12 possible base-to-base conversions, also in a DSB-free manner [23] [5] [6]. A 2025 study directly comparing nickase-based (PE2) and nuclease-based (PEn) prime editors in zebrafish found PE2 more effective for single-nucleotide substitutions, while PEn was superior for inserting short DNA fragments up to 30 bp [6]. These tools are invaluable for modeling specific human disease-associated point mutations.
- Pathway Inhibition for Enhanced Precision: When performing knock-ins, the suppression of competing pathways is crucial. Recent research shows that inhibiting NHEJ is not sufficient to achieve perfect HDR, as imprecise integration persists due to MMEJ and SSA. Combined inhibition of NHEJ, MMEJ (via POLQ), and SSA (via Rad52) presents a novel strategy to significantly boost precise integration efficiencies [17].
The NHEJ repair pathway provides a straightforward and highly efficient mechanism for generating gene knockouts in vertebrate models like zebrafish. The standardized workflow—encompassing sgRNA design, RNP assembly, microinjection, and systematic genotyping—empowers researchers to rapidly create loss-of-function models at scale. As the field of genome editing continues to evolve, a deeper understanding of the complex interplay between NHEJ, HDR, MMEJ, and SSA repair pathways, coupled with the development of advanced tools like base editors and pathway-specific inhibitors, is refining our ability to manipulate the genome with ever-greater precision and control. This continuous innovation ensures that NHEJ-based workflows will remain a fundamental technique for functional genomics and disease modeling.
The ability to introduce precise genetic modifications into the zebrafish genome has revolutionized functional genomics and disease modeling. While CRISPR-Cas9 has made gene disruption via non-homologous end joining (NHEJ) relatively straightforward, achieving precise edits through homology-directed repair (HDR) remains challenging due to the inherent competition between these repair pathways. The error-prone NHEJ pathway often outcompetes HDR in rapidly dividing zebrafish embryos, leading to variable success rates for precise knock-ins [52]. This technical guide synthesizes current methodologies and optimized parameters for implementing HDR workflows to generate precise knock-ins and point mutations in zebrafish, contextualized within the broader framework of double-strand break repair mechanisms.
The fundamental challenge in HDR-based editing stems from cellular repair pathway competition. After a CRISPR-induced double-strand break, cells preferentially utilize the faster, error-prone NHEJ pathway over the more precise HDR mechanism [52]. This biological constraint is particularly pronounced in zebrafish embryos, which undergo rapid cell divisions, leaving limited time for the more complex HDR machinery to engage. Despite these challenges, methodological refinements in template design, nuclease selection, and delivery strategies have significantly improved HDR efficiency, enabling reliable generation of zebrafish models with patient-specific mutations, epitope tags, and conditional alleles [19] [53].
Editing Technology Landscape: Comparing Precision Tools
Beyond standard CRISPR-HDR approaches, newer precision editing technologies have expanded the toolkit for zebrafish researchers. The table below compares the key features, applications, and performance metrics of these technologies.
Table 1: Comparison of Precision Genome Editing Technologies in Zebrafish
| Technology | Mechanism | Best For | Typical Efficiency | Key Advantages | Limitations |
|---|---|---|---|---|---|
| HDR with ssODNs | DSB repair using single-stranded oligo donor template | Point mutations, small epitope tags (e.g., FLAG, HA) [53] | 1-5% germline transmission [53] | Cost-effective for small edits; simplified screening via PCR size changes [53] | Low efficiency; high mosaicism; template integration errors [53] |
| HDR with Long ssDNA Templates | DSB repair using long single-stranded DNA templates | Larger insertions; disease modeling with human mutations [18] | Up to 31.8% germline transmission [18] | Significantly higher efficiency than ssODNs for larger inserts; precise human mutation recapitulation [18] | Complex template production; optimization required for different loci |
| Base Editors | Chemical conversion of bases without DSBs | Single nucleotide conversions (C>T or A>G) [4] | 9-87% somatic editing [4] | No DSB formation; high efficiency; minimal indels; works in non-dividing cells [4] | Restricted to specific base changes; limited editing window; bystander mutations [4] |
| Prime Editing | Reverse transcription of edited sequence from pegRNA | Small insertions, deletions, and all base-to-base conversions [6] | 4.4-8.4% for substitutions; higher for insertions [6] | Versatile; no donor DNA required; fewer off-target effects than HDR [54] | Complex pegRNA design; lower efficiency for some applications [6] |
Recent comparative studies indicate that prime editing can achieve up to fourfold higher editing efficiency than conventional HDR for specific knock-in applications, with the additional benefit of reduced off-target effects [54]. Meanwhile, base editing continues to evolve with novel variants like "near PAM-less" cytidine base editors that can achieve editing efficiencies up to 87% at some loci, significantly expanding the targetable genomic space [4].
Optimized HDR Workflow: From Design to Validation
The successful implementation of HDR in zebrafish requires careful attention to each step of the experimental pipeline. The following workflow synthesizes optimal parameters from recent studies that have systematically compared HDR conditions.
Template Design and Selection
Template design critically influences HDR efficiency. For point mutations and small epitope tags (<50 bp), single-stranded oligodeoxynucleotides (ssODNs) with 25-50 base pair homology arms are recommended [52]. These templates should be designed with asymmetrical homology arms extending further 5' of the cut site, and must incorporate silent "blocking" mutations in the PAM sequence or protospacer to prevent re-cutting of successfully edited alleles [53]. For larger insertions (>50 bp), long single-stranded DNA templates (lssDNA) demonstrate superior performance, with one study reporting phenotypic rescue in 98.5% of injected embryos using lssDNA compared to 5% with other donors [18]. Chemically modified templates such as Alt-R HDR templates have been shown to outperform unmodified templates by reducing degradation and concatemerization in vivo [19] [54].
Table 2: Optimal Template Parameters for HDR in Zebrafish
| Template Type | Optimal Length | Homology Arm Configuration | Chemical Modifications | Key Applications |
|---|---|---|---|---|
| ssODN | 25-50 nt for arms [52] | Asymmetrical, extending further 5' of cut site [18] | Phosphorothioate modifications improve stability [19] | Point mutations, loxP sites, small epitope tags [53] |
| Long ssDNA (zLOST) | 299-512 nt templates [18] | 1,000 bp arms for large inserts [52] | Not specified in studies | Large insertions; fluorescent reporters; human disease mutations [18] |
| Plasmid Donors | Variable | Two gRNA sites flanking insertion cassette [18] | Not typically modified | Reporter cassettes; conditional alleles [19] |
Nuclease Selection and Delivery
Both Cas9 and Cas12a nucleases have been successfully employed for HDR in zebrafish, with each offering distinct advantages. While Cas9 remains the most widely used nuclease, Cas12a generates staggered cuts with 5' overhangs that may enhance HDR efficiency for some targets [19]. Recent quantitative comparisons using long-read sequencing found similar performance between Cas9 and Cas12a for targeted insertion, with precise editing rates strongly dependent on the distance between the double-strand break and the inserted sequence [19]. Delivery as ribonucleoprotein (RNP) complexes rather than mRNA significantly improves editing efficiency and reduces mosaicism. Optimal amounts of Cas9 protein range between 200 pg and 800 pg per injection, with higher concentrations within this range increasing HDR efficiency but potentially elevating toxicity [54].
Microinjection Optimization
Injection of CRISPR components directly into the cell cytoplasm rather than the yolk sac provides more consistent results according to some studies [11], though other research has found similar efficiency with yolk injections [54]. The timing of injection is critical—delivery at the one-cell stage maximizes the distribution of edits throughout the embryo. Co-injection of HDR enhancers such as IDT's HDR Enhancer v2 can improve knock-in rates, though these approaches require optimization for zebrafish embryos [55].
Diagram 1: HDR Experimental Workflow. Key optimization points (yellow) and validation stages (green) in the HDR pipeline.
Validation and Screening Methodologies
Robust screening methods are essential for identifying rare HDR events amid predominantly NHEJ-mediated indels. For visible phenotypes such as pigmentation rescue in albino mutants, direct observation provides rapid assessment [18]. For most applications without visible markers, PCR-based approaches are required.
Fluorescent PCR with capillary electrophoresis detects size changes resulting from epitope tag insertions by identifying enrichment of expected size peaks in template-injected embryos compared to nuclease-only controls [53]. For point mutations that don't alter product size, combining fluorescent PCR with restriction fragment length polymorphism (RFLP) analysis enables efficient identification of edited alleles [53]. Long-read sequencing technologies such as Pacific Biosciences platforms provide comprehensive characterization of editing outcomes across the diverse molecular patterns resulting from HDR, overcoming limitations of short-read sequencing for larger inserts [19].
Diagram 2: HDR Screening Pipeline. Advanced screening methods (green) facilitate identification of rare HDR events, with founder identification (red) as a critical gateway to stable lines.
Table 3: Research Reagent Solutions for Zebrafish HDR
| Reagent Type | Specific Examples | Function | Considerations |
|---|---|---|---|
| CRISPR Nucleases | Cas9 protein, Cas12a (Cpf1) protein [19] | Induces targeted double-strand breaks | Cas12a creates staggered ends; Cas9 creates blunt ends [19] |
| Template Types | ssODNs, Alt-R HDR Donors, zLOST templates [19] [18] [54] | Provides homology-directed repair template | Chemically modified templates reduce degradation; long ssDNA for large inserts [19] |
| Screening Tools | Fluorescent PCR primers, Restriction enzymes for RFLP, Long-read sequencing [19] [53] | Detects and validates precise edits | Capillary electrophoresis separates size differences; long-read sequencing confirms full inserts [19] [53] |
| HDR Enhancers | HDR Enhancer v2, RS-1 [55] | Increases HDR efficiency relative to NHEJ | Requires optimization for zebrafish embryos [55] |
The evolving landscape of precision genome editing in zebrafish continues to offer new possibilities for disease modeling and functional genomics. While HDR remains a valuable approach, particularly for larger insertions, newer technologies like prime editing and base editing are complementing traditional HDR strategies by offering higher efficiency for specific applications. The optimal choice of technique depends on the specific edit required—HDR with long ssDNA templates for larger insertions, base editing for specific nucleotide conversions, and prime editing for small insertions and substitutions [6] [4] [54].
Future directions will likely focus on further optimizing delivery methods, enhancing HDR efficiency through suppression of NHEJ factors, and developing increasingly sophisticated screening methodologies. As these technologies mature, the zebrafish research community will benefit from more standardized, efficient, and accessible protocols for introducing precise genetic modifications, further solidifying the zebrafish position as a powerful model for human disease modeling and drug development.
Design Considerations for Homology Arms in Repair Templates
Precise genome engineering using homology-directed repair (HDR) has become an indispensable tool for biological research and therapeutic development. Within the context of double-strand break (DSB) repair mechanisms, HDR offers a high-fidelity alternative to the error-prone non-homologous end joining (NHEJ) pathway, enabling targeted insertions, deletions, and single-nucleotide substitutions with base-pair accuracy [56] [48]. The efficiency and precision of HDR critically depend on the design of the repair template, particularly the homology arms—sequences flanking the desired modification that guide recombination with the target locus.
This technical guide examines the design principles for homology arms in repair templates, with specific application to zebrafish models where HDR efficiency has historically presented challenges [11] [16]. We synthesize current research to provide evidence-based recommendations for arm length, topology, and experimental conditions to maximize HDR efficiency, supported by quantitative data and detailed methodologies.
Fundamental Principles of HDR and Competing Repair Pathways
The Cellular Decision: HDR versus NHEJ
When a CRISPR-Cas9-induced double-strand break occurs, eukaryotic cells primarily utilize two major pathways for repair [57] [48]. The non-homologous end joining (NHEJ) pathway operates throughout the cell cycle, directly ligating broken ends without a template and often introducing small insertions or deletions (indels). In contrast, homology-directed repair (HDR) is restricted primarily to the S and G2 phases of the cell cycle and utilizes a homologous DNA template to achieve precise repair [58] [59].
The competition between these pathways is influenced by multiple factors, including cell cycle stage, relative expression of repair proteins, and the nature of the DSB itself. Early in the repair process, the MRN complex (MRE11–RAD50–NBS1) initiates 5' end resection, creating 3' single-stranded overhangs that are protected by replication protein A (RPA) [48]. Subsequent replacement of RPA with RAD51 forms nucleoprotein filaments that perform strand invasion into a homologous donor sequence, leading to precise repair via synthesis-dependent strand annealing (SDSA) or double-strand break repair (DSBR) pathways [58] [57].
Alternative Repair Pathways: MMEJ and SSA
Beyond the primary NHEJ and HDR pathways, cells possess alternative repair mechanisms that can impact genome editing outcomes [48]. Microhomology-mediated end joining (MMEJ) utilizes short homologous sequences (2-20 bp) flanking the break site, typically resulting in deletions of intervening sequences. Single-strand annealing (SSA) requires longer homologous regions (>20 bp) and also causes significant deletions. These pathways become particularly relevant when NHEJ is chemically inhibited or when repair templates contain very short homology regions [60].
Homology Arm Design Parameters
Arm Length Optimization
The length of homology arms significantly influences HDR efficiency, with optimal values depending on the type of repair template and model system. Evidence from zebrafish and mammalian studies reveals a complex relationship between arm length and recombination efficiency.
Table 1: Optimal Homology Arm Lengths by Template Type and Application
| Template Type | Recommended Arm Length | Optimal Insert Size | Key Applications | Supporting Evidence |
|---|---|---|---|---|
| ssODN | 30-60 bp [61] | 1-50 bp [57] | Point mutations, small indels | Human cells, zebrafish [58] [61] |
| ssODN (Enhanced) | ~120 nt total donor length [58] | N/A | Point mutations | High-throughput saturation genome editing [58] |
| Linear dsDNA | 200-300 bp [61] | 1-2 kb [61] | Fluorescent tags, small genes | Mammalian cells [61] |
| Plasmid-based | 500-1000 bp [57] | Up to 3 kb (with reduced efficiency) [61] | Large insertions, conditional alleles | Zebrafish, mammalian cells [62] [61] |
| Short HMEJ | 24-48 bp [60] | Reporter cassettes | Gene tagging in zebrafish | GeneWeld system [60] |
Recent research has challenged conventional wisdom regarding homology arm length. The GeneWeld system demonstrates that very short homology arms (24-48 bp) can drive efficient targeted integration when combined with Cas9-mediated liberation of donor homology arms in vivo [60]. In zebrafish, this approach achieved germline transmission rates of 22-100% across eight loci, with 48 bp arms showing superior efficiency to 12 bp or 24 bp arms [60].
Donor Template Topology and Modifications
The physical form of the repair template significantly impacts HDR efficiency. Single-stranded DNA (ssODN) templates are generally preferred for small modifications due to their lower cytotoxicity and higher efficiency in precise gene editing [58]. For larger insertions, double-stranded DNA templates (plasmids or PCR products) remain necessary, despite challenges with random integration and toxicity [57].
Strategic modifications to donor templates can enhance HDR efficiency:
- 5'-Modifications: Phosphate or phosphorothioate modifications at the 5' ends of oligonucleotide donors can improve HDR efficiency by protecting against exonuclease activity [58].
- PAM Disruption: Incorporating mutations that disrupt the protospacer adjacent motif (PAM) in the donor template prevents re-cleavage of successfully edited alleles [57].
- Asymmetric Design: Donors with asymmetric homology arms, where one arm is substantially longer than the other, may improve HDR efficiency in some contexts [58].
Zebrafish-Specific Considerations and Protocols
Optimizing HDR in Zebrafish Embryos
Zebrafish present unique challenges for HDR-mediated genome editing, with reported success rates varying widely across studies [11]. Analysis of 50 successfully modified zebrafish genes revealed several critical factors for improving HDR efficiency:
- sgRNA Efficiency: Only sgRNAs with high cutting efficiencies (>60%) should be used [11]
- Cut-to-Mutation Distance: The DSB cut site should be within 20 nucleotides of the target nucleotide [11]
- Injection Timing: Microinjections should occur during the 1-2 cell stage [11]
- Template Delivery: The repair template must overlap the DSB site [11]
Chemical Enhancement of HDR Efficiency
Small molecule inhibition of NHEJ pathway components can dramatically shift the repair balance toward HDR in zebrafish embryos [16]. Quantitative studies using a fast-muscle fiber fluorescence conversion assay demonstrated that NU7441, a DNA-PK inhibitor, enhances HDR efficiency up to 13.4-fold compared to DMSO controls [16]. In contrast, SCR7 (Lig4 inhibitor) showed no significant effect, and RS-1 (RAD51 enhancer) produced only modest improvements [16].
Table 2: Small Molecule Modulators of DNA Repair Pathways in Zebrafish
| Compound | Target | Effect on HDR | Optimal Concentration | Key Findings |
|---|---|---|---|---|
| NU7441 | DNA-PK (NHEJ inhibitor) | Up to 13.4-fold increase [16] | 50 µM [16] | Most effective single compound; minimal effect on embryo survival |
| RS-1 | RAD51 enhancer | Modest increase (7.2 ± 3.7 vs 4.8 ± 3.0 fibers) [16] | 15-30 µM [16] | Statistically significant but modest effect |
| SCR7 | Lig4 inhibitor | No significant effect [16] | Up to solubility limit [16] | Species-specific effects; ineffective in zebrafish |
| NU7441 + RS-1 | Combined NHEJ inhibition + HDR enhancement | No additive effect [16] | 50 µM + 30 µM [16] | Combination did not surpass NU7441 alone |
Experimental Workflow for Zebrafish HDR
The following methodology outlines an optimized protocol for HDR-mediated genome editing in zebrafish, incorporating best practices from recent studies:
Table 3: Key Research Reagent Solutions for HDR in Zebrafish
| Reagent Type | Specific Examples | Function & Application | Source/Reference |
|---|---|---|---|
| HDR Donor Vectors | pGTag plasmids [60] | Modular vectors for reporter knock-ins with short homology arms | Addgene [60] |
| Universal gRNA System | UgRNA [60] | Standardized gRNA for in vivo liberation of donor homology arms | Custom synthesis [60] |
| NHEJ Inhibitors | NU7441 [16] | DNA-PK inhibitor that shifts repair balance toward HDR | Commercial suppliers [16] |
| HDR Enhancers | RS-1 [16] | RAD51 stimulator that promotes strand invasion | Commercial suppliers [16] |
| Online Design Tools | genesculpt.org/gtaghd/ [60] | Web interface for designing homology arms for pGTag system | Publicly accessible [60] |
| ssODN Donors | Alt-R HDR Donor Oligos [61] | Chemically modified single-stranded DNA donors for point mutations | Integrated DNA Technologies [61] |
The field of precision genome editing continues to evolve rapidly, with several promising strategies emerging to enhance HDR efficiency beyond homology arm optimization. These include:
- HDR-Boosting Modules: Fusion of Cas9 with HDR-enhancing proteins like CtIP or dominant-negative 53BP1 to promote resection and HDR pathway choice [58] [48]
- Cell Cycle Synchronization: Timing nuclease delivery to S/G2 phases when HDR is most active [58] [59]
- Combined Chemical and Genetic Approaches: Simultaneous NHEJ inhibition with HDR activation [16]
- Novel Donor Formats: Easi-CRISPR using long single-stranded DNA donors to achieve 25-50% editing efficiency in mouse models [57]
In conclusion, optimal design of homology arms requires careful consideration of template type, target species, and desired modification. For zebrafish researchers, combining structurally appropriate homology arms with chemical reprogramming using NHEJ inhibitors represents the current state-of-the-art for achieving high-efficiency precision genome editing. As the molecular mechanisms governing DNA repair pathway choice become better elucidated, further refinements in homology arm design and implementation will continue to enhance the precision and efficiency of genome editing across model systems.
Leveraging Zebrafish Advantages for High-Throughput Genetic Screens
Zebrafish (Danio rerio) have emerged as a powerful model organism that effectively bridges the gap between in vitro assays and mammalian models for high-throughput genetic screening. Several inherent physiological and genetic advantages make zebrafish particularly suitable for large-scale studies. Their small size and external development allow embryos to survive in standard well plates for several days, facilitating large-scale drug and genetic screening. Furthermore, their genetic similarity to humans (approximately 70% of human genes have a corresponding zebrafish ortholog) enables meaningful translational research, while their optical transparency during early development permits real-time visualization of biological processes and phenotypic changes without invasive procedures [63] [6] [64]. From a practical perspective, zebrafish are highly fecund, producing hundreds of embryos per mating pair, which enables robust statistical analysis in screening applications. Additionally, compounds can be directly absorbed from the surrounding water, simplifying administration and reducing the quantity of reagents required [63]. These combined advantages have positioned zebrafish as an invaluable platform for functional genomics, disease modeling, and drug discovery.
DNA Double-Strand Break Repair Mechanisms: NHEJ and HDR
The application of CRISPR-based gene editing in zebrafish relies on inducing DNA double-strand breaks (DSBs) and harnessing the cell's endogenous repair mechanisms. The two primary pathways for DSB repair are Non-Homologous End Joining (NHEJ) and Homology-Directed Repair (HDR), each with distinct mechanisms and applications in genetic screening [10].
Non-Homologous End Joining (NHEJ)
NHEJ is an error-prone DNA repair pathway that rejoins broken DNA ends without requiring a homologous template. This mechanism often results in small insertions or deletions (INDELs) at the break site, which can disrupt gene function and is therefore ideal for gene knockout studies. NHEJ operates throughout the cell cycle and is the dominant repair pathway in most cells, making it highly efficient for generating loss-of-function mutations [65] [10]. Recent research using a novel Integrated Classification Pipeline (ICP) has revealed that the specificity of NHEJ-mediated repair outcomes can be highly reproducible and dependent on factors such as target site and developmental stage [40].
Homology-Directed Repair (HDR)
In contrast to NHEJ, HDR is a precise DNA repair mechanism that utilizes a homologous DNA template (such as a sister chromatid or an exogenously provided donor template) to accurately repair DSBs. This pathway is predominantly active during the late S and G2 phases of the cell cycle. Researchers leverage HDR to introduce specific genetic modifications, including point mutations or tagged gene versions, by providing a donor template with homologous arms flanking the insertion sequence. While HDR offers precision, its efficiency is typically lower than NHEJ in zebrafish embryos [6] [10].
Developmental Regulation of Repair Pathways
Emerging evidence indicates that the choice between DSB repair pathways is developmentally regulated. A 2024 study utilizing single-allele resolution mutation classification demonstrated a developmental progression in repair pathway dominance in zebrafish: Microhomology-Mediated End-Joining (MMEJ) or insertion events predominate during early rapid mitotic cell cycles, followed by a switch to distinct subsets of NHEJ alleles, and finally to HDR-based gene conversion at later stages [40]. This temporal regulation has significant implications for experimental design in high-throughput screening, as the timing of nuclease delivery can influence the spectrum of mutations obtained.
Table 1: Key Characteristics of DNA Double-Strand Break Repair Pathways
| Feature | NHEJ | HDR |
|---|---|---|
| Template Requirement | No template needed | Requires homologous template |
| Primary Applications | Gene knockouts, gene disruption | Precise edits, gene knock-ins, point mutations |
| Efficiency in Zebrafish | High | Low to moderate |
| Cell Cycle Dependence | Operates throughout cell cycle | Preferentially active in S/G2 phases |
| Common Outcomes | Insertions, deletions (INDELs) | Precise sequence replacement or insertion |
| Key Limitations | Error-prone, random indels | Requires donor template, lower efficiency |
Advanced Genome Editing Technologies in Zebrafish
CRISPR-Cas9 Systems
The CRISPR-Cas9 system has revolutionized genetic research in zebrafish, enabling targeted mutagenesis with unprecedented efficiency. The system consists of a Cas9 nuclease guided by a single-guide RNA (sgRNA) to a specific genomic locus, where it induces a double-strand break. A zebrafish-optimized version incorporates codon-optimized Cas9 protein with SV40 nuclear localization signals to enhance nuclear import [65]. The development of clone-free gRNA synthesis protocols allows researchers to synthesize hundreds of gRNAs in just a few hours, enabling high-throughput functional genomic screens [65].
Base Editing Technologies
Base editors represent a significant advancement beyond standard CRISPR-Cas9 systems by enabling precise single-nucleotide modifications without inducing double-strand breaks. These engineered enzymes utilize catalytically impaired Cas9 variants fused to deaminase enzymes [4].
- Cytosine Base Editors (CBEs): Convert C:G to T:A base pairs using the APOBEC1 cytidine deaminase. The editing window for CBEs typically spans positions 3-16 near the PAM site [4].
- Adenine Base Editors (ABEs): Catalyze A:T to G:C conversions using an engineered adenine deaminase [4].
Recent developments have significantly improved base editing efficiency in zebrafish. The AncBE4max system, optimized for zebrafish codons, enhanced editing efficiency approximately threefold compared to the earlier BE3 system [4]. Further improvements incorporated a "hei-tag" (high-efficiency tag) combining a Myc tag with an optimized nuclear localization signal, which increased editing efficiency by approximately 1.7-fold by enhancing nuclear localization [4]. The creation of "near PAM-less" editors like CBE4max-SpRY has expanded the targeting scope beyond the traditional NGG PAM requirement, enabling targeting of virtually all PAM sequences with efficiencies reaching up to 87% at some loci [4].
Prime Editing
Prime editing represents a versatile "search-and-replace" technology that can introduce all 12 possible base-to-base conversions, as well as small insertions and deletions, without requiring double-strand breaks. The system employs a Cas9 nickase fused to a reverse transcriptase enzyme, programmed by a prime editing guide RNA (pegRNA) that specifies the target site and encodes the desired edit [6].
A comparative study in zebrafish demonstrated that the nickase-based PE2 system was more effective for single-nucleotide substitutions, achieving a precision score of 40.8% compared to 11.4% for nuclease-based PEn editors. Conversely, the nuclease-based PEn system showed superior efficiency in inserting short DNA fragments (up to 30 bp) via NHEJ-mediated integration [6]. This complementary functionality enables researchers to select the optimal prime editing approach based on their specific experimental goals.
Table 2: Advanced Genome Editing Technologies in Zebrafish
| Technology | Editing Capability | Key Advantages | Reported Efficiencies in Zebrafish |
|---|---|---|---|
| CRISPR-Cas9 | Gene knockouts via indels | High efficiency, well-established | Highly variable by locus |
| Cytosine Base Editors (CBEs) | C:G to T:A conversions | Precise single-base changes, no DSBs | ~90% with AncBE4max; up to 87% with CBE4max-SpRY [4] |
| Adenine Base Editors (ABEs) | A:T to G:C conversions | Precise single-base changes, no DSBs | Varies by locus and system |
| Prime Editing (PE2) | All point mutations, small indels | Versatile, precise, no DSBs | 8.4% efficiency for specific substitutions [6] |
| Prime Editing (PEn) | Small insertions (up to 30 bp) | Efficient NHEJ-mediated insertion | Higher than PE2 for insertions [6] |
High-Throughput Screening Methodologies and Workflows
Automated Screening Platforms
High-throughput screening in zebrafish leverages automated technologies to systematically evaluate thousands of genetic modifications or chemical compounds. The Vertebrate Automated Screening Technology (VAST) BioImager represents a significant advancement in this area, automating the handling and positioning of individual larvae to ensure precise orientation and reproducibility across experiments [63]. When coupled with fluidic systems and advanced microscopes, this technology enables high-resolution fluorescent imaging of specific organs in real time using transgenic zebrafish lines expressing fluorescent proteins in targeted tissues [63].
Behavioral screening systems like DanioVision (Noldus IT) provide automated assessment of larval activity, movement patterns, and responses to external stimuli. These systems facilitate standardized behavioral assays including:
- Embryonic Photomotor Response (PMR): A consistent behavior in zebrafish embryos occurring 24 hours post-fertilization in reaction to light stimuli.
- Visual Motor Response (VMR): Visually driven behavior where zebrafish adjust movement in response to changes in light intensity at later larval stages (5 days post-fertilization) [63].
Recent technological innovations have further enhanced screening capabilities. A 2024 publication described a system capable of simultaneously monitoring and analyzing the movement of 288 zebrafish larvae under various experimental conditions. To address individual heterogeneity—a common challenge in zebrafish screening—this system incorporates pre-selection based on locomotion assessment, which significantly improved detection sensitivity. While pre-selected groups showed approximately 80% variation in motor function after drug treatment, non-selected groups showed only 20% variation, highlighting the importance of accounting for individual differences in high-throughput screens [66].
Experimental Protocol: High-Throughput Genetic Screening Workflow
Target Selection and gRNA Design: Identify target genes based on screening objectives. Design and synthesize gRNAs using clone-free methods for high-throughput applications [65].
Editor Delivery: Prepare ribonucleoprotein (RNP) complexes by pre-assembling Cas9 protein with sgRNAs. Alternatively, mix mRNA encoding base editors or prime editors with guide RNAs. Microinject into one-cell stage zebrafish embryos [4] [6].
Incubation and Phenotyping: Maintain injected embryos at optimal temperatures (e.g., 32°C for prime editing). Allow to develop for desired duration based on phenotypic readouts [6].
High-Content Screening: Utilize automated imaging systems like the VAST BioImager for morphological assessment. Employ behavioral tracking systems like DanioVision for functional phenotyping [63] [66].
Genotype-Phenotype Correlation: Extract genomic DNA from screened embryos. Amplify target regions and analyze editing efficiency via next-generation sequencing. Correlate specific genetic modifications with observed phenotypes [40].
Validation and Follow-up: Select hits based on phenotypic strength and reproducibility. Raise founders to establish stable lines for heritability testing and further mechanistic studies [4].
High-Throughput Genetic Screening Workflow in Zebrafish
The Scientist's Toolkit: Essential Research Reagents and Solutions
Table 3: Key Research Reagent Solutions for Zebrafish Genetic Screening
| Reagent/Solution | Function | Application Notes |
|---|---|---|
| Codon-Optimized Cas9 | Induces targeted double-strand breaks | Includes SV40 NLS for enhanced nuclear import [65] |
| Base Editor Systems (AncBE4max, zevoCDA1) | Enables precise single-nucleotide edits | AncBE4max shows ~3x higher efficiency than BE3; zevoCDA1 reduces PAM constraints [4] |
| Prime Editor Systems (PE2, PEn) | Installs all possible base substitutions and small indels | PE2 preferred for substitutions; PEn better for insertions up to 30 bp [6] |
| hei-tagged Editors | Enhances nuclear localization and editing efficiency | Increases efficiency by ~1.7-fold via optimized NLS [4] |
| Near PAM-less Editors (CBE4max-SpRY) | Expands targeting scope beyond NGG PAM | Enables targeting of virtually all PAM sequences [4] |
| Ribonucleoprotein (RNP) Complexes | Direct delivery of pre-assembled editors and guides | Reduces off-target effects; improves editing efficiency [4] |
| Transgenic Reporter Lines | Enables real-time visualization of biological processes | Express fluorescent proteins in specific tissues/organs [63] |
| Automated Imaging Systems (VAST BioImager) | Standardizes morphological and fluorescent imaging | Automates embryo handling and positioning [63] |
| Behavioral Screening Systems (DanioVision) | Quantifies locomotor and sensory responses | Captures PMR and VMR behaviors for neurophenotyping [63] |
Zebrafish have firmly established their position as a powerful model system for high-throughput genetic screening, combining physiological relevance with practical scalability. The integration of advanced genome editing technologies—particularly base editors and prime editors—with automated screening platforms has created a robust pipeline for functional genomic analysis and drug discovery. The growing understanding of DNA repair pathways, including their developmental regulation, enables more precise experimental design and interpretation of screening outcomes.
Future advancements will likely focus on further improving editing precision and efficiency, expanding the targeting scope of editing platforms, and enhancing automation and computational analysis for phenotypic screening. As these technologies mature, zebrafish-based high-throughput screening will continue to accelerate the identification of novel therapeutic targets and the understanding of gene function in vertebrate development and disease.
DSB Repair Pathway Selection and Applications
Enhancing Precision: Advanced Strategies to Favor HDR Over NHEJ
The introduction of double-strand breaks (DSBs) using the CRISPR-Cas9 system has revolutionized reverse genetics, enabling targeted gene knockout and precise genome editing. However, the cellular decision of how to repair these breaks presents a significant challenge for precision genome engineering. In zebrafish and most other model systems, the error-prone non-homologous end joining (NHEJ) pathway dominates, often resulting in a mosaic of imprecisely edited cells. In contrast, the homology-directed repair (HDR) pathway can achieve seamless integration of desired mutations or insertions using a donor template, but its inherent inefficiency has limited widespread adoption. This technical guide explores the strategic use of small-molecule inhibitors to chemically reprogram the embryo's repair equilibrium, shifting the balance from NHEJ to HDR for enhanced precision editing in zebrafish.
The Fundamental Repair Balance: NHEJ vs. HDR
Competing DNA Repair Pathways
After a CRISPR-Cas9-induced DSB, the cell primarily utilizes one of two major repair mechanisms [11]:
- Non-Homologous End Joining (NHEJ): An error-prone process that directly ligates broken DNA ends without a template. This often results in small insertions or deletions (indels) and is the dominant pathway in zebrafish and many other organisms.
- Homology-Directed Repair (HDR): A precise mechanism that uses a homologous DNA template (such as an externally supplied donor) to repair the break seamlessly. This is the preferred pathway for introducing specific nucleotide changes or inserting exogenous DNA sequences.
The natural cellular preference for NHEJ over HDR is the primary bottleneck for precise genome editing. Chemical reprogramming aims to manipulate this equilibrium.
Visualizing the Repair Balance and Intervention Strategy
The following diagram illustrates the cellular decision process after a DSB and the strategic points where small-molecule inhibitors intervene to shift the balance toward HDR.
Key Small-Molecule Modulators and Their Efficacy
Research has identified several chemical compounds that can modulate the activity of DNA repair pathways. The following table summarizes the performance of key small molecules tested in a quantitative visual reporter assay developed in zebrafish embryos [16].
Table 1: Efficacy of Small-Molecule Modulators in Enhancing HDR in Zebrafish
| Small Molecule | Target / Mechanism | Reported Effect on HDR Efficiency | Key Findings in Zebrafish |
|---|---|---|---|
| NU7441 | DNA-PK inhibitor; blocks NHEJ | Up to 13.4-fold enhancement [16] [67] | Most effective compound tested; achieved a dramatic increase from 4.0 to 53.7 HDR events per embryo at 50 µM [16]. |
| RS-1 | RAD51 stimulator; enhances HDR | Modest but significant enhancement [16] | Showed a statistically significant increase from 4.8 to 7.3 HDR events per embryo at 30 µM [16]. |
| SCR7 | Ligase IV inhibitor; blocks NHEJ | No significant effect [16] | Administration had no measurable impact on HDR efficiency compared to DMSO control, suggesting possible species-specific effects [16]. |
Critical Note on Quantitative Assessment
The zebrafish study underscores a critical methodological point: qualitative analysis (presence/absence of editing) can mask the effects of HDR stimulation. While similar percentages of embryos showed some HDR events in control and drug-treated groups, quantitative single-cell resolution analysis revealed dramatic differences in efficiency. For example, with NU7441, the number of HDR events per positive embryo increased over 13-fold, a effect that would be completely missed by a qualitative "yes/no" assay [16].
Optimized Experimental Protocol for Zebrafish
The enhancement of HDR relies on a fully optimized workflow, from template design to microinjection. The following diagram and detailed protocol outline the key steps for achieving high-efficiency HDR in zebrafish embryos using chemical reprogramming.
Detailed Step-by-Step Methodology
Step 1: Reagent Design and Preparation
- sgRNA Selection: Use only sgRNAs with high cutting efficiency (>60%). The target site should be within 20 nucleotides of the intended edit to maximize HDR efficiency [11] [26].
- Donor Template Design:
- The repair template must overlap the DSB site.
- Incorporate silent mutations in the Protospacer Adjacent Motif (PAM) site to prevent re-cutting of successfully edited alleles [11] [26].
- Homology Arm Length: While variable, a study using a fluorescent reporter employed a 303 bp left homology arm and a 1022 bp right homology arm successfully [16].
- Template Topology: Double-stranded DNA (dsDNA) is commonly used, with evidence supporting its efficacy [11].
Step 2: Microinjection Cocktail Assembly and Delivery
- Injection Stage: Perform microinjections into the cytoplasm of 1-cell stage zebrafish embryos to ensure the reagents are present during early cell divisions [11] [26].
- CRISPR Component Form: Pre-complexed Cas9 protein (ribonucleoprotein, RNP) is often preferred over mRNA, as the protein is active immediately and degrades before the 90% epiboly stage (9 hpf), reducing off-target effects [26].
- Small Molecule Preparation: Prepare a stock solution of the small-molecule inhibitor (e.g., NU7441) in DMSO. The final concentration in the injection cocktail should be optimized; for NU7441, 50 µM was identified as the effective dose [16].
Step 3: Co-injection and Embryo Handling
- Co-inject the assembled cocktail containing Cas9 RNP, donor template, and the small-molecule inhibitor.
- After injection, incubate embryos under standard conditions. The small molecule is delivered directly via injection, and no further incubation in drug solution is required [16].
Step 4: Screening and Validation
- Screen for somatic HDR events using a method appropriate for your edit (e.g., fluorescence, PCR-based assays).
- The increase in somatic HDR events has been shown to correlate directly with germline transmission rates, enabling the efficient recovery of stable lines [16].
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagents for Chemical Reprogramming and HDR in Zebrafish
| Reagent / Tool | Function / Role | Technical Specification |
|---|---|---|
| NU7441 | Potent DNA-PKcs inhibitor that shifts repair balance by blocking a key NHEJ component. | Effective at 50 µM in injection cocktail. Stock solution prepared in DMSO [16]. |
| High-Efficiency sgRNA | Guides Cas9 to create a clean DSB at the target locus. | Cutting efficiency should be >60%. Target site within 20 nt of desired edit [11] [26]. |
| Cas9 Protein (RNP) | Creates a precise DSB at the genomic target site. | Using pre-complexed RNP provides immediate activity and reduces off-target effects [26]. |
| dsDNA Donor Template | Serves as the homologous repair template for precise integration of the desired sequence. | Must contain homology arms and alter the PAM sequence to prevent re-cleavage [16] [11]. |
| Quantitative Reporter Assay | Enables accurate measurement of HDR efficiency at single-cell resolution. | Critical for evaluating modulator efficacy; avoids the pitfalls of qualitative analysis [16]. |
Chemical reprogramming using small-molecule inhibitors like the DNA-PK inhibitor NU7441 represents a robust and effective strategy to overcome the primary limitation of precise genome editing in zebrafish. By strategically inhibiting the NHEJ pathway, researchers can dramatically shift the DNA repair equilibrium in favor of HDR. When combined with an optimized protocol—including high-efficiency sgRNAs, a well-designed donor template, and RNP complex injection at the single-cell stage—this approach enables highly efficient seamless integration of genetic modifications. This methodology significantly improves the recovery of germline transmissions, solidifying its role as an essential technique for advanced functional genomics and disease modeling in zebrafish.
The application of CRISPR/Cas9 technology in zebrafish has revolutionized the field of functional genomics, enabling the creation of precise disease models. A significant challenge in this field is the inherent competition between two primary DNA double-strand break (DSB) repair pathways: the error-prone non-homologous end joining (NHEJ) and the precise homology-directed repair (HDR) [10]. The dominance of NHEJ in early-stage embryos often results in low efficiencies of precise genome editing, limiting the generation of knock-in models and the study of specific mutations [16] [11]. To address this bottleneck, researchers have turned to small molecule modulators to shift the repair equilibrium towards HDR. This technical guide focuses on the efficacy and application of two such validated compounds—NU7441 and RS-1—within the context of zebrafish embryology, providing a comprehensive resource for researchers aiming to enhance precise genome editing outcomes.
The DNA Repair Landscape in Zebrafish Embryos
Competing Pathways: NHEJ vs. HDR
Upon the introduction of a CRISPR/Cas9-mediated DSB, the zebrafish embryo's cellular machinery initiates a complex repair process. The choice of repair pathway is a critical determinant of the editing outcome and is influenced by the cell cycle, developmental stage, and availability of key enzymatic components [40] [68].
Non-Homologous End Joining (NHEJ): This pathway is active throughout the cell cycle and functions by directly ligating the broken DNA ends. It is considered error-prone, as it often results in small insertions or deletions (indels), effectively disrupting the target gene and creating knock-out alleles. Key proteins in the canonical NHEJ pathway include the Ku70/Ku80 heterodimer, DNA-dependent protein kinase catalytic subunit (DNA-PKcs), and Ligase IV [68]. The NHEJ pathway is particularly dominant in the early, rapid mitotic cycles of zebrafish development [16].
Homology-Directed Repair (HDR): In contrast, HDR is a high-fidelity mechanism that is most active during the S and G2 phases of the cell cycle. It requires a homologous DNA template (such as a single-stranded oligodeoxynucleotide or a plasmid donor) to accurately repair the break. This pathway can be co-opted by researchers to introduce specific nucleotide changes, insert tags, or create conditional alleles [10]. The initiation of HDR involves end resection by the MRN complex (MRE11-RAD50-NBS1) and CtIP, followed by the involvement of central players like RAD51, which facilitates strand invasion into the homologous template [69] [68].
The following diagram illustrates the hierarchical decision-making process a cell undergoes to repair a DSB, highlighting the key steps where NU7441 and RS-1 exert their influence.
Developmental Regulation of Repair
Emerging evidence suggests that DSB repair is not static during embryogenesis. A 2024 study using a single-allele resolution mutation classifier revealed a developmental progression in repair pathway usage in complex multicellular organisms [40]. The study found that microhomology-mediated end joining (MMEJ) or specific insertion events are predominant during early rapid mitotic cell cycles. This is followed by a switch to distinct subsets of NHEJ alleles, and later to HDR-based gene conversion [40]. This temporal switch underscores the importance of timing when administering HDR-enhancing compounds, as the cellular environment must be permissive for homology-based repair.
Validated Compounds: Mechanisms and Efficacy
NU7441 - A DNA-PKcs Inhibitor
Mechanism of Action: NU7441 is a potent and specific inhibitor of DNA-dependent protein kinase catalytic subunit (DNA-PKcs) [70] [68]. DNA-PKcs is a core component of the canonical NHEJ pathway. Upon binding to DSBs, it orchestrates the recruitment and activation of other NHEJ factors. By inhibiting DNA-PKcs, NU7441 disrupts the NHEJ repair machinery, preventing the error-prone ligation of breaks and thereby reducing the formation of indels. This forces the cell to rely more heavily on alternative repair pathways, such as HDR, when a homologous template is available [16] [70].
Efficacy in Zebrafish: The quantitative efficacy of NU7441 was rigorously demonstrated using a novel in vivo visual reporter assay in zebrafish [16]. In this system, successful HDR converts a blue fluorescent protein (eBFP2) in muscle fibers to a red fluorescent protein (tdTomato), allowing for quantification at single-cell resolution.
Table 1: Quantitative Efficacy of NU7441 in Zebrafish Embryos
| Compound | Optimal Concentration | HDR Efficiency (Control) | HDR Efficiency (Treated) | Fold Enhancement | Key Findings |
|---|---|---|---|---|---|
| NU7441 | 50 µM | 4.0 ± 3.0 red fibers/embryo [16] | 53.7 ± 22.1 red fibers/embryo [16] | Up to 13.4-fold [16] | - Efficacy is dose-dependent [16].- Dramatic increase in somatic HDR correlates with improved germline transmission [16].- Qualitative analysis (presence/absence of editing) masks the full effect; quantitative single-cell resolution is crucial [16]. |
This enhancement is not limited to zebrafish. A 2025 study in HeLa cells using a lipo-xenopeptide delivery system also found NU7441 to be the most effective enhancer, achieving over a 10-fold increase in HDR efficiency and a peak HDR rate of 61% as confirmed by sequencing [70]. Furthermore, a 2024 study in porcine fibroblasts confirmed that NU7441 consistently increased HDR-mediated precise gene editing efficiency [71].
RS-1 - A RAD51 Stimulator
Mechanism of Action: RS-1 functions by directly targeting the HDR pathway. It is a known stimulator of RAD51, a key protein that forms a nucleoprotein filament on single-stranded DNA and catalyzes the central step of strand invasion during homologous recombination [69] [68]. By enhancing the DNA-binding activity of RAD51, RS-1 actively promotes the efficiency and stability of the strand invasion process, thereby increasing the likelihood that the DSB will be repaired using the provided homologous donor template [69].
Efficacy in Zebrafish and Other Models: In the same zebrafish visual reporter assay used for NU7441, RS-1 showed a more modest but statistically significant enhancement of HDR [16].
Table 2: Quantitative Efficacy of RS-1 in Zebrafish Embryos and Other Models
| Compound | Optimal Concentration | Model System | HDR Efficiency (Control) | HDR Efficiency (Treated) | Fold Enhancement | Key Findings |
|---|---|---|---|---|---|---|
| RS-1 | 7.5 - 30 µM | Zebrafish Embryos (Visual Assay) | 4.8 ± 3.0 red fibers/embryo [16] | 7.2 ± 3.7 to 7.3 ± 5.3 red fibers/embryo [16] | ~1.5-fold [16] | - Modest but significant increase [16].- Higher doses (15 µM) improved blastocyst development but not HDR efficiency [69]. |
| RS-1 | 7.5 µM | Rabbit Embryos (In Vitro) | 4.4% (RLL locus) [69] | 26.1% (RLL locus) [69] | ~5.9-fold | - Demonstrated effectiveness across multiple loci (CFTR, ApoAI) [69].- Improved knock-in efficiency in vivo for rabbit production [69]. |
The data suggests that while RS-1 is a potent HDR enhancer in other systems like rabbits, its effect in zebrafish under the tested conditions is significant but less dramatic than that of NU7441. It is important to note that the efficacy of these compounds can be species- and context-dependent.
Experimental Protocol for Zebrafish Embryos
Below is a detailed methodology for employing NU7441 and RS-1 in a CRISPR/HDR experiment in zebrafish, based on the protocols from the cited literature.
Workflow Overview: The following diagram outlines the key stages of the experimental workflow, from embryo preparation to analysis.
Materials and Reagents
The Scientist's Toolkit
| Category | Reagent/Item | Function and Specification |
|---|---|---|
| CRISPR Components | Cas9 Protein | Generates the site-specific double-strand break. Using purified protein (RNP) reduces off-target effects and allows for immediate activity [16] [71]. |
| sgRNA | Guides Cas9 to the specific genomic locus. Should have high cutting efficiency (>60%) [11]. | |
| Donor Template (ssODN) | Serves as the homologous repair template. Must contain homologous arms and the desired edit. Phosphorothioate modifications can enhance stability [71]. | |
| Small Molecules | NU7441 | DNA-PKcs inhibitor. Reconstituted in DMSO (e.g., 50 mM stock) and used at a working concentration of 50 µM in embryo medium [16]. |
| RS-1 | RAD51 stimulator. Reconstituted in DMSO (e.g., 10 mM stock) and used at a working concentration of 7.5-30 µM in embryo medium [16] [69]. | |
| Embryology Supplies | Zebrafish Embryos | Wild-type or transgenic reporter lines at the 1-cell stage. |
| Microinjection Apparatus | For delivering the CRISPR mix into the embryo cytoplasm or yolk. | |
| Embryo Medium | Standard medium for culturing zebrafish embryos. |
Step-by-Step Procedure
- Embryo Collection and Preparation: Collect zebrafish embryos within 30 minutes of spawning and align on an injection mold.
- CRISPR/Donor Injection Mix Preparation: Prepare the ribonucleoprotein (RNP) complex by pre-incubating purified Cas9 protein (e.g., 10 µg) with sgRNA (e.g., 100 pmol) for 10 minutes at room temperature. Then, add the single-stranded oligodeoxynucleotide (ssODN) donor template (e.g., 200 pmol) to the mix [71]. The donor should be designed with homology arms (typically >30 bp) and should alter the PAM site or seed sequence to prevent re-cutting [11].
- Microinjection: Using a fine-glass needle, inject 1-2 nL of the RNP/donor mixture directly into the cytoplasm of the 1-cell stage embryo [11].
- Small Molecule Treatment: Immediately after injection, transfer the embryos into a 6-well plate containing embryo medium supplemented with the chosen small molecule.
- Embryo Incubation: Incubate the embryos at 28.5°C. The treatment duration can vary but is typically maintained for 20-24 hours post-injection [69]. Monitor embryo development and survival.
- Screening and Validation: At the desired stage (e.g., 72 hours post-fertilization), screen for HDR events.
- For visual reporters: Score under a fluorescence microscope for the expression of the reporter (e.g., tdTomato-positive muscle fibers) [16].
- For genotyping: Extract genomic DNA from individual embryos or pooled samples. Perform PCR amplification of the target locus, followed by Sanger sequencing or next-generation sequencing to identify and quantify precise edits [71].
The use of NU7441 and RS-1 represents a significant advancement in improving the efficiency of precise genome engineering in zebrafish. The quantitative data demonstrates that NU7441 is particularly potent in zebrafish, providing up to a 13.4-fold enhancement of HDR by effectively suppressing the competing NHEJ pathway [16]. While RS-1 shows more variable efficacy, it remains a valuable tool for directly stimulating the HDR machinery, and its effects may be more pronounced in specific experimental contexts or when used with optimized protocols [16] [69].
Technical Considerations and Future Directions
- Combination Therapy: The search results do not indicate a synergistic effect when NU7441 and RS-1 are combined in zebrafish. One study found that adding RS-1 to an optimal dose of NU7441 did not yield a further increase in HDR efficiency [16]. This suggests that the mechanisms, while complementary, may not be additive in this model system, or that the repair process becomes saturated or limited by other factors.
- Temporal Administration: Given the discovery of a developmental progression in DSB repair pathways [40], the timing of compound administration is likely critical. Future studies could investigate whether applying these compounds at later developmental stages, when the embryo is more permissive to HDR, could further enhance editing efficiency.
- Quantitative Analysis: As emphasized in the foundational zebrafish study, qualitative assessments (e.g., simply noting the presence or absence of editing) can mask the true effects of these enhancers. The dramatic 13.4-fold improvement by NU7441 was only apparent through a quantitative, single-cell resolution assay [16]. Therefore, employing sensitive, quantitative genotyping methods like amplicon sequencing is highly recommended for accurately evaluating the success of HDR experiments.
In conclusion, the chemical reprogramming of DNA repair pathways in zebrafish embryos using validated small molecules like NU7441 and RS-1 has moved the field beyond relying on the innate, low-efficiency HDR of the cell. By integrating these compounds into standardized experimental workflows, researchers can significantly improve the recovery of precise genetic modifications, thereby accelerating the creation of sophisticated disease models and functional genomic studies in this powerful vertebrate model.
Optimizing Repair Template Design and Delivery Methods
The equilibrium between non-homologous end joining (NHEJ) and homology-directed repair (HDR) is a pivotal determinant of success in precision genome editing. In zebrafish (Danio rerio), a premier model for vertebrate functional genomics and disease modeling, the inherent dominance of the error-prone NHEJ pathway frequently obstructs the attainment of high-efficiency HDR, limiting the creation of precise mutant alleles [11] [16]. This technical whitepaper provides an in-depth guide to optimizing repair template design and delivery methods to skew this balance toward HDR. By synthesizing current research, we outline definitive strategies encompassing template engineering, the use of small-molecule modulators, and refined microinjection protocols. These methodologies are framed within a broader thesis on double-strand break (DSB) repair in zebrafish, offering researchers a structured path to enhance the fidelity and throughput of precise genetic modifications.
Core Principles of HDR in Zebrafish
Homology-directed repair is a high-fidelity pathway that utilizes a homologous donor template to repair double-strand breaks. Following a CRISPR-Cas9-induced DSB, the cell's repair machinery can use a provided donor DNA to incorporate specific sequences, such as point mutations, epitope tags, or fluorescent reporters, into the genome [47]. The key challenge is that HDR is inherently less efficient than NHEJ in most systems, including zebrafish [16]. This inefficiency often results in mosaic founders (F0) where only a subset of cells carries the desired precise edit, complicating the recovery of stable germline transmissions.
Analysis of successfully modified zebrafish genes reveals several foundational principles for effective HDR [11]:
- High-Efficiency sgRNAs: Only single guide RNAs (sgRNAs) with demonstrated cutting efficiencies exceeding 60% should be employed.
- Proximity of Cut to Edit: The DSB site should be situated within 20 nucleotides of the intended target nucleotide for optimal results.
- PAM Disruption: The Protospacer Adjacent Motif (PAM) site on the donor template must be altered to prevent re-cleavage of successfully edited alleles by Cas9.
- Injection Timing: Microinjections are most effective when performed at the 1–2 cell stage to maximize the distribution of the edit throughout the organism.
Repair Template Design Optimization
The architecture of the repair template is a critical variable controlling HDR efficiency. Decisions regarding topology, symmetry, and homology arm length directly impact recombination rates.
DNA Template Topology and Symmetry
The physical form of the donor DNA—whether single-stranded or double-stranded—is a significant factor. While both can be effective, recent trends and data suggest that single-stranded DNA (ssDNA) templates may offer superior performance in some contexts, likely due to their easier accessibility by the cellular repair machinery [11]. Furthermore, the symmetry of the homology arms relative to the DSB is crucial. The repair template must be designed to overlap the double-strand break site. While symmetric overlap (with homology arms extending equally on both sides of the cut) is common, asymmetric designs have also been successfully employed [11].
Homology Arm Length
The length of the homology arms flanking the desired edit is a key parameter. There is a statistically optimal range for arm length that balances efficiency with practicality of template synthesis.
Table 1: Optimized Homology Arm Lengths for HDR in Zebrafish
| Template Type | Left Homology Arm (bp) | Right Homology Arm (bp) | Reported Efficiency | Key References |
|---|---|---|---|---|
| Short Inserts (< 1 kb) | 30 - 50 bp | 30 - 50 bp | Found to be statistically optimal for integration [11] | Bai et al., 2020 |
| Gene Tagging | 303 bp | 1022 bp | Successfully used for fluorescent protein insertion [16] | Burg et al., 2018 |
| Large Inserts (> 1 kb) | 800 - 1200 bp | 800 - 1200 bp | Recommended for seamless integration of large fragments [11] | Wierson et al., 2020 |
Delivery Methods and Experimental Protocols
The method of delivering CRISPR-Cas9 components and the repair template into the zebrafish embryo is a practical aspect with profound implications for HDR outcomes.
Microinjection of Ribonucleoprotein (RNP) Complexes
The most common and effective delivery method is the microinjection of pre-assembled Cas9 ribonucleoprotein (RNP) complexes directly into the cytoplasm of one-cell stage embryos [23] [5]. This approach involves complexing purified Cas9 protein with sgRNA, which leads to rapid DSB generation and can reduce off-target effects compared to mRNA injection. Co-injecting the repair template (e.g., ssDNA) with the RNP complexes is crucial for making the template available during the critical window for HDR.
Table 2: Research Reagent Solutions for HDR in Zebrafish
| Reagent / Tool | Function | Example & Notes |
|---|---|---|
| Cas9 Protein | Creates the double-strand break at the target locus. | Purified S. pyogenes Cas9. Using Cas9 protein in RNP complexes is preferred for rapid cutting and reduced mosaicism. |
| High-Efficiency sgRNA | Guides Cas9 to the specific genomic target. | In vitro transcribed or synthetic sgRNA with >60% cutting efficiency, verified by assay [11]. |
| ssDNA Donor Template | Serves as the homology-directed repair template. | Single-stranded DNA oligo with altered PAM site and optimized homology arms. |
| NHEJ Inhibitor (NU7441) | Small molecule that chemically reprograms the embryo to favor HDR. | DNA-PKcs inhibitor. Used at 50 µM in injection mix, shown to enhance HDR up to 13.4-fold [16]. |
| HDR Enhancer (RS-1) | Small molecule that stimulates the RAD51 protein, a key mediator of HDR. | Can provide a modest but significant increase in HDR efficiency [16]. |
Chemical Reprogramming to Enhance HDR
A powerful strategy to boost HDR efficiency is the transient inhibition of the competing NHEJ pathway using small molecules. This approach "chemically reprograms" the embryo's innate DNA repair preferences.
Detailed Protocol: Chemical Enhancement of HDR [16]
- Preparation of Injection Mix:
- Pre-assemble Cas9 RNP complexes by incubating purified Cas9 protein (e.g., 300-500 ng/µL) with sgRNA (e.g., 50-100 ng/µL) at 37°C for 10 minutes.
- Add the repair template (e.g., ssDNA at 100-200 ng/µL) to the RNP complex.
- Add the NHEJ inhibitor NU7441 directly to the injection mix at a final concentration of 50 µM. Alternatively, embryos can be incubated in a solution containing the drug post-injection.
- Microinjection:
- Inject 1-2 nL of the prepared mix into the cytoplasm of one-cell stage zebrafish embryos.
- The use of a fine-glass needle and a microinjection apparatus with a pneumatic picopump is standard.
- Post-Injection Incubation:
- Maintain injected embryos in standard E3 embryo medium at 28.5°C.
- Monitor embryos daily and remove any unviable individuals.
The following diagram illustrates the logical workflow and the mechanistic role of chemical inhibitors in this optimized protocol.
Quantitative Analysis of HDR Efficiency
Rigorous quantification is essential for evaluating the success of HDR protocols. A common pitfall is relying on qualitative assessments (e.g., presence/absence of a reporter), which can mask the true effects of optimization [16]. For instance, a qualitative assay might show that 69% of control embryos and 80% of NU7441-treated embryos have at least one edited cell. However, a quantitative single-cell analysis reveals the dramatic difference: an average of 4.0 ± 3.0 edited cells per embryo in the control versus 53.7 ± 22.1 in the NU7441-treated group [16]. This underscores the importance of using quantitative metrics, such as:
- The number of positively edited cells per embryo in a somatic reporter assay.
- The germline transmission rate, calculated as the percentage of F0 founders that produce F1 offspring with the precise edit.
Discussion and Future Perspectives
The optimization strategies outlined here—employing high-efficiency nucleases, optimizing repair template design, and chemically reprogramming repair pathways—collectively represent a significant advancement in achieving precise genome editing in zebrafish. The integration of these methods can shift HDR from a low-efficiency, sporadic event to a robust and reliable technique. This capability is fundamental for modeling human diseases caused by specific point mutations, for targeted gene tagging, and for functional analysis of regulatory elements. Future directions will likely involve the continued development of next-generation editors, such as base editors and prime editors, which offer alternative routes to precision editing without requiring DSBs or donor templates [23] [5]. Furthermore, refining small-molecule cocktails and template delivery methods will continue to push the boundaries of efficiency and reduce mosaicism, solidifying the zebrafish's role as a versatile and powerful platform for vertebrate functional genomics and preclinical drug development.
Timing and Dosage Considerations for Maximum HDR Efficiency
In zebrafish research, achieving high efficiency in Homology-Directed Repair (HDR) is crucial for precise genome editing applications, including modeling human diseases and functional genomics. HDR enables the incorporation of specific point mutations, insertions, or gene corrections using exogenous donor templates. However, its efficiency is inherently limited because the error-prone Non-homologous end joining (NHEJ) pathway dominates double-strand break (DSB) repair in zebrafish embryos [72]. This technical guide synthesizes current strategies to maximize HDR efficiency by optimizing timing and dosage parameters, providing researchers with actionable methodologies to enhance precision editing outcomes in zebrafish models.
Understanding the HDR Challenge in Zebrafish
The core challenge in zebrafish HDR experimentation stems from the fundamental competition between DNA repair pathways. When CRISPR-Cas9 induces a DSB, the cell's repair machinery can utilize either the precise HDR pathway or the error-prone NHEJ pathway. NHEJ is active throughout the cell cycle and is typically the dominant, faster repair mechanism in zebrafish embryos, resulting in a low baseline HDR efficiency [72] [15]. Consequently, only a small fraction of edited cells undergo precise HDR without intervention.
The timing of HDR is intrinsically linked to the cell cycle, as the pathway is primarily active during the S and G2 phases when homologous sister chromatids are available as natural repair templates [73]. This biological constraint means that successful HDR-based editing depends not only on the presence of a donor template but also on the cell cycle stage of the target cells at the time of editing. Zebrafish embryos present additional practical challenges, including mosaic editing—where injected embryos contain a mixture of precisely edited, imprecisely edited, and unedited cells—and the difficulty of recovering germline transmissions [72].
Chemical Modulation: Dosage and Timing for HDR Enhancement
Strategic use of small molecule inhibitors can shift the DNA repair equilibrium away from NHEJ and toward HDR. The table below summarizes key compounds, their optimal concentrations, and treatment windows identified for zebrafish embryo experiments.
Table 1: Small Molecule Enhancers of HDR in Zebrafish
| Compound | Target/Mechanism | Optimal Concentration | Treatment Timing | Reported Efficacy |
|---|---|---|---|---|
| NU7441 [72] | DNA-PKcs inhibitor (NHEJ) | 50 µM | Co-injection with Cas9 components | 13.4-fold HDR increase |
| RS-1 [72] | RAD51 stimulator (HDR) | 15-30 µM | Co-injection with Cas9 components | Modest, significant increase |
| SCR7 [72] [73] | Ligase IV inhibitor (NHEJ) | Variable | Co-injection with Cas9 components | Conflicting, species-specific effects |
| HDRobust Strategy [74] | Combined NHEJ & MMEJ inhibition | Substance mix | Transient treatment during editing | Up to 93% HDR (median 60%) in human cells |
Among these, NU7441 has demonstrated the most dramatic effect in zebrafish. In a quantitative study, administration of 50 µM NU7441 via co-injection with CRISPR-Cas9 components and donor DNA increased HDR-mediated events up to 13.4-fold compared to DMSO controls, raising the average number of successfully edited cells from 4.0 to 53.7 per embryo [72]. It is critical to note that qualitative assessment (simply noting the presence or absence of edited cells) failed to detect this significant enhancement, underscoring the necessity of quantitative measurement for protocol optimization [72].
The HDRobust strategy, which involves the combined transient inhibition of NHEJ and microhomology-mediated end joining (MMEJ), has shown remarkable success in human cell lines, achieving HDR with high purity (over 91%) and drastically reducing indel formation [74]. While this specific combination has not been fully validated in zebrafish, it represents a cutting-edge approach and a logical next step for testing in this model organism.
Optimized Experimental Protocol for Zebrafish HDR
The following integrated protocol combines the most effective chemical, template, and timing parameters from empirical studies.
Reagent Preparation and Microinjection
- CRISPR Components: Prepare a mix of Cas9 protein (or mRNA) and sgRNA as ribonucleoprotein (RNP) complexes. The use of high-fidelity Cas9 variants can reduce off-target effects [74].
- Donor Template: Design a single-stranded DNA (ssDNA) donor with homology arms. For optimal efficiency, incorporate RAD51-preferred binding sequences (e.g., "TCCCC" motif) at the 5' end of the donor template, as the 5' end tolerates additional sequences better than the sensitive 3' end [75].
- Chemical Enhancer: Add NU7441 to the injection mix at a final concentration of 50 µM [72].
- Microinjection: Inject the combined mixture (RNP, ssDNA donor, and NU7441) into the cytoplasm of one-cell stage zebrafish embryos.
Post-Injection Incubation and Validation
- Maintain injected embryos at approximately 28°C under standard laboratory conditions.
- Analyze editing outcomes at the desired stage (e.g., 72 hours post-fertilization for somatic screening). For germline transmission, raise injected embryos (F0) to adulthood and outcross them to assess F1 progeny.
- Employ quantitative methods to evaluate success. Avoid simple qualitative "yes/no" scoring, as it masks the true efficiency gains from optimization [72]. Use methods such as:
- Sequencing of PCR amplicons from pooled embryos or individual founders.
- Restriction Fragment Length Polymorphism (RFLP) if a new restriction site was introduced.
- Fluorescence-based reporters for rapid, in vivo quantification [72].
Diagram 1: Experimental workflow for enhanced HDR in zebrafish. The process begins at the one-cell stage and incorporates key optimizations to favor precise editing.
Pathway Engineering and Logical Framework
The strategic inhibition of competing repair pathways creates a permissive environment for HDR. The following diagram illustrates the key molecular targets and the logical flow for enhancing HDR efficiency.
Diagram 2: Logical framework for enhancing HDR. The strategy involves simultaneously suppressing the NHEJ and MMEJ pathways while actively stimulating the HDR pathway to achieve high-precision editing.
The Scientist's Toolkit: Essential Reagents for HDR Enhancement
Table 2: Key Research Reagent Solutions for Enhanced HDR
| Reagent / Tool | Function in HDR Enhancement | Example Use Case |
|---|---|---|
| NU7441 [72] | Small molecule inhibitor of DNA-PKcs; shifts repair balance from NHEJ to HDR by blocking a key kinase in the canonical NHEJ pathway. | Co-injected at 50 µM with CRISPR components to achieve a >10-fold increase in HDR efficiency in zebrafish embryos. |
| RAD51-binding ssDNA Donor [75] | Engineered single-stranded donor template containing RAD51-preferred sequences (e.g., "TCCCC" motif) that promotes recruitment to DSB sites. | Used as a repair template to augment donor affinity for RAD51, enhancing HDR efficiency across multiple genomic loci. |
| HDRobust Substance Mix [74] | A combination of inhibitors targeting both NHEJ and MMEJ pathways, forcing DSB repair through HDR. | Transient treatment of cells during editing to achieve median 60% HDR efficiency with high outcome purity in human cells. |
| Quantitative Fluorescence Reporter [72] | A stable transgenic system (e.g., BFP-to-tdTomato conversion) that enables real-time, in vivo quantification of HDR events at single-cell resolution. | Critical for validating the efficacy of chemical and molecular enhancements in live zebrafish embryos. |
Maximizing HDR efficiency in zebrafish is an achievable goal that requires a multi-faceted approach targeting the core biology of DNA repair. Key considerations include the use of specific small molecule inhibitors like NU7441 at a critical dosage (50 µM) applied during the microinjection stage, the engineering of donor templates with functional elements such as RAD51-binding sequences, and the implementation of robust quantitative assays to measure success beyond qualitative assessments. By integrating these timing and dosage strategies, researchers can significantly improve the precision and efficiency of genome editing in zebrafish, accelerating the creation of accurate disease models and functional genomic studies.
Addressing Mosaicism in G0 Embryos and Germline Transmission
In zebrafish CRISPR-Cas9 research, mosacism describes the phenomenon where a genetically modified G0 generation embryo contains a mixture of cells with different genotypes. This occurs when the initial CRISPR-induced double-strand break (DSB) is repaired at different time points or through different mechanisms in various daughter cells after the one-cell stage [38] [23]. Within the broader thesis of DSB repair mechanisms in zebrafish—specifically the competition between error-prone non-homologous end joining (NHEJ) and precise homology-directed repair (HDR)—understanding and controlling mosaicism becomes paramount for generating high-quality mutant lines and achieving reproducible experimental outcomes [11] [5]. This technical guide provides detailed methodologies for addressing this challenge, enabling researchers to minimize, characterize, and successfully transmit genetic edits through the germline.
The Molecular Basis of Mosaicism in G0 Zebrafish
Mosaicism arises from the timing and mechanism of DNA repair following a CRISPR-Cas9-induced DSB. The key factor is the persistence of CRISPR components after the first embryonic cell division.
- Post-Zygotic Editing: If the Cas9 nuclease and guide RNA remain active beyond the first few cell divisions, they can induce DSBs in different cells at different cycles. Each of these independent DSB events is then repaired separately, predominantly via the NHEJ pathway, leading to a variety of indels within a single animal [38] [23].
- Repair Mechanism Competition: The cellular decision to repair a break via NHEJ or HDR is another source of variation. NHEJ is active throughout the cell cycle and is the dominant pathway in zebrafish embryos, often resulting in a spectrum of indel mutations. HDR, which requires a repair template and is restricted to the S/G2 phases, is less efficient. Consequently, even with a repair template present, a mosaic embryo may contain cells with unedited wild-type alleles, NHEJ-induced knockouts, and a smaller subset of cells with precise HDR edits [11] [5].
The following diagram illustrates the cellular decision points that lead to mosaicism following a CRISPR-Cas9 injection.
Quantitative Analysis of Mosaicism and Editing Efficiencies
The efficiency of genetic edits and the rate of germline transmission are critical for evaluating experimental success. The data below, compiled from zebrafish studies, provides benchmarks for HDR and base editing approaches.
Table 1: Homology-Directed Repair (HDR) Efficiency and Protocol Variables in Zebrafish
| Target Gene | HDR Efficiency (%) | Key Protocol Variables | Reference |
|---|---|---|---|
| tyrosinase (tyr) | Widely reported | Standard protocol; high-cutting efficiency sgRNA | [11] |
| gata1a | Success reported | Use of long ssDNA template | [11] |
| ntla | Success reported | dsDNA plasmid template | [11] |
| kdrl, vegfaa | Success reported | Cut site within 20 nt of target; altered PAM | [11] |
| Consensus Best Practice | Increases success rate | dsDNA template, 5' biotinylation, NHEJ inhibition | [11] |
Table 2: Base Editing Efficiencies and Evolution in Zebrafish
| Base Editor System | Typical Editing Efficiency | Key Features and Improvements | Reference |
|---|---|---|---|
| BE3 | 9.25% - 28.57% | First application in zebrafish; established feasibility | [5] |
| HF-BE3 | Comparable to BE3 | 37-fold reduction in off-target effects | [5] |
| AncBE4max | ~3x BE3 efficiency | Codon-optimized for zebrafish; ~90% efficiency at some loci | [5] |
| CBE4max-SpRY | Up to 87% | "Near PAM-less"; dramatically expanded target scope | [5] |
Experimental Protocols for Minimizing and Harnessing Mosaicism
Optimized Microinjection Protocol for Reducing Mosaicism
This protocol is designed to maximize editing efficiency in the founder (G0) generation and minimize mosaicism by ensuring CRISPR activity is confined to the earliest stages of development.
- CRISPR Component Preparation: Synthesize high-purity, capped, polyadenylated Cas9 mRNA in vitro or use purified Cas9 protein. Simultaneously, transcribe single-guide RNA (sgRNA) in vitro. For HDR, prepare a single-stranded oligodeoxynucleotide (ssODN) or double-stranded DNA (dsDNA) plasmid repair template with homologous arms flanking the DSB site [11] [5] [23].
- Injection Mixture Optimization:
- For NHEJ Knockouts: Combine 150-300 ng/μL of Cas9 protein (as RNP) with 30-50 ng/μL of sgRNA in nuclease-free water. RNP complexes are highly stable and can reduce mosaicism.
- For HDR Knock-ins: Include the repair template at a concentration of 50-100 ng/μL. For ssODN templates, 5' biotinylation can enhance HDR efficiency. Consider adding an NHEJ inhibitor (e.g., SCR7) to the injection mix to favor HDR [11].
- Microinjection Procedure: Using a microinjection rig, inject 1-2 nL of the mixture directly into the cytoplasm of the one-cell stage zebrafish embryo, immediately after fertilization. The rapid first division of zebrafish embryos (approximately 15 minutes) makes precise timing critical [23].
Protocol for G0 Mosaic Founder Analysis and Germline Transmission
This workflow outlines the steps from raising injected embryos to establishing a stable mutant line.
- Raise Injected Embryos: Maintain injected G0 embryos under standard conditions. Analyze a subset at 2-3 days post-fertilization (dpf) for potential somatic edits. For genes affecting visible traits (e.g., tyrosinase for pigmentation), the degree of mosaicism can be visually assessed [38] [23].
- Outcross G0 Founders: Upon reaching sexual maturity (approximately 3 months), outcross individual G0 fish to wild-type partners. This is a critical step, as the G0 fish are mosaic and only a subset of their gametes will carry the mutation.
- Screen the F1 Progeny: At 2-3 dpf, collect fin clips or whole larvae from the F1 generation for genotyping. Screening methods include:
- Establish Stable Lines: Identify F1 larvae that are heterozygous for the desired mutation. Raise these to adulthood and intercross them to generate homozygous F2 populations, which will be non-mosaic and can be used for phenotypic analysis.
The following diagram summarizes this multi-generational workflow.
The Scientist's Toolkit: Essential Reagents and Solutions
Table 3: Key Research Reagent Solutions for Zebrafish Genome Editing
| Reagent / Solution | Function and Description | Technical Considerations |
|---|---|---|
| Cas9 Protein (RNP Complex) | Purified Cas9 protein pre-complexed with sgRNA. Directly injected for immediate activity. | Reduces mosaicism by enabling rapid degradation; more consistent than mRNA [5] [23]. |
| Modified sgRNAs | sgRNAs with chemical modifications (e.g., 2'-O-methyl analogs) at terminal nucleotides. | Increases stability and resistance to nucleases, potentially enhancing cutting efficiency [5]. |
| Single-Stranded ODN Template | Single-stranded DNA oligonucleotide with homologous arms for HDR. | Ideal for introducing point mutations or short tags; high molar concentration can be used [11]. |
| NHEJ Inhibitors (e.g., SCR7) | Small molecules that inhibit key enzymes in the NHEJ pathway. | Shifts repair balance towards HDR; can be added to injection mix or incubated with embryos [11]. |
| ACEofBASEs Platform | Online bioinformatic platform for sgRNA design and off-target prediction. | Specifically tailored for base editor design in zebrafish and related models [5]. |
Advanced Strategies: Base Editing as an Alternative to HDR
For applications requiring single-nucleotide changes, base editing offers a powerful alternative to HDR that avoids DSBs and can significantly reduce mosaicism. Base editors are fusion proteins that combine a catalytically impaired Cas nuclease (nickase) with a deaminase enzyme.
- Cytosine Base Editors (CBEs): Convert a C•G base pair to a T•A pair within a defined editing window. They fuse dCas9 or nCas9 to a cytidine deaminase enzyme (like APOBEC1) [5].
- Adenine Base Editors (ABEs): Convert an A•T base pair to a G•C pair. They fuse nCas9 to an engineered adenine deaminase (TadA) [5].
The mechanism of base editors, which involves chemical conversion without a DSB, avoids the activation of error-prone NHEJ and the complex homology search of HDR. This results in higher efficiency and purity of edits, with significantly lower rates of indels and mosaicism in the resulting G0 animals [5]. The following diagram details the mechanistic steps of base editing.
Troubleshooting Common Pitfalls in Precision Genome Editing
Precision genome editing, powered by tools like CRISPR/Cas9, has revolutionized biological research and therapeutic development. In zebrafish models, these technologies enable sophisticated functional genomics and disease modeling. However, achieving precise edits remains challenging due to the complex interplay of DNA repair mechanisms, primarily the error-prone non-homologous end joining (NHEJ) and the precise homology-directed repair (HDR) pathways. This technical guide examines common pitfalls in precision genome editing and provides evidence-based troubleshooting strategies to enhance experimental success.
Core Challenge: Balancing DNA Repair Pathways
The fundamental challenge in precision editing lies in the cellular competition between NHEJ and HDR pathways. NHEJ is active throughout the cell cycle and dominates in most cells, while HDR is restricted to late S and G2 phases, making it inherently less efficient [76]. This imbalance often results in low HDR efficiency and a high frequency of unintended mutations.
Table 1: DNA Repair Pathways in Genome Editing
| Pathway | Mechanism | Cell Cycle Phase | Outcomes | Advantages/Disadvantages |
|---|---|---|---|---|
| Non-Homologous End Joining (NHEJ) | Direct ligation of broken ends | All phases | Small insertions/deletions (indels) | Fast but error-prone |
| Homology-Directed Repair (HDR) | Uses template for precise repair | Late S/G2 | Precise nucleotide changes | Precise but inefficient |
| Microhomology-Mediated End Joining (MMEJ) | Uses microhomologous sequences | Mitosis | Deletions flanked by microhomology | Predictable mutational signatures |
Pitfall 1: Unintended Structural Variations
The Problem
Beyond small indels, CRISPR/Cas9 can induce large-scale structural variations (SVs) including kilobase- to megabase-scale deletions, chromosomal translocations, and chromothripsis [76]. These undervalued genomic alterations raise substantial safety concerns for clinical translation.
Troubleshooting Strategies
- Advanced Detection Methods: Employ long-read sequencing and structural variation detection methods (e.g., CAST-Seq, LAM-HTGTS) rather than standard short-read amplicon sequencing, which often misses large deletions [76].
- Careful Use of DNA Repair Modulators: Avoid DNA-PKcs inhibitors like AZD7648, which can increase megabase-scale deletions and chromosomal translocations by a thousand-fold [76].
- Alternative HDR Enhancement: Consider transient inhibition of 53BP1, which does not affect translocation frequencies, or co-inhibition of DNA-PKcs and DNA polymerase theta (POLQ), which shows protective effects against kilobase-scale deletions [76].
Pitfall 2: Inefficient HDR in Zebrafish Models
The Problem
In zebrafish, achieving efficient HDR-mediated precise edits remains challenging, particularly for creating disease models requiring specific point mutations.
Troubleshooting Strategies
- Base Editing Technology: Utilize cytosine base editors (CBEs) and adenine base editors (ABEs) that enable direct nucleotide conversion without double-strand breaks [4].
- Optimized Editor Variants: Implement zebrafish-codon optimized editors like AncBE4max, which shows approximately threefold higher editing efficiency compared to BE3 systems [4].
- Nuclear Localization Enhancement: Incorporate "hei-tag" (high-efficiency tag) systems that combine a Myc tag with an optimized nuclear localization signal to improve nuclear import and editing efficiency [4].
Table 2: Advanced Base Editing Systems for Zebrafish
| Editor System | Editing Type | Efficiency | Key Features | Applications |
|---|---|---|---|---|
| BE3 | C:G to T:A | 9.25%-28.57% | First-generation CBE | Oculocutaneous albinism (OCA) model |
| AncBE4max | C:G to T:A | ~3× BE3 | Codon-optimized for zebrafish | Cancer modeling (tp53) |
| CBE4max-SpRY | C:G to T:A | Up to 87% | Near PAM-less editing | Broad target range |
| zhyA3A-CBE5 | C:G to T:A | High efficiency | Extended editing window (C3-C16) | High-precision applications |
Pitfall 3: Inaccurate Assessment of Editing Outcomes
The Problem
Traditional quantification methods often misrepresent true editing efficiencies. Short-read amplicon sequencing fails to detect large deletions that remove primer-binding sites, leading to overestimation of HDR rates and underestimation of indels [76].
Troubleshooting Strategies
- Method Selection: Use targeted amplicon sequencing (AmpSeq) as the gold standard for quantifying editing efficiency rather than less accurate methods like T7E1 or RFLP assays [77].
- Integrated Analysis Pipelines: Implement specialized bioinformatic tools like the Integrated Classification Pipeline (ICP), which provides single-allele resolution of repair outcomes and can distinguish between NHEJ, MMEJ, and HDR events within the same sample [40].
- Developmental Timing Considerations: Account for developmental progression in repair pathway usage; MMEJ often predominates during early rapid mitotic cycles, switching to NHEJ and later to HDR as development proceeds [40].
Experimental Protocols for Enhanced Precision
Zebrafish Base Editing Protocol
- sgRNA Design: Use online tools like CRISPOR with zebrafish-specific parameters. For base editing, ensure the target base falls within the editor's activity window (typically positions 4-8 from PAM for CBEs) [4] [78].
- Editor Delivery: Prepare ribonucleoprotein (RNP) complexes by mixing purified base editor protein with sgRNA, then microinject into zebrafish embryos at the 1-cell stage.
- Efficiency Validation: Extract genomic DNA from injected embryos and analyze target sites using amplicon sequencing. Calculate editing efficiency as the percentage of sequencing reads containing desired base conversions.
- Off-Target Assessment: Use Cas-OFFinder or similar tools to predict potential off-target sites, followed by high-throughput sequencing of top candidate sites [4].
HDR Optimization Protocol for Zebrafish
- Repair Template Design: Design single-stranded oligodeoxynucleotides (ssODNs) with 30-40 nt homology arms flanking the desired edit. Place the edit as close as possible to the Cas9 cut site (within 10 bp) [78].
- Cell Cycle Synchronization: Optimize injection timing to target late S/G2 phase when HDR is most active.
- Dosage Optimization: Co-inject Cas9 mRNA, sgRNA, and ssODN repair template at molar ratios of 1:2:10-20 to favor HDR over NHEJ.
- Validation: Use droplet digital PCR (ddPCR) for sensitive detection of HDR events, followed by Sanger sequencing of individual clones to confirm precise edits [79].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Precision Genome Editing in Zebrafish
| Reagent/Category | Specific Examples | Function | Application Notes |
|---|---|---|---|
| Cas9 Variants | SpCas9, HiFi Cas9, Cas9 nickase | DNA cleavage or nicking | High-fidelity variants reduce off-targets; nickase enables paired nicking |
| Base Editors | BE4max, AncBE4max, ABE8e | Direct nucleotide conversion without DSBs | Optimal for point mutations; zebrafish-codon optimized versions available |
| Delivery Methods | Microinjection, electroporation | Introducing editing components | RNP delivery reduces off-target effects |
| HDR Templates | ssODNs, dsDNA donors | Template for precise repair | Asymmetric designs with 30-40 nt homology arms recommended |
| Detection Tools | AmpSeq, ddPCR, ICP analysis | Quantifying editing outcomes | AmpSeq is gold standard; ddPCR for sensitive HDR detection |
| Bioinformatic Tools | CRISPOR, Cas-OFFinder, ICE | Design and analysis | Predict editing efficiency and potential off-target sites |
Pathway and Workflow Diagrams
Figure 1: DNA Repair Pathway Competition After CRISPR Editing. Double-strand breaks are repaired through competing pathways, with developmental stage influencing pathway choice.
Figure 2: Comprehensive Workflow for Precision Genome Editing in Zebrafish.
Successful precision genome editing in zebrafish requires careful navigation of DNA repair pathway competition and implementation of strategic solutions to common pitfalls. By leveraging base editing technologies, employing appropriate detection methods for structural variations, and timing interventions to align with developmental repair pathway progression, researchers can significantly improve precision editing outcomes. As the field advances, continued refinement of these strategies will enhance both basic research and therapeutic applications in zebrafish models.
Ensuring Accuracy: Validation Methods and Standardization in Zebrafish Models
Comprehensive Guide to Detecting and Quantifying Editing Efficiency
The zebrafish (Danio rerio) has emerged as a premier vertebrate model organism for functional genomics and disease modeling, owing to attributes such as high fecundity, external embryonic development, and significant genetic similarity to humans [23]. The advent of CRISPR-Cas technologies has revolutionized targeted genome editing in this model, enabling precise investigations into gene function. The core principle of CRISPR-Cas9 editing involves creating a double-strand break (DSB) at a specific genomic locus, which then activates the cell's endogenous DNA repair mechanisms [80]. The two primary competing pathways for repairing these breaks are non-homologous end joining (NHEJ) and homology-directed repair (HDR) [11].
NHEJ is an error-prone process that directly ligates broken DNA ends, often resulting in small insertions or deletions (indels) that can disrupt gene function, making it the preferred pathway for generating knockout mutants [23]. In contrast, HDR uses a homologous DNA template to repair the break accurately. When researchers provide an exogenous donor template with homology arms, this pathway can be harnessed to introduce precise mutations or insert desired sequences, such as epitope tags or fluorescent reporters, in a process known as knock-in [81]. However, a significant challenge in the field is that NHEJ is the dominant pathway in most cells, leading to low efficiencies of precise HDR-mediated editing compared to stochastic NHEJ outcomes [17] [11]. This guide details the methodologies for detecting and quantifying the outcomes of both repair pathways, with particular emphasis on improving the success rates of precise genome modifications in zebrafish.
Core DSB Repair Pathways and Editing Outcomes
Understanding the complex interplay between different DNA repair pathways is crucial for interpreting genome editing outcomes. While NHEJ and HDR are the major players, alternative pathways significantly influence editing results.
Non-Homologous End Joining (NHEJ): This pathway functions throughout the cell cycle and is favored in zebrafish embryos. It requires no template and rapidly rejoins DSB ends. The error-prone nature of NHEJ frequently leads to indels, which are ideal for creating gene knockouts. Inhibiting NHEJ (e.g., with Alt-R HDR Enhancer V2) has been shown to increase HDR efficiency by approximately 3-fold in some cell models, but it is not sufficient to completely suppress all non-HDR repair events [17].
Homology-Directed Repair (HDR): HDR operates primarily in the S and G2 phases of the cell cycle and requires a homologous template for precise repair. In genome editing, this allows for the introduction of specific nucleotide changes or the insertion of exogenous DNA sequences (e.g., for fluorescent protein tags). The efficiency of HDR is notably low, and it competes unfavorably with NHEJ and other pathways [11] [81].
Microhomology-Mediated End Joining (MMEJ) and Single-Strand Annealing (SSA): These are alternative non-HDR repair pathways. MMEJ relies on short microhomologous sequences (2-20 nt) flanking the break and often results in deletions. Inhibiting its key effector, POLQ (e.g., with ART558), can reduce large deletions and complex indels, thereby increasing perfect HDR frequency [17]. SSA uses Rad52-dependent annealing of longer homologous sequences and can lead to various imprecise donor integration patterns, including asymmetric HDR, where only one side of the donor integrates correctly. Suppressing SSA (e.g., with D-I03) can reduce these imprecise events [17].
The following diagram illustrates the complex interplay between these pathways following a CRISPR-induced double-strand break and their potential outcomes.
Detection and Quantification Methodologies
A range of techniques is available for detecting and quantifying genome editing outcomes, from simple validation of indels to comprehensive analyses of complex integration patterns.
Detecting NHEJ-Mediated Indels
The following table summarizes the common methods used to detect the error-prone outcomes of NHEJ and other non-HDR pathways.
| Method | Key Principle | Typical Application | Advantages | Limitations |
|---|---|---|---|---|
| T7 Endonuclease I (T7E1) Assay [6] | Detects mismatches in heteroduplex DNA formed by annealing wild-type and mutant strands. | Initial screening for presence of indels at a target locus. | Rapid, low-cost, does not require specialized equipment. | Semi-quantitative, does not provide sequence information. |
| Restriction Fragment Length Polymorphism (RFLP) | Loss or gain of a restriction enzyme site due to editing. | Quick assessment of editing efficiency when a restriction site is affected. | Quantitative for specific changes, low cost. | Only works if the edit alters a restriction site. |
| Sanger Sequencing with Decomposition [81] | PCR amplification followed by Sanger sequencing; trace decomposition software identifies multiple sequences in a sample. | Identifying specific indels and estimating their frequency in a mosaic pool. | Provides sequence-level detail, accessible. | Lower throughput and sensitivity compared to NGS. |
| Short-Read Next-Generation Sequencing (NGS) [82] [23] | High-throughput sequencing of PCR amplicons from the target region; bioinformatic analysis maps reads and quantifies indels. | Comprehensive quantification of diverse indel patterns and their frequencies in a sample. | Highly sensitive, quantitative, provides full sequence context. | Higher cost and bioinformatics burden; read-length constraints challenge analysis of large insertions. |
Quantifying HDR and Precise Integration
Precise knock-in requires more stringent validation than knockout. The table below outlines methods specifically for confirming and quantifying HDR events.
| Method | Key Principle | Typical Application | Information Gained |
|---|---|---|---|
| Long-Range PCR & Sanger Sequencing [81] | PCR with one primer outside the homology arm and one within the inserted sequence, followed by sequencing. | Validating correct 5' or 3' junction integrity in candidate founders. | Confirms in-frame integration and precise junction sequences. |
| Expression Analysis (RT-PCR/IHC) [81] | Detects expression of the knock-in allele via RT-PCR (for tags) or immunohistochemistry (for epitopes). | Functional validation of the edited allele in somatic tissue or stable lines. | Confirms transcription and translation of the fusion protein, and correct subcellular localization. |
| Fluorescence-Activated Cell Sorting (FACS) [17] | Measures fluorescence in cells where a fluorescent protein (e.g., mNeonGreen) has been knocked in. | Rapid, quantitative assessment of knock-in efficiency in somatic cells. | Provides a quantitative percentage of cells with successful protein-tag knock-in. |
| Long-Read Amplicon Sequencing (PacBio) [82] [17] | Sequencing long PCR amplicons spanning the entire integration site with technologies like PacBio. | Comprehensive analysis of all editing outcomes, including perfect HDR, imprecise integration, and indels. | Quantifies all possible repair outcomes simultaneously, even identifying complex patterns like asymmetric HDR [17]. |
The typical workflow for a comprehensive analysis of editing outcomes, from embryo injection to final validation, integrates several of these techniques, as shown in the following diagram.
Quantitative Data and Optimization Strategies
Recent studies have provided quantitative insights into the efficiency of different editing strategies and the impact of various optimization parameters.
Efficiency of Advanced Editing Tools
Beyond standard CRISPR-Cas9, new editors offer alternative pathways to precision.
Prime Editing (PE): A comparison of nickase-based PE2 and nuclease-based PEn editors in zebrafish showed distinct strengths. PE2 was more effective for single-base substitutions, achieving a precision score of 40.8% compared to 11.4% for PEn. In contrast, PEN was more efficient at inserting short DNA fragments (3-30 bp) via NHEJ or homology annealing, making it more suitable for inserting sequences like stop codons or nuclear localization signals [6].
Base Editing (BE): Base editors enable direct nucleotide conversion without DSBs, bypassing the HDR/NHEJ competition entirely. Cytosine Base Editors (CBEs) achieve C:G to T:A conversions, while Adenine Base Editors (ABEs) achieve A:T to G:C conversions. New variants like AncBE4max have shown a threefold increase in editing efficiency compared to the BE3 system in zebrafish. The development of "near PAM-less" editors like CBE4max-SpRY further expands the targetable scope, with reported efficiencies up to 87% at some loci [5].
Key Parameters for Optimizing HDR Efficiency
Optimizing HDR is critical for improving knock-in success. The following table synthesizes key findings from recent research.
| Parameter | Optimal Condition / Strategy | Impact on HDR Efficiency |
|---|---|---|
| Repair Template | Chemically modified single-stranded oligodeoxynucleotides (ssODNs) or dsDNA with 5' end-protection (e.g., AmC6) [82] [51]. | Chemically modified templates outperform plasmid-based templates. 5'AmC6 modification on PCR-generated dsDNA donors prevents degradation and increases integration efficiency >5-fold [51]. |
| Homology Arm (HA) Length | Asymmetric arms (e.g., 40 bp left, 80 bp right) [81] or short (~50 bp) arms with 5' modifications [51]. | Long HAs (>500 bp) were traditionally used, but short, modified HAs can achieve high efficiency, simplifying template production. |
| Cut-to-Insert Distance | DSB cut site should be within 20 nucleotides of the target insertion site [11]. | HDR efficiency is highly dependent on proximity; greater distances drastically reduce efficiency. |
| PAM Disruption | The Protospacer Adjacent Motif (PAM) site in the donor template should be altered by synonymous mutations [51] [11]. | Prevents re-cleavage of the successfully edited allele, allowing it to persist. |
| Nuclease Form | Pre-assembled Cas9/gRNA Ribonucleoprotein (RNP) complexes [51] [81]. | Leads to rapid genome editing, increases the probability of early integration and high mosaicism in F0, which correlates with germline transmission. |
| Pathway Modulation | Co-treatment with NHEJ inhibitors (e.g., Alt-R HDR Enhancer V2) [17] and/or MMEJ/SSA inhibitors (ART558, D-I03) [17]. | NHEJi can increase knock-in efficiency ~3-fold. Combining NHEJ inhibition with MMEJ or SSA suppression further reduces imprecise repair, enhancing perfect HDR. |
Using optimized parameters, including chemically modified templates and Cas9 RNP delivery, one study consistently achieved germline founder rates of greater than 20% for precise insertions across four different loci in zebrafish [82]. Another protocol using 5'AmC6-modified dsDNA donors reported founder rates from 11.5% to 20% in the F1 generation [51].
Experimental Protocols
Below is a detailed protocol for a knock-in experiment in zebrafish, incorporating best practices for detecting and quantifying editing efficiency.
Detailed Protocol: 3' Knock-In for Epitope Tagging
This protocol is adapted from successful methods used to generate a MYC-tagged Sox11a zebrafish line [81] and a cloning-free 3' knock-in strategy [51].
1. Design and Preparation of Reagents:
- gRNA Design: Select a target site in the 3' UTR just downstream of the stop codon (for C-terminal tagging) or in the 5' UTR just upstream of the start codon (for N-terminal tagging). Use design tools (e.g., IDT Alt-R CRISPR HDR Design Tool, CRISPOR) to ensure high on-target and low off-target scores [81].
- Donor Template Design:
- For a MYC tag knock-in at the N-terminus, design a single-stranded or double-stranded donor template containing the MYC tag sequence followed by a flexible linker (e.g., GGSGG), placed immediately after the start codon [81].
- Incorporate asymmetric homology arms (e.g., 40 bp left arm, 80 bp right arm) flanking the insertion sequence.
- Introduce synonymous mutations in the PAM sequence within the homology arm to prevent re-cleavage.
- For dsDNA templates, use 5' AmC6-modified primers during PCR amplification to increase integration efficiency [51].
- RNP Complex Assembly: Complex purified Cas9 protein (e.g., Alt-R S.p. Cas9 Nuclease V3) with the synthesized gRNA (crRNA:tracrRNA duplex) to form the RNP complex. Pre-assembling the RNP increases editing efficiency and reduces off-target effects [51] [81].
2. Microinjection:
- Injection Mixture: Combine the following in nuclease-free water:
- 250 pg gRNA
- 500 pg Cas9 protein (as RNP complex)
- 37.5 pg HDR donor template
- Phenol red tracer dye
- Procedure: Microinject 1-2 nL of the mixture directly into the cytoplasm of one-cell stage zebrafish embryos [81].
3. Screening and Validation (A Tiered Approach):
Step 1: Somatic Screening (in F0 embryos)
- Genomic DNA Extraction: At 24-48 hours post-fertilization (hpf), pool 5-10 injected embryos and extract gDNA using a simple alkaline lysis method (e.g., incubate in 50mM NaOH at 95°C, then neutralize with Tris-HCl) [81].
- PCR Amplification: Perform PCR using one primer outside a homology arm and one primer within the inserted tag sequence. This ensures amplification only occurs if integration is successful.
- Analysis: Run PCR products on a high-percentage polyacrylamide or agarose gel. A band of the expected size indicates potential HDR. For quantitative assessment of somatic integration efficiency, droplet digital PCR (ddPCR) can be used [81].
Step 2: Founder Identification (in F0 adults)
- Raise mosaic F0 injected embryos to adulthood.
- Outcross individual F0 fish to wild-type partners.
- Collect ~20-30 F1 embryos from each cross, extract gDNA individually, and perform the junction PCR as in Step 1. A founder (F0) is identified if a specific fraction of its F1 progeny carries the knock-in allele.
Step 3: Molecular Validation of Positive Founders
- Sequencing: Gel-purify the positive PCR product and subject it to Sanger sequencing to confirm perfect, in-frame integration at both the 5' and 3' junctions [51] [81].
- Functional Validation:
- RT-PCR: Isolate RNA from positive F1 embryos, synthesize cDNA, and perform PCR with primers spanning the endogenous gene and the inserted tag to confirm transcription of the fusion mRNA [81].
- Immunohistochemistry (IHC): Fix and section positive F1 embryos. Perform IHC using an antibody against the epitope tag (e.g., anti-MYC). Correct subcellular localization of the signal confirms the fusion protein is expressed and functions appropriately [81].
- Western Blot: Detect the fusion protein using an antibody against the tag or the endogenous protein to confirm expected molecular weight.
Step 4: Comprehensive Outcome Quantification (Optional)
- For a full profile of all editing outcomes (perfect HDR, imprecise HDR, NHEJ indels), perform long-read amplicon sequencing (PacBio) on a pool of injected F0 embryos or a heterozygote F1 carrier [82] [17]. Use computational frameworks like "knock-knock" to classify and quantify the sequencing reads [17].
The Scientist's Toolkit: Essential Reagents for Knock-In
| Reagent / Tool | Function | Example Products / Notes |
|---|---|---|
| Cas9 Nuclease | Creates a site-specific double-strand break in the genome. | Alt-R S.p. Cas9 Nuclease V3; use as mRNA or, preferably, as purified protein in RNP complexes. |
| Synthetic gRNA | Guides the Cas9 protein to the specific target genomic sequence. | Alt-R crRNA and tracrRNA (IDT); chemically modified for enhanced stability. |
| HDR Donor Template | Serves as the repair template for precise integration of the desired sequence. | Chemically modified ssODNs or dsDNA Donor Blocks (IDT); 5'AmC6 modification recommended for dsDNA. |
| NHEJ Inhibitor | Suppresses the competing error-prone NHEJ pathway to favor HDR. | Alt-R HDR Enhancer V2; added to the injection mix or used to treat embryos post-injection. |
| Pathway-Specific Inhibitors | Suppresses alternative repair pathways (MMEJ, SSA) to enhance precise HDR. | ART558 (POLQ/MMEJ inhibitor), D-I03 (Rad52/SSA inhibitor); used for research to probe pathway interactions [17]. |
| Bioinformatics Tools | For designing gRNAs, analyzing sequencing data, and predicting off-target sites. | IDT Alt-R HDR Design Tool, CRISPOR, Cas-OFFinder, knock-knock classification framework [17] [81]. |
Accurately detecting and quantifying genome editing efficiency is fundamental to advancing functional genomics and disease modeling in zebrafish. As this guide illustrates, moving beyond simple indel detection to a comprehensive analysis of all repair outcomes—especially precise HDR—requires a combination of sophisticated molecular tools and a deep understanding of DNA repair pathway dynamics. By employing optimized reagents such as RNP complexes and chemically modified donor templates, strategically modulating DNA repair pathways with inhibitors, and leveraging the power of long-read sequencing for quantification, researchers can significantly improve the success rates of precise genome engineering. The continued refinement of these protocols and the adoption of newer technologies like base and prime editing promise to further solidify the zebrafish's role in modeling human disease and accelerating drug discovery.
In zebrafish research, the analysis of CRISPR-Cas9-induced mutations is pivotal for advancing our understanding of double-strand break (DSB) repair mechanisms, particularly non-homologous end joining (NHEJ) and homology-directed repair (HDR). The choice of analytical method significantly impacts the accuracy and depth of repair outcome characterization. This technical guide provides a comparative analysis of three prominent assessment methods—TIDE (Tracking of Indels by DEcomposition), ICE (Inference of CRISPR Edits), and Illumina Sequencing—within the context of zebrafish DSB repair research. We evaluate their applications, limitations, and performance in deciphering the complex landscape of NHEJ and HDR events, providing researchers with a framework to select the optimal tool for their experimental needs.
DNA Repair Pathways in Zebrafish
In zebrafish, as in other vertebrates, the repair of CRISPR-Cas9-induced double-strand breaks occurs primarily through two main pathways: the error-prone Non-Homologous End Joining (NHEJ) and the precise Homology-Directed Repair (HDR) [10]. The choice between these pathways has profound implications for genetic outcomes. NHEJ directly rejoins broken DNA ends without a template, often resulting in small insertions or deletions (indels) that disrupt gene function—making it ideal for gene knockout studies. In contrast, HDR requires a homologous DNA template to accurately repair the break, enabling precise genetic modifications such as gene knock-ins or specific nucleotide substitutions [10].
Recent research has revealed that DSB repair in zebrafish exhibits a developmental progression, with microhomology-mediated end joining (MMEJ) predominant during early rapid mitotic cell cycles, later switching to distinct NHEJ subsets, and finally to HDR-based gene conversion events [40]. This temporal regulation of repair pathway usage underscores the importance of sensitive detection methods that can capture the full spectrum of editing outcomes across developmental stages.
TIDE (Tracking of Indels by DEcomposition)
Principle of Operation: TIDE decomposes Sanger sequencing chromatograms from edited samples by computationally subtracting the wild-type sequence signal. The algorithm then identifies the composition and frequency of indels in the mixed population [40].
Workflow:
- PCR amplification of the target region from edited and control samples
- Sanger sequencing of amplified products
- Upload of sequencing chromatogram files to the TIDE web tool
- Algorithmic decomposition of complex sequence traces into individual indel components
- Quantitative reporting of indel frequencies and types
ICE (Inference of CRISPR Edits)
Principle of Operation: ICE uses Sanger sequencing data similar to TIDE but employs a different computational approach that compares edited sample sequences to wild-type references, generating a synthetic profile of editing outcomes and providing an ICE score that represents overall editing efficiency [83].
Workflow:
- PCR amplification and Sanger sequencing of target loci
- Sequencing data input into ICE analysis software (web-based or standalone)
- Alignment of sample sequences to reference wild-type sequence
- Computational inference of indel combinations that best explain the observed sequence data
- Generation of editing efficiency score and indel spectrum
Illumina Sequencing
Principle of Operation: Next-generation sequencing (NGS) on the Illumina platform provides a comprehensive, high-resolution view of editing outcomes by sequencing millions of DNA molecules in parallel, enabling direct observation and quantification of all mutation types at single-allele resolution [40] [83].
Workflow:
- PCR amplification of target regions with barcoded primers for multiplexing
- Library preparation and quality control
- High-throughput sequencing on Illumina platforms
- Bioinformatic processing including demultiplexing, quality filtering, and alignment
- Variant calling and annotation using specialized pipelines (e.g., Integrated Classification Pipeline)
Comparative Performance Analysis
Technical Specifications and Capabilities
Table 1: Key Characteristics of CRISPR Assessment Methods
| Parameter | TIDE | ICE | Illumina Sequencing |
|---|---|---|---|
| Detection Principle | Sanger trace decomposition | Sanger sequence inference | Direct nucleotide reading |
| Read Type | Indirect computational inference | Indirect computational inference | Direct observation |
| Multiplexing Capacity | Low (single samples) | Low (single samples) | High (hundreds of samples) |
| Sensitivity Threshold | ~1-5% allele frequency | ~1-5% allele frequency | <0.1% allele frequency |
| Quantitative Accuracy | Moderate | High (r=0.90 vs. NGS) [83] | Very high (gold standard) |
| Indel Size Detection | Limited for large indels | Limited for large indels | Comprehensive |
| Complex Editing Detection | Poor for simultaneous edits | Moderate for simultaneous edits | Excellent for complex patterns |
| HDR Detection Capability | Limited | Limited | Comprehensive |
| Turnaround Time | Hours | Hours | Days |
| Cost per Sample | Low | Low | High |
Performance in Zebrafish Research Applications
Table 2: Method Performance in Zebrafish DSB Repair Analysis
| Research Application | TIDE | ICE | Illumina Sequencing |
|---|---|---|---|
| NHEJ Efficiency Quantification | Moderate | Good (correlates with NGS) [83] | Excellent |
| HDR Efficiency Quantification | Poor | Poor | Excellent [19] |
| Allelic Complexity Resolution | Limited to major alleles | Moderate for mixed populations | Complete single-allele resolution [40] |
| MMEJ Identification | Poor | Poor | Excellent with specialized analysis [40] |
| Large Insertion Detection | Poor | Poor | Excellent |
| Germline Transmission Assessment | Indirect inference | Indirect inference | Direct quantification |
| Founder Line Establishment | Limited utility | Moderate utility | High utility for selection |
Somatic Editing Analysis: In zebrafish studies, ICE has demonstrated strong correlation with Illumina sequencing for indel quantification (Pearson's r = 0.90, p ≤ 0.001) [83], making it a cost-effective option for initial screening. However, both TIDE and ICE show limitations in detecting small (1-2 bp) indels, with CRISPR-STAT (TIDE-based) particularly underestimating efficiency at lower percentages [83].
Complex Repair Outcome Analysis: For comprehensive DSB repair fingerprinting, Illumina sequencing coupled with specialized bioinformatic pipelines like the Integrated Classification Pipeline (ICP) enables categorization of mutations into distinct repair pathways including PEPPR (PAM-End Proximal Protected Repair), MMEJ, DELET, and INSRT classes [40]. This granular classification reveals highly reproducible lineage-specific mutation fingerprints in individual organisms that are inaccessible to Sanger-based methods.
Experimental Protocols for Zebrafish DSB Repair Analysis
Sample Preparation Workflow
Figure 1: Experimental workflow for zebrafish CRISPR editing assessment, from embryo injection to data analysis.
Protocol for Comprehensive Editing Assessment
Step 1: Zebrafish Embryo Injection and Sampling
- Inject one-cell stage zebrafish embryos with CRISPR-Cas9 components (ribonucleoprotein complexes recommended for higher efficiency [83])
- Include repair templates for HDR experiments (single-stranded oligodeoxynucleotides with non-target asymmetric PAM-distal conformation show superior performance [83])
- Raise embryos to 72-96 hours post-fertilization (hpf)
- For somatic editing analysis: extract genomic DNA from pooled embryos (n=10-20) or individual embryos using minimally invasive methods like the Zebrafish Embryo Genotyper (ZEG) device [83]
- For germline transmission analysis: raise injected embryos to adulthood and cross with wild-types, then genotype F1 progeny
Step 2: Target Amplification and Library Preparation
- Design PCR primers flanking the target site with overhangs compatible with Illumina sequencing adapters
- Amplify target regions using high-fidelity DNA polymerase to minimize PCR errors
- For Illumina sequencing: incorporate dual indices and sequencing adapters via a second PCR step
- Quality control and quantify amplicon libraries using fragment analysis or bioanalyzer
Step 3: Sequencing and Data Analysis
- For TIDE/ICE: Sanger sequence PCR products and analyze using respective web platforms with default parameters
- For Illumina sequencing: sequence on appropriate platform (MiSeq for small panels, NovaSeq for large studies)
- Process NGS data using specialized pipelines (e.g., ICP for single-allele resolution classification [40])
- Quantify editing efficiencies: % indels, % HDR, allele-specific frequencies, and repair pathway signatures
Research Reagent Solutions for Zebrafish Genome Editing
Table 3: Essential Reagents for Zebrafish CRISPR-DSB Repair Studies
| Reagent Category | Specific Examples | Function & Application | Performance Notes |
|---|---|---|---|
| CRISPR Nucleases | SpCas9, Cas12a, SpRY [19] [84] | Induce targeted DSBs | SpRY offers relaxed PAM requirements [84] |
| Efficiency Enhancers | hei-tag [85], aNLS [84] | Boost nuclear import and editing | hei-tag increases bi-allelic editing by 70% [85] |
| HDR Templates | ssODNs (NAD conformation) [83], dsDNA with chemical modifications [19] | Provide repair homology | Chemically modified templates outperform plasmid-based [19] |
| Analysis Kits | T7 Endonuclease I [6], Library Prep Kits | Detect and prepare editing events | T7E1 for initial screening of modifications [6] |
| Delivery Methods | mRNA, RNP complexes [83] | Introduce editing components | RNP with Cas9 protein superior to mRNA for HDR [83] |
The selection of assessment methodology for zebrafish DSB repair studies should align with experimental objectives and resource constraints. TIDE offers rapid, cost-effective screening for preliminary experiments where high sensitivity is not critical. ICE provides more reliable quantification of editing efficiency, with demonstrated correlation to NGS data, making it suitable for standard knockout generation and efficiency optimization. Illumina sequencing remains the gold standard for comprehensive characterization of complex editing outcomes, particularly for HDR quantification, precise allele structure determination, and investigation of fundamental repair mechanisms.
For researchers exploring the intricacies of NHEJ and HDR in zebrafish, the integration of early NGS-based genotyping with the ZEG device enables efficient selection of high-efficiency founders, potentially increasing germline transmission rates by nearly 17-fold [83]. As zebrafish continue to serve as a vital model for functional genomics and disease modeling, appropriate method selection will remain crucial for advancing our understanding of DNA repair mechanisms and their applications in biomedical research.
Evaluating On-target and Off-target Effects in Zebrafish
The zebrafish (Danio rerio) has emerged as a preeminent model organism in genetic research and drug discovery, bridging the gap between in vitro studies and mammalian models. Its genome shares approximately 70% homology with humans, with 82% of human disease-associated genes having clear zebrafish orthologs [86] [87]. For research focusing on double-strand break (DSB) repair mechanisms, zebrafish offer unique advantages including high fecundity, external development, transparent embryos, and ease of genetic manipulation. Understanding the balance between non-homologous end joining (NHEJ) and homology-directed repair (HDR) is crucial for evaluating on-target and off-target effects in genome editing experiments.
DSB repair pathways compete to repair CRISPR-induced breaks, leading to diverse editing outcomes. NHEJ is the dominant pathway in zebrafish and other vertebrates, operating throughout the cell cycle but predominantly during G1 phase [16] [88]. This error-prone pathway directly ligates broken DNA ends, often resulting in small insertions or deletions (indels). In contrast, HDR is largely restricted to S and G2 phases when sister chromatids are available as repair templates [88]. HDR can precisely incorporate desired genetic changes when an exogenous donor template is provided. Recent research has revealed that alternative repair pathways, including microhomology-mediated end joining (MMEJ) and single-strand annealing (SSA), also contribute significantly to editing outcomes, complicating the prediction of on-target effects [17].
DNA Repair Mechanisms: NHEJ, HDR, and Alternative Pathways
Competing Double-Strand Break Repair Pathways
The following diagram illustrates the major DNA repair pathways that compete to resolve CRISPR-Cas9-induced double-strand breaks, leading to diverse genomic outcomes:
Pathway Characteristics and Functional Roles
Table 1: Key Characteristics of Major DNA Double-Strand Break Repair Pathways
| Pathway | Template Requirement | Key Molecular Players | Fidelity | Primary Functional Role |
|---|---|---|---|---|
| NHEJ | None | Ku70/80, DNA-PKcs, Ligase IV, XRCC4 | Error-prone | Rapid repair throughout cell cycle; dominant in zebrafish |
| HDR | Homologous template | RAD51, BRCA1/2, RAD52 | High-fidelity | Precise repair during S/G2 phases using sister chromatid |
| MMEJ | Microhomology (2-20 bp) | POLθ, PARP1, FEN1 | Error-prone | Backup pathway generating deletions |
| SSA | Long homologies (>20 bp) | RAD52, ERCC1, XPF | Mutagenic | Processes repeats causing genomic rearrangements |
The interplay between these pathways creates complex editing outcomes that must be carefully evaluated. Research in human cell lines suggests approximately 40% of DSBs are available for HDR when a donor template is present [88]. However, even with NHEJ inhibition, perfect HDR events may account for less than half of all integration events, with imprecise repair persisting through alternative pathways [17]. In zebrafish, chemical inhibition of NHEJ with NU7441 has been shown to enhance HDR efficiency up to 13.4-fold [16], demonstrating the potential for pathway manipulation to favor precise editing.
Evaluating On-Target Effects
Comprehensive Analysis of Intended Modifications
On-target effects refer to the genetic alterations occurring precisely at the intended CRISPR target site. A sophisticated evaluation strategy must distinguish between precise HDR events and various forms of imprecise repair. Long-read amplicon sequencing combined with computational frameworks like knock-knock enables comprehensive categorization of editing outcomes [17]. This approach can differentiate between perfect HDR, partial HDR, asymmetric HDR (where only one side integrates precisely), and various indel patterns.
Recent studies have revealed that large, mono-allelic genomic deletions and loss-of-heterozygosity can occur in up to 40% of edited clones, often escaping standard quality controls [89]. These substantial rearrangements represent significant on-target effects that could compromise experimental results or therapeutic applications. To address this challenge, quantitative genotyping PCR (qgPCR) and SNP genotyping-based tools have been developed as sensitive detection methods [89].
Experimental Protocol: Quantitative HDR Assessment in Zebrafish
The following workflow details a robust method for quantifying HDR efficiency in zebrafish embryos using a visual reporter system:
Table 2: Protocol for Quantitative HDR Assessment in Zebrafish Muscle Fibers
| Step | Description | Key Parameters | Purpose |
|---|---|---|---|
| 1. Transgenic Line Preparation | Use acta1:eBFP2; smyhc1:eGFP double transgenic zebrafish | Stable transgenic lines with muscle-specific expression | Visual tracking of editing efficiency in distinct muscle populations |
| 2. Repair Template Design | Create tdTomato donor with homology arms flanking Cas9 target site | 303 bp left homology arm, 1022 bp right homology arm; target site embedded in homology arm | Template for HDR-mediated conversion of BFP to tdTomato |
| 3. Microinjection | Co-inject Cas9 protein, sgRNA targeting eBFP2, and donor template into 1-2 cell stage embryos | Injection into cell rather than yolk; Cas9 protein instead of mRNA for rapid activity | Ensure delivery before primordial germ cell specification (before 4 hpf) |
| 4. Small Molecule Treatment | Apply NHEJ inhibitors (NU7441) or HDR enhancers (RS-1) immediately after injection | NU7441 at 50 μM dissolved in DMSO; monitor embryo survival | Shift repair balance toward HDR and away from NHEJ |
| 5. Phenotypic Quantification | Image embryos at 72 hpf using fluorescence microscopy | Count tdTomato-positive fast muscle fibers per embryo; minimum 40 embryos per condition | Single-cell resolution quantification of HDR efficiency |
| 6. Data Analysis | Compare treatment groups to DMSO controls; statistical analysis with t-tests | Calculate fibers per embryo rather than binary positive/negative assessment | Maintain dynamic range and avoid masking treatment effects |
This protocol leverages the optical transparency of zebrafish embryos and muscle-specific transgene expression to quantify HDR events at single-cell resolution. The system converts fast-muscle fibers from blue to red fluorescence upon successful HDR, enabling rapid visual assessment [16]. Critical considerations include the use of sgRNAs with high cutting efficiencies (>60%), repair templates that overlap the DSB site, and modification of the PAM site to prevent re-cutting of successfully edited loci [90].
Advanced Detection Methods for Complex On-Target Effects
Beyond the visual reporter approach, several molecular methods provide comprehensive analysis of on-target editing:
High-Resolution Melting Analysis (HRMA): This technique detects sequence variations by analyzing the melting behavior of PCR amplicons. HRMA is particularly sensitive to heteroduplex formation in mosaic embryos and can identify NHEJ-derived indels with high throughput [88].
TaqMan qPCR Assays: Using probes situated across the CRISPR cut site, this method quantitatively discriminates between wild-type and modified sequences. The approach can be enhanced with a second probe outside the primary target to measure HDR contribution specifically [88].
Long-Range PCR and Sequencing: Amplification of the entire target region with subsequent long-read sequencing (PacBio, Nanopore) enables detection of large deletions and complex rearrangements that would be missed by short-read approaches [17].
The experimental workflow below illustrates the integrated approach for comprehensive on-target effect evaluation:
Assessing Off-Target Effects
Prediction and Detection of Unintended Editing
Off-target effects refer to CRISPR-induced modifications at genomic loci other than the intended target site, resulting from guide RNA binding to sequences with partial complementarity. In zebrafish, several strategies have been developed to identify and minimize these effects:
In Silico Prediction Tools: Computational algorithms identify potential off-target sites based on sequence similarity to the guide RNA. However, these predictions may miss true off-target sites due to the complexity of genomic context and chromatin accessibility.
Whole-Genome Sequencing: The most comprehensive approach involves sequencing the entire genome of edited zebrafish. While cost-prohibitive for large numbers of samples, it provides unbiased detection of all variants.
GUIDE-seq and Related Methods: These molecular techniques capture genome-wide Cas9 cleavage sites by integrating oligonucleotides at DSB locations, providing empirical data on off-target activity.
RNA-seq Analysis: Transcriptome sequencing can identify unintended splicing alterations or gene expression changes resulting from off-target editing.
Recent research emphasizes that chromatin accessibility significantly influences off-target susceptibility, with open chromatin regions being more vulnerable to Cas9 cleavage [90]. Interestingly, a 2015 study suggested zebrafish may not adhere to the same chromatin restrictions as mammalian cells [26], highlighting the need for species-specific off-target assessment methods.
The Scientist's Toolkit: Essential Reagents and Solutions
Table 3: Key Research Reagents for Zebrafish Genome Editing Studies
| Reagent Category | Specific Examples | Function/Application | Considerations for Zebrafish Research |
|---|---|---|---|
| NHEJ Inhibitors | NU7441, SCR7, Alt-R HDR Enhancer V2 | Shift repair balance toward HDR by blocking NHEJ pathway | NU7441 shows 13.4-fold HDR enhancement in zebrafish; SCR7 effects are species-specific |
| HDR Enhancers | RS-1 (RAD51 stimulator) | Promote RAD51-mediated strand invasion during HDR | Modest effect alone; may combine with NHEJ inhibitors |
| MMEJ Inhibitors | ART558 (POLQ inhibitor) | Suppress microhomology-mediated end joining | Reduces large deletions and complex indels at target site |
| SSA Inhibitors | D-I03 (RAD52 inhibitor) | Block single-strand annealing pathway | Reduces asymmetric HDR and imprecise donor integration |
| Editing Reagents | Cas9 protein, sgRNAs, donor templates with homology arms | Create targeted DSBs and provide repair template | Cas9 protein provides immediate activity; long homology arms (≥90 bp) improve HDR |
| Detection Tools | HRMA assays, TaqMan probes, long-read sequencing platforms | Identify and quantify editing outcomes | Combination approach recommended for comprehensive assessment |
Evaluation of on-target and off-target effects remains a critical challenge in zebrafish genome engineering. The complex interplay between competing DNA repair pathways necessitates multi-faceted assessment strategies that capture the full spectrum of editing outcomes. While NHEJ inhibition significantly improves HDR efficiency, recent research reveals that suppressing alternative pathways like MMEJ and SSA can further enhance precise editing by reducing imprecise integration events [17].
The future of precise genome editing in zebrafish will likely involve combined pathway modulation alongside continued refinement of detection methodologies. As CRISPR applications expand in disease modeling and drug discovery, comprehensive effect evaluation will be essential for generating reliable models and interpreting phenotypic outcomes. The standardization of assessment protocols across the zebrafish research community will enhance reproducibility and translational potential of findings from this versatile model organism.
Standardization Protocols for Reproducible Drug Screening
The zebrafish has emerged as a premier model organism for high-throughput drug screening and functional genomics, bridging the gap between cell culture studies and mammalian models. Its high physiological conservation with humans, transparency during embryonic development, and high fecundity make it ideal for large-scale chemical toxicity screens and prioritization of drugs for testing in mammals [16] [91]. However, the full potential of this model is hampered by significant intra- and inter-laboratory variability in experimental outcomes. Recent systematic evaluations have identified multiple sources of this variability, including the use of static versus static renewal exposures, presence or absence of the chorion, and critically, the ionic composition and strength of exposure media [92] [91]. This technical guide establishes a comprehensive framework for standardizing protocols with a specific focus on how manipulation of DNA repair pathways—particularly the balance between error-prone non-homologous end joining (NHEJ) and precise homology-directed repair (HDR)—can enhance the rigor and reproducibility of drug screening in zebrafish models.
Standardizing Exposure Media: A Foundational Element
The Problem of Media-Dependent Variability
Despite advancements in zebrafish husbandry and experimentation, exposure media has remained a significant "blind spot" in standardization efforts. The OECD Test No. 236 (Fish Embryo Acute Toxicity Test) provides guidance on water quality parameters but lacks specific recommendations for solute concentrations within exposure media [91]. Consequently, laboratories worldwide utilize media with varying ionic composition and strength, leading to substantial variability in chemical potency assessments.
Research has demonstrated that exposure media can dramatically influence phenotypic outcomes. Studies with triphenyl phosphate (TPHP) revealed that its cardiotoxicity is dependent on epidermal injury, disruption of embryonic osmoregulation, and pericardial edema formation—effects that are exacerbated by increased ionic strength, particularly high chloride concentrations [91]. Similar media-dependent effects have been observed for other chemical classes, including TCDD and tricresyl phosphate isomers, suggesting a broad phenomenon beyond organophosphate esters [91].
Recommended Standardization Parameters
Table 1: Key Parameters for Standardizing Zebrafish Exposure Media
| Parameter | Current Variability | Recommended Standardization | Impact on Assay Results |
|---|---|---|---|
| Ionic Composition | E2, E3, Hanks' Balanced Salt Solution, and other formulations | Development of consensus formulation | Directly affects chemical uptake, osmoregulation, and toxicity manifestations |
| Chloride Concentration | Varies significantly between media types | Standardization to optimal range | High chloride exacerbates pericardial edema for certain toxicants |
| Osmolarity | Laboratory-specific formulations | Defined osmolarity range | Maintains embryonic osmoregulation and prevents artifactual edema |
| Exposure Method | Static vs. static renewal | Protocol-specific standardization | Renewal prevents chemical degradation; static may underestimate potency |
| Chorion Status | Intact vs. dechorionated | Endpoint-driven recommendation | Chorion can barrier to chemical uptake; removal increases sensitivity |
The critical need for media standardization was highlighted by the Systematic Evaluation of the Application of Zebrafish in Toxicology (SEAZIT) program, which found interlaboratory variation in benchmark concentrations (BMCs) of up to two orders of magnitude for 39 test substances depending on exposure conditions [91]. This degree of variability can lead to significant errors in hit identification, chemical prioritization, and human health risk characterization.
Manipulating NHEJ and HDR Balance for Enhanced Genome Editing
The efficiency of precise genome editing in zebrafish is fundamentally limited by the competition between two major DNA double-strand break (DSB) repair pathways: the error-prone non-homologous end joining (NHEJ) and the precise homology-directed repair (HDR). In most organisms, including zebrafish, NHEJ dominates, resulting in low HDR efficiency that has hampered the generation of precise mutant lines [11] [16]. Strategic manipulation of this balance represents a powerful approach for improving reproducible genome editing in drug screening applications.
Small Molecule Modulation of Repair Pathways
Chemical inhibition of NHEJ and stimulation of HDR has emerged as a highly effective strategy for enhancing precise genome editing efficiency. Using a quantitative in vivo reporter assay in zebrafish fast-muscle fibers, researchers have systematically evaluated small molecule modulators:
Table 2: Small Molecule Modulators of DNA Repair Pathways
| Compound | Target | Effect on HDR | Optimal Dose | Mechanism of Action |
|---|---|---|---|---|
| NU7441 | DNA-PK inhibitor (NHEJ) | 13.4-fold enhancement | 50 µM | Blocks NHEJ by inhibiting DNA-PK, shifting balance to HDR |
| RS-1 | RAD51 stimulator (HDR) | Modest increase (1.5-fold) | 15-30 µM | Stimulates RAD51-mediated strand invasion in HDR |
| SCR7 | Ligase IV inhibitor (NHEJ) | No significant effect | Up to solubility limit | Proposed NHEJ inhibition; species-specific effects |
| NU7441 + RS-1 | Combined approach | No additive benefit | 50 µM + 30 µM | Combined inhibition lacks synergistic effect |
The most dramatic enhancement was achieved with NU7441, which increased HDR efficiency up to 13.4-fold at 50 µM concentration without affecting embryo survival [16]. This approach directly translates to improved germline transmission, permitting efficient recovery of large seamlessly integrated DNA fragments—a critical advancement for creating stable transgenic lines for drug screening.
Critical Factors for Successful HDR in Zebrafish
Analysis of 50 successfully HDR-modified zebrafish genes reveals consistent patterns in successful protocols [11]:
- sgRNA Efficiency: Use only sgRNAs with high cutting efficiencies (>60-70%)
- DSB-Target Proximity: Double-strand break site should be within 20 nucleotides of the target nucleotide
- Developmental Timing: Microinjections must occur during the 1–2 cell stage
- PAM Site Alteration: Protospacer adjacent motif site must be modified to prevent re-cutting of successfully repaired targets
- Template Design: Repair template must overlap the DSB site (asymmetrically or symmetrically)
Protocol variations that significantly impact HDR success rates include template type (single-stranded vs. double-stranded DNA), length of homology arms (25-40 bp optimal for MMEJ), symmetry of repair template, and choice of endonuclease (Cas9 mRNA or protein) [11] [93].
Optimized Workflows for Enhanced Reproducibility
Standardized HDR Enhancement Workflow
The following diagram illustrates an optimized, standardized workflow for enhancing HDR efficiency in zebrafish embryos through chemical reprogramming:
Media Standardization Framework
Standardizing exposure media requires a systematic approach to identify and control critical variables:
The Scientist's Toolkit: Essential Reagents and Materials
Table 3: Research Reagent Solutions for Standardized Zebrafish Research
| Reagent Category | Specific Examples | Function & Application | Optimization Notes |
|---|---|---|---|
| NHEJ Inhibitors | NU7441 (50 µM), KU0060648 | Shifts repair balance toward HDR; enhances precise editing | Dose optimization critical; embryo toxicity screening required |
| HDR Stimulators | RS-1 (15-30 µM) | Enhances RAD51-mediated strand invasion | Modest effects alone; test combinations |
| Exposure Media | Standardized E3, E2, or Hanks' formulations | Consistent chemical exposure across experiments | Ionic strength and chloride concentration critical parameters |
| CRISPR Components | High-efficiency sgRNA (>60%), Cas9 protein/mRNA | Induces targeted double-strand breaks | sgRNA efficiency correlates with HDR success rate |
| Repair Templates | S-25 donor (25-bp homology arms), 5'-modified dsDNA | Provides homology for precise repair | Asymmetric/symmetric design; PAM site alteration essential |
| Detection Systems | Fluorescent reporters, junction PCR assays | Quantifies editing efficiency and germline transmission | Quantitative single-cell analysis preferred over qualitative |
Standardization of exposure media and DNA repair manipulation protocols represents a critical advancement for enhancing the rigor and reproducibility of zebrafish-based drug screening. The integration of chemical reprogramming approaches with standardized environmental conditions addresses two major sources of variability in experimental outcomes. As the field moves toward more sophisticated screening paradigms, including organoid-based systems and AI-driven data analysis, establishing robust foundational protocols becomes increasingly important [94]. The recommended frameworks and optimized workflows presented in this guide provide actionable strategies for researchers to implement in their drug discovery pipelines, ultimately leading to more reliable hit identification, improved chemical prioritization, and enhanced translatability of findings from zebrafish models to mammalian systems and clinical applications. Future efforts should focus on international harmonization of these standards, similar to the SEAZIT program, but with explicit consideration of exposure media composition and DNA repair modulation protocols as critical variables rather than secondary concerns.
Genetic Background Considerations and Strain Selection
In zebrafish research, the genetic background of the model organism and the specific strain selected are critical considerations that significantly influence the outcome of double-strand break (DSB) repair experiments. The efficiency of both non-homologous end joining (NHEJ) and homology-directed repair (HDR) pathways varies substantially based on the genetic context, which directly impacts the success of functional genomics studies, disease modeling, and therapeutic development [11] [23]. Understanding these variables is essential for designing robust experiments, particularly as CRISPR-based technologies enable increasingly precise genomic modifications in zebrafish models.
Zebrafish have emerged as a pivotal model organism for studying DSB repair mechanisms due to their genetic similarity to humans, with approximately 70% of human genes having a corresponding ortholog in zebrafish [6] [23]. The zebrafish model combines the advantages of a vertebrate system with experimental flexibility, including transparent embryos for visual screening, rapid development, and high fecundity. However, the efficiency of precise genome editing remains challenging due to the complex interplay between genetic background variables and the cellular decision-making processes that determine whether NHEJ or HDR pathways will predominate following DSB induction [11] [16].
This technical guide examines how genetic background considerations and strategic strain selection can optimize DSB repair outcomes in zebrafish research, with particular emphasis on enhancing HDR efficiency for precise genome modifications.
DNA Repair Pathways in Zebrafish: NHEJ vs. HDR
When CRISPR/Cas9 introduces a double-strand break in zebrafish DNA, the cell employs primarily one of two major repair pathways: non-homologous end joining (NHEJ) or homology-directed repair (HDR). These pathways compete within the cell, with NHEJ typically dominating in most zebrafish embryos and somatic cells [10] [16] [15].
Diagram 1: CRISPR/Cas9-Induced DNA Repair Pathways. Following a double-strand break, cells primarily utilize either the error-prone NHEJ pathway or the precise HDR pathway, each yielding distinct genetic outcomes.
Non-Homologous End Joining (NHEJ): The Default Pathway
NHEJ is an error-prone repair mechanism that directly ligates broken DNA ends without requiring a homologous template. This pathway is active throughout the cell cycle and represents the dominant DSB repair mechanism in zebrafish embryos [10] [15]. The NHEJ process often results in small insertions or deletions (indels) at the repair site, which typically disrupt gene function by causing frameshift mutations or premature stop codons. This makes NHEJ particularly suitable for gene knockout studies where complete loss of function is desired [10]. The efficiency of NHEJ-mediated knockout generation in zebrafish is remarkably high, with success rates of 75-99% reported across multiple loci [23] [18].
Homology-Directed Repair (HDR): The Precision Pathway
HDR is a precise repair mechanism that uses homologous sequences (such as sister chromatids or exogenous donor templates) as a blueprint for error-free repair. Unlike NHEJ, HDR is restricted primarily to the S and G2 phases of the cell cycle when homologous templates are available [15]. In zebrafish research, HDR can be harnessed to introduce specific genetic modifications—including single-nucleotide changes, epitope tags, or conditional alleles—by providing an exogenous donor template with homology to the target locus [11] [95]. However, HDR efficiency in zebrafish is substantially lower than NHEJ, presenting a significant technical challenge that can be addressed through strategic experimental design, including optimal strain selection [16] [18].
Quantitative Comparison of DSB Repair Outcomes in Zebrafish
Table 1: Efficiency Comparison of DNA Repair Pathways and Genome Editing Technologies in Zebrafish
| Technology/Approach | Typical Efficiency Range | Key Applications | Genetic Background Considerations |
|---|---|---|---|
| NHEJ (Knockout) | 75-99% [23] [18] | Gene disruption, loss-of-function studies | Consistent across most genetic backgrounds; minimal strain-dependent variation |
| HDR (Knock-in) | 2-10% (standard) [11] [18] | Point mutations, epitope tagging, precise edits | Highly variable; depends on target locus, strain, and template design |
| Enhanced HDR (zLOST) | Up to 31.8% germline transmission [18] | Precise knock-in of larger fragments | Long single-stranded DNA templates improve efficiency across backgrounds |
| Chemical Enhancement (NU7441) | Up to 13.4-fold improvement [16] | HDR-mediated precise editing | NHEJ inhibition effectiveness may vary with genetic background |
| Base Editing | 9-28% (C->T, A->G conversions) [5] | Specific nucleotide transitions | PAM sequence requirements limit targetable sites |
| Prime Editing | 4.4-8.4% (substitution) [6] | Precise edits without donor templates | Newer technology with background-dependent efficiency still being characterized |
Table 2: Successful HDR-Targeted Genes in Zebrafish and Efficiency Factors
| Gene | HDR Efficiency | Critical Optimization Factors | Reference |
|---|---|---|---|
| tyrosinase (tyr) | ~98% somatic (zLOST method) [18] | Long single-stranded DNA templates, visible phenotypic selection | Bai et al., 2020 |
| sox11a | 4.3-10.6% (MYC tag insertion) [95] | Chemically modified donor templates, RNP complex delivery | Krueger & Morris, 2022 |
| rors | Varies by approach | Distance between DSB and insertion site, template design | Multiple studies |
| Multiple loci (50 genes) | Wide variation reported [11] | sgRNA cutting efficiency >60%, PAM site alteration, proximity of DSB to target | Burg et al., 2020 |
Genetic Background Variables Affecting DSB Repair Outcomes
Target Locus Characteristics
The genomic context of the target site significantly influences DSB repair outcomes. Studies analyzing 50 successfully modified genes in zebrafish identified that the cutting efficiency of the sgRNA is a primary determinant, with efficiencies exceeding 60% being essential for successful HDR [11]. The proximity of the double-strand break to the target nucleotide also critically impacts HDR success, with optimal distances typically within 20 nucleotides of the target site [11]. Additionally, altering the protospacer adjacent motif (PAM) site in the repair template prevents re-cutting of successfully repaired targets, thereby enriching for precisely edited cells [11].
Strain Selection Considerations
The selection of appropriate zebrafish strains can dramatically impact the efficiency of precise genome editing. Strains with defined genetic backgrounds, such as the AB strain commonly used in HDR experiments, provide more consistent results compared to genetically heterogeneous populations [95]. Specialized reporter strains, such as the acta1:eBFP2 transgenic line used for quantitative HDR assessment, enable rapid screening and quantification of editing efficiency through visual phenotyping [16]. For disease modeling, strains with specific sensitized backgrounds (e.g., tyr mutants with albino phenotype for visual tracking of repair) can significantly streamline the identification of successful editing events [18].
Epigenetic and Chromatin Landscape
The local chromatin environment and epigenetic modifications at the target locus influence the accessibility to CRISPR-Cas9 machinery and consequently affect editing efficiency. While the search results do not provide zebrafish-specific data on this aspect, general principles from mammalian systems suggest that euchromatic regions with open chromatin configurations are more amenable to efficient editing compared to heterochromatic regions. Strategic sgRNA design that avoids densely packed chromatin regions can improve consistency across different genetic backgrounds.
Experimental Protocols for Optimized DSB Repair
Enhanced HDR Efficiency Using zLOST Methodology
The zebrafish Long Single-Stranded DNA Template (zLOST) method represents a significant advancement for achieving precise genome modifications through HDR. This protocol employs long single-stranded DNA donors to dramatically improve knock-in efficiency [18]:
Donor Template Design: Generate single-stranded DNA templates containing 299-512 nucleotide sequences with the desired modification flanked by homology arms symmetrical to the target locus. The optimal length depends on the specific application, with longer templates (∼500 nt) showing superior efficiency for larger insertions [18].
Microinjection Mixture Preparation: Co-inject zCas9 mRNA, target-specific gRNA, and the zLOST donor template into one-cell stage zebrafish embryos. For the tyr locus, this approach achieved phenotypic rescue in 98.5% of injected embryos, with precise HDR-dependent repair confirmed by sequencing [18].
Screening and Validation: For visible phenotypes (e.g., pigmentation restoration in tyr mutants), conduct initial visual screening at 3 days post-fertilization. Follow with PCR amplification of the target region and sequencing validation to confirm precise integration. This method has demonstrated germline transmission rates up to 31.8% [18].
Chemical Reprogramming to Favor HDR
Small molecule inhibition of NHEJ pathways can shift the repair equilibrium toward HDR, significantly enhancing precise editing efficiency [16]:
Compound Selection: Prepare working solutions of NU7441 (DNA-PK inhibitor) at 50μM concentration in DMSO. This specific inhibitor demonstrated a 13.4-fold enhancement of HDR efficiency in zebrafish embryos, while other inhibitors (SCR7, RS-1) showed minimal or modest effects [16].
Embryo Treatment: Microinject the compound simultaneously with CRISPR components during the 1-2 cell stage. The treatment window is critical, as early inhibition of NHEJ factors redirects repair toward HDR pathways during initial cell divisions.
Efficiency Quantification: Use quantitative reporter systems (e.g., fast-muscle fiber fluorescence conversion) for precise measurement of HDR events at single-cell resolution. Qualitative assessment (presence/absence of editing) masks the substantial enhancement effects, with quantitative analysis revealing increases from 4.0±3.0 to 53.7±22.1 red fibers per embryo with NU7441 treatment [16].
RNP Complex Delivery with Modified Donors
Ribonucleoprotein (RNP) complex delivery combined with chemically modified donors provides an efficient approach for epitope tagging and precise insertion [95]:
RNP Complex Formation: Pre-complex synthetic crRNA:tracrRNA with Cas9 protein to form RNP complexes, which show higher mutagenesis efficiency compared to mRNA injection.
Donor Template Modification: Utilize chemically modified single-stranded DNA donors containing phosphorothioate linkages at the ends to enhance stability and HDR efficiency. This approach achieved 4.3-10.6% HDR efficiency for MYC tag integration at the sox11a locus [95].
Germline Transmission Screening: Raise injected embryos to adulthood and outcross to identify founders transmitting the precise modification. Subsequent generations should be validated for stable inheritance, expression, and functionality of the edited allele.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for DSB Repair Studies in Zebrafish
| Reagent/Category | Specific Examples | Function/Application | Optimization Notes |
|---|---|---|---|
| CRISPR Nucleases | Cas9 nuclease, Cas9 nickase, Cas12a [82] | DSB induction, target DNA cleavage | Cas9 and Cas12a show similar insertion efficiency; Cas9 nickase used in prime editors |
| Repair Templates | ssODN, dsDNA, zLOST (long ssDNA) [18] | Homology-directed repair template | Long ssDNA (zLOST) dramatically outperforms other templates (98.5% vs 5% efficiency) |
| NHEJ Inhibitors | NU7441 (DNA-PK inhibitor) [16] | Shifts repair balance toward HDR | 50μM concentration optimal; 13.4-fold HDR enhancement |
| HDR Enhancers | RS-1 (RAD51 stimulator) [16] | Promotes RAD51-mediated strand invasion | Modest effect (7.2±3.7 vs 4.8±3.0 fibers/embryo) |
| Delivery Tools | RNP complexes, chemically modified gRNAs [95] | Efficient component delivery | RNP complexes with synthetic crRNA:tracrRNA improve efficiency |
| Strain Resources | AB strain, transgenic reporter lines [16] [95] | Consistent genetic background, efficiency screening | Reporter strains enable quantitative single-cell HDR assessment |
Decision Framework for Strain Selection and Experimental Design
Diagram 2: Strain Selection and Experimental Design Framework. A decision pathway for selecting appropriate zebrafish strains and methods based on specific research goals, highlighting the connection between objectives and optimal genetic backgrounds.
Genetic background considerations and strategic strain selection are fundamental to successful double-strand break repair studies in zebrafish. The inherent competition between NHEJ and HDR pathways necessitates careful experimental design that accounts for locus-specific characteristics, strain genetics, and template optimization. The development of enhanced methods like zLOST for HDR and chemical inhibition of NHEJ has dramatically improved precise editing efficiency, enabling more reliable generation of zebrafish models with specific genetic modifications.
As CRISPR technologies continue to evolve, with base editors and prime editors offering new possibilities for precision genome engineering [5] [6], understanding the interplay between these tools and genetic background variables becomes increasingly important. By applying the principles and protocols outlined in this technical guide, researchers can optimize their strain selection and experimental approaches to maximize the success of their functional genomics studies and disease modeling efforts in zebrafish.
Benchmarking Zebrafish HDR Efficiency Against Other Model Systems
Homology-Directed Repair (HDR) enables precise genome editing for inserting specific DNA sequences, correcting point mutations, and generating knock-in models. However, its efficiency remains a significant challenge across model organisms. This technical review quantitatively benchmarks HDR efficiency in zebrafish against other common models, including mammalian cell lines and plants. We systematically analyze factors influencing HDR outcomes—including template design, nuclease selection, and experimental conditioning—and provide optimized protocols for achieving germline transmission rates exceeding 20% in zebrafish. The data presented establishes zebrafish as a competitive vertebrate model for precise genome editing while highlighting system-specific considerations for cross-species experimental design.
The CRISPR/Cas9 system has revolutionized genetic research by enabling targeted DNA double-strand breaks (DSBs), but the resulting edits are determined by cellular repair mechanisms rather than the cutting machinery itself [10] [15]. Two primary pathways compete to repair these breaks: Non-Homologous End Joining (NHEJ) and Homology-Directed Repair (HDR).
NHEJ is an error-prone repair pathway that directly ligates broken DNA ends without requiring a template, often resulting in small insertions or deletions (indels) that disrupt gene function [10] [15]. This pathway is active throughout the cell cycle and is generally more efficient than HDR, making it ideal for gene knockout studies [15].
In contrast, HDR is a precise repair mechanism that uses homologous DNA sequences (such as sister chromatids or exogenously supplied donor templates) as templates for error-free repair [10]. This pathway enables precise genetic modifications, including gene corrections, insertions of specific sequences, and creation of point mutations [15]. However, HDR is restricted to the S and G2 phases of the cell cycle when homologous templates are available, and its efficiency is typically substantially lower than NHEJ across most systems [96] [16].
The competitive balance between these pathways presents a fundamental challenge for precision genome editing. Understanding their dynamics across model systems is essential for designing effective gene editing strategies.
Figure 1: DNA Repair Pathways Following CRISPR/Cas9-Induced Double-Strand Breaks. HDR enables precise editing but requires a donor template, while NHEJ generates random indels suitable for gene knockouts.
Quantitative Benchmarking of HDR Efficiency Across Models
Direct comparison of HDR efficiency across experimental systems reveals significant variation dependent on organism, cell type, target locus, and experimental parameters.
Zebrafish HDR Efficiency Metrics
Recent advances have substantially improved HDR efficiency in zebrafish. A 2025 study demonstrated that optimized parameters consistently achieved germline founder rates exceeding 20% for precise insertions across four different loci [19]. This represents a significant improvement over earlier reports and establishes zebrafish as a viable platform for precise genome engineering.
Chemical enhancement strategies have further boosted HDR outcomes in zebrafish. Inhibition of NHEJ with NU7441 enhanced HDR-mediated repair efficiency up to 13.4-fold in zebrafish embryos, while RS-1 showed more modest improvement (approximately 1.5-fold increase) [16]. Importantly, the quantitative relationship between somatic HDR events and germline transmission was directly correlated, enabling predictive screening [16].
Cross-System HDR Efficiency Comparison
Table 1: HDR Efficiency Benchmarks Across Model Systems
| Model System | Typical HDR Efficiency Range | Key Influencing Factors | Notable Achievements |
|---|---|---|---|
| Zebrafish | 5-20% (germline transmission) [19] | Template design, NHEJ inhibition, nuclease selection | >20% germline transmission with optimized parameters; 13.4-fold enhancement with NU7441 [19] [16] |
| Mammalian Cell Cultures (HEK293T, HeLa) | HDR can exceed NHEJ under specific conditions [96] | Locus dependence, nuclease platform, cell type | HDR>NHEJ ratio highly variable (0.1 to >10) depending on conditions [96] |
| Human iPSCs | Variable; often lower than cell lines [96] | Cell cycle synchronization, delivery method | Successful HDR with allele-specific disruption common [96] |
| Plants (Rice, Tobacco) | Extremely low without specialized systems [97] | Tissue type, transformation method | cgRNA and CRISPEY strategies show limited success [97] |
| Prime Editing (Zebrafish) | 4.4-8.4% (precise substitution) [6] | Editor type (PE2 vs PEn), target site | PE2 superior for single-base substitutions; PEn better for insertions [6] |
Comparative analysis reveals that HDR/NHEJ ratios are highly dependent on gene locus, nuclease platform, and cell type [96]. Contrary to the common assumption that NHEJ generally predominates, studies in mammalian cells have found that HDR can exceed NHEJ under multiple conditions, though this balance varies significantly across systems [96].
Experimental Protocols for Optimized Zebrafish HDR
Template Design and Selection
The choice of HDR template significantly impacts editing efficiency. For zebrafish knock-in experiments, comparative studies have identified optimal approaches:
- Chemically modified double-stranded DNA templates outperform templates released in vivo from plasmids [19]
- Single-stranded oligodeoxynucleotides (ssODNs) are effective for introducing sequence variants, with established protocols achieving >5% founder rates across many loci [19]
- Homology arm optimization is critical, with precise editing rates dependent on the distance between the double-strand break and the inserted sequence [19]
- Elimination of non-homologous base pairs in homology templates significantly improves precise editing rates [19]
Table 2: Essential Research Reagents for Zebrafish HDR Experiments
| Reagent Category | Specific Examples | Function/Application | Optimization Notes |
|---|---|---|---|
| Nucleases | Cas9, Cas12a (Cpf1) [19] | Induce DSBs at target sites | Cas12a creates 5-nt 5' overhangs; may improve HDR in some contexts [19] |
| Template Types | ssODNs, dsODNs, chemically modified templates [19] | Provide repair homology | Chemical modifications reduce degradation/concatemerization [19] |
| HDR Enhancers | NU7441, RS-1, SCR7 [16] | Modulate repair pathway choice | NU7441 (50µM) shows dramatic HDR enhancement [16] |
| Delivery Tools | Microinjection apparatus [5] [6] | Introduce editing components | RNP complex delivery can improve efficiency [5] |
| Detection Assays | Long-read sequencing (Pacific Biosciences) [19] | Quantify editing outcomes | Overcomes size bias limitations of Illumina sequencing [19] |
Nuclease Selection and Delivery
Both Cas9 and Cas12a nucleases have been successfully employed for HDR in zebrafish, with each offering distinct advantages:
- Cas9 generates blunt-end DSBs and remains the most widely used nuclease platform [19]
- Cas12a recognizes T-rich PAM sequences and creates single-strand overhangs that may facilitate HDR in some contexts [19]
- Ribonucleoprotein (RNP) complex delivery via microinjection provides immediate activity and potentially reduced off-target effects [5]
- Nuclease performance is locus-dependent, requiring empirical testing for optimal results [19]
Chemical Modulation of Repair Pathways
Small molecule inhibition of NHEJ represents a powerful strategy for enhancing HDR efficiency:
- NU7441 (DNA-PK inhibitor): Demonstrated the most dramatic effect, increasing HDR efficiency up to 13.4-fold at 50µM concentration [16]
- RS-1 (RAD51 stimulator): Showed modest but significant improvement (approximately 1.5-fold increase) [16]
- SCR7 (Lig4 inhibitor): Displayed negligible effects in zebrafish, highlighting species-specific responses to chemical modulators [16]
- Treatment timing: Co-injection with editing components is critical for effective pathway modulation [16]
Figure 2: Optimized Workflow for Efficient HDR in Zebrafish. Critical optimization parameters at the assembly stage significantly impact final HDR efficiency and germline transmission rates.
Technical Considerations and Advanced Approaches
Assessment Methodologies
Accurate quantification of HDR outcomes requires specialized approaches:
- Long-read sequencing (Pacific Biosciences) enables comprehensive analysis of precise knock-in events by spanning entire insertion sequences and homology regions, overcoming limitations of short-read platforms [19]
- Digital PCR (ddPCR) assays provide highly sensitive quantification of both HDR and NHEJ events simultaneously at endogenous loci [96]
- In vivo visual reporters enable rapid quantification of HDR-mediated events at single-cell resolution in living zebrafish embryos [16]
- Germline transmission validation remains the gold standard, with somatic editing rates serving as a reliable proxy for predicting heritable edits [19] [16]
Alternative Precision Editing Platforms
While HDR remains a cornerstone of precise genome editing, emerging technologies offer complementary approaches:
- Prime Editing: PE2 (nickase-based) shows higher precision for single-nucleotide substitutions (8.4% efficiency vs 4.4% for PEn), while PEn (nuclease-based) more efficiently inserts short DNA fragments up to 30bp [6]
- Base Editing: Enables direct single-nucleotide conversions without double-strand breaks, with cytosine base editors (CBEs) achieving C:G to T:A conversions and adenine base editors (ABEs) facilitating A:T to G:C conversions [5]
- Hybrid Approaches: Combining HDR with alternative editing technologies can leverage the strengths of each platform for specific applications
Zebrafish represents a robust model system for HDR-mediated precise genome editing, with recent optimizations enabling germline transmission rates competitive with other vertebrate models. The demonstrated efficiency exceeding 20% at multiple loci, coupled with chemical enhancement strategies that boost HDR outcomes more than 13-fold, positions zebrafish as a premier system for precise genetic modeling. The cross-system benchmarking presented here provides a framework for selecting appropriate models and optimization strategies based on specific research objectives. Continued refinement of template design, nuclease selection, and pathway modulation will further enhance the precision and efficiency of zebrafish genome editing, solidifying its role in functional genomics and disease modeling.
Conclusion
The strategic manipulation of NHEJ and HDR pathways in zebrafish represents a powerful approach for advancing functional genomics and therapeutic discovery. By understanding the fundamental mechanisms, implementing optimized protocols, employing chemical enhancement strategies, and adhering to rigorous validation standards, researchers can significantly improve precision genome editing outcomes. Future directions include developing more specific small-molecule modulators of DNA repair, refining high-throughput screening methodologies, and translating findings from zebrafish models to clinical applications, particularly for neuromuscular diseases and cancer research. The continued standardization of zebrafish drug testing parameters will further enhance the model's utility in identifying novel therapeutic compounds and advancing personalized medicine approaches.
References
- 1. DNA double strand break repair pathway choice in somatic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7315405/]
- 2. Double-strand break repair model [https://en.wikipedia.org/wiki/Double-strand_break_repair_model]
- 3. Methods Favoring Homology-Directed Repair Choice in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7555059/]
- 4. Base editors in zebrafish: a new era for functional ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12133889/]
- 5. Base editors in zebrafish: a new era for functional ... [https://www.frontiersin.org/journals/genome-editing/articles/10.3389/fgeed.2025.1598887/full]
- 6. Optimised genome editing for precise DNA insertion and ... [https://elifesciences.org/reviewed-preprints/107475]
- 7. Genome editing with the HDR-enhancing DNA-PKcs ... [https://www.nature.com/articles/s41587-024-02488-6]
- 8. DNA Double-Strand Break Repair by Non-Homologous ... [https://geneglobe.qiagen.com/us/knowledge/pathways/dna-double-strand-break-repair-by-non-homologous-end-joining]
- 9. Non-homologous end joining [https://en.wikipedia.org/wiki/Non-homologous_end_joining]
- 10. HDR vs. NHEJ: CRISPR Gene Editing Techniques [https://www.zeclinics.com/blog/hdr-vs-nhej-in-crispr-gene]
- 11. Homology-Directed Repair in Zebrafish: Witchcraft and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7793982/]
- 12. Inhibiting NHEJ in HNSCC cell lines by the ligase IV ... [https://www.nature.com/articles/s41598-025-03159-5]
- 13. DNA repair by nonhomologous end joining and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2754209/]
- 14. Uncovering the Molecular Drivers of NHEJ DNA Repair ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10606680/]
- 15. HDR vs NHEJ: DNA Repair Pathway Comparison [https://invivobiosystems.com/crispr/hdr-vs-nhej/]
- 16. Chemical reprogramming enhances homology-directed ... [https://www.nature.com/articles/s42003-019-0444-0]
- 17. Comparative analysis of multiple DNA double-strand break ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12075812/]
- 18. CRISPR/Cas9-mediated precise genome modification by a ... [https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-020-6493-4]
- 19. Identifying optimal conditions for precise knock-in of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12211562/]
- 20. CRISPR/Cas9-mediated homology-directed repair by ... [https://pubmed.ncbi.nlm.nih.gov/30355591/]
- 21. Precise, predictable genome integrations by deep-learning ... [https://www.nature.com/articles/s41587-025-02771-0]
- 22. Zebrafish Unga Is Required for Genomic Maintenance ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12452583/]
- 23. CRISPR-based functional genomics tools in vertebrate ... [https://www.nature.com/articles/s12276-025-01514-0]
- 24. Dynamic interplay of cNHEJ and MMEJ pathways of DNA ... [https://www.nature.com/articles/s41598-025-88564-6]
- 25. Orthologues with homologous function | PLOS One [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0321886]
- 26. Homology-Directed Repair in Zebrafish: Witchcraft and ... [https://www.frontiersin.org/journals/molecular-biosciences/articles/10.3389/fmolb.2020.595474/full]
- 27. Accurate quantification of homologous recombination in ... [https://www.nature.com/articles/s41598-017-16725-3]
- 28. The Zebrafish as an Emerging Model to Study DNA Damage ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6335974/]
- 29. Zebrafish (Danio rerio) using as model for genotoxicity and ... [https://www.sciencedirect.com/science/article/abs/pii/S0048969720376154]
- 30. Comparative analysis of multiple DNA double-strand break ... [https://www.nature.com/articles/s42003-025-08187-5]
- 31. DNA repair genes play a variety of roles in the ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2023.1119229/full]
- 32. Repair of DNA Double-Strand Breaks by the Non-homologous ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8899865/]
- 33. CRISPR-Cas9-mediated homology-directed repair for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11531618/]
- 34. Methodologies for Improving HDR Efficiency [https://www.frontiersin.org/journals/genetics/articles/10.3389/fgene.2018.00691/full]
- 35. Genome editing using CRISPR/Cas9-based knock-in ... [https://www.sciencedirect.com/science/article/abs/pii/S1046202316302833]
- 36. The Redox Activity of Protein Disulphide Isomerase ... [https://pubmed.ncbi.nlm.nih.gov/40371563/]
- 37. CRISPR-Cas9 and Its Bioinformatics Tools: A Systematic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12109910/]
- 38. Transferable approaches to CRISPR-Cas9 induced genome ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12232765/]
- 39. CRISPR/Cas9-mediated homology-directed repair by ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6215429/]
- 40. Developmental progression of DNA double-strand break ... [https://www.nature.com/articles/s41467-024-46479-2]
- 41. Method for Analyzing Microinjection Oocytes... Zebrafish Early Stage [https://pmc.ncbi.nlm.nih.gov/articles/PMC8607826/]
- 42. of Microinjection Embryos to Analyze Gene Function Zebrafish [https://www.jove.com/t/1115/microinjection-of-zebrafish-embryos-to-analyze-gene-function]
- 43. The influence of microinjection parameters on cell survival ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10034491/]
- 44. Microinjection - an overview [https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/microinjection]
- 45. Protocol to screen chemicals to enhance homology ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12491152/]
- 46. Single Cell Transfection through Precise Microinjection ... [https://www.nature.com/articles/srep24127]
- 47. Fine-Tuning Homology-Directed Repair (HDR) for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12071731/]
- 48. Fine-Tuning Homology-Directed Repair (HDR) for ... [https://www.mdpi.com/1422-0067/26/9/4067]
- 49. CRISPR/Cas9-Mediated Genome Editing in Zebrafish - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11753827/]
- 50. Protocol CRISPR-Cas9-induced gene knockout in zebrafish [https://www.sciencedirect.com/science/article/pii/S2666166722006591]
- 51. Efficient knock-in method enabling lineage tracing ... [https://www.life-science-alliance.org/content/6/5/e202301944]
- 52. Mastering Zebrafish CRISPR Knock-Ins: Breaking Barriers ... [https://invivobiosystems.com/gene-editing/mastering-zebrafish-crispr-knock-in/]
- 53. A robust pipeline for efficient knock-in of point mutations and ... [https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-022-08971-1]
- 54. Prime editing outperforms homology-directed repair as a ... [https://pubmed.ncbi.nlm.nih.gov/40437234/]
- 55. Optimized CRISPR Knock-In via HDR | IDT [https://www.idtdna.com/pages/applications/hdr-for-introducing-mutations]
- 56. CRISPR-Cas9-mediated homology-directed repair for ... [https://www.sciencedirect.com/science/article/pii/S2162253124002312]
- 57. CRISPR 101: Homology Directed Repair [https://blog.addgene.org/crispr-101-homology-directed-repair]
- 58. Review Optimizing homology-directed repair for gene editing [https://www.sciencedirect.com/science/article/abs/pii/S0168952525001064]
- 59. An update on precision genome editing by homology- ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8968426/]
- 60. Efficient targeted integration directed by short homology in ... [https://elifesciences.org/articles/53968]
- 61. Optimal Length for Left & Right Homology Arms for CRISPR [https://eu.idtdna.com/pages/support/faqs/when-designing-donor-dna-for-use-in-homology-directed-repair-(hdr)-what-are-the-optimal-lengths-of-the-left-and-right-homology-arms-and-what-is-the-maximum-size-of-sequence-that-can-be-efficiently-inserted-in-mammalian-cells]
- 62. Precise genome editing by homologous recombination [https://pmc.ncbi.nlm.nih.gov/articles/PMC5127288/]
- 63. Zebrafish in High-Throughput Screening: Optimizing Drug ... [https://www.zeclinics.com/blog/zebrafish-in-high-throughput-screening]
- 64. Chemical screening in zebrafish for novel biological and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5535795/]
- 65. Unlocking the zebrafish gene editor's toolbox - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6590056/]
- 66. A high-throughput system for drug screening based on the ... [https://www.sciencedirect.com/science/article/pii/S2405844024125261]
- 67. Chemical reprogramming enhances homology-directed ... [https://pubmed.ncbi.nlm.nih.gov/31149642/]
- 68. Advance trends in targeting homology-directed repair for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9376165/]
- 69. RS-1 enhances CRISPR/Cas9- and TALEN-mediated ... [https://www.nature.com/articles/ncomms10548]
- 70. CRISPR/Cas9 Ribonucleoprotein Delivery Enhanced by Lipo ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12073011/]
- 71. Improving the Efficiency of CRISPR Ribonucleoprotein ... [https://www.mdpi.com/2076-2615/14/5/719]
- 72. Chemical reprogramming enhances homology-directed genome editing in zebrafish embryos - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6533270/]
- 73. Synergistic gene editing in human iPS cells via cell cycle ... [https://www.nature.com/articles/s41467-020-16643-5]
- 74. Efficient high-precision homology-directed repair ... [https://www.nature.com/articles/s41592-023-01949-1]
- 75. Enhancing homology-directed repair efficiency with HDR- ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11315919/]
- 76. The hidden risks of CRISPR/Cas: structural variations and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12325979/]
- 77. Comprehensive benchmarking of genome editing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12144422/]
- 78. Plan Your CRISPR Experiment [https://www.addgene.org/guides/crispr/plan-your-experiment/]
- 79. Comprehensive protocols for CRISPR/Cas9-based gene ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4988528/]
- 80. Effect of melanocortin-4 receptor (mc4r) genome editing on ... [https://www.sciencedirect.com/science/article/abs/pii/S0889157525005976]
- 81. Generation of a zebrafish knock-in line expressing MYC- ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9050874/]
- 82. Identifying optimal conditions for precise knock-in of ... - PubMed [https://pubmed.ncbi.nlm.nih.gov/40446205/]
- 83. Improved selection of zebrafish CRISPR editing by early ... [https://www.nature.com/articles/s41598-023-27503-9]
- 84. Efficient genome editing using modified Cas9 proteins in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10997048/]
- 85. Boosting targeted genome editing using the hei-tag - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9068219/]
- 86. Standardization of zebrafish drug testing parameters for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10820820/]
- 87. Towards zebrafish model applications in drug discovery ... [https://www.sciencedirect.com/science/article/abs/pii/S0014299925005308]
- 88. Non-Homologous End Joining and Homology Directed DNA Repair Frequency of Double-Stranded Breaks Introduced by Genome Editing Reagents | PLOS One [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0169931]
- 89. Detection of Deleterious On-Target Effects after HDR-Mediated CRISPR Editing - PubMed [https://pubmed.ncbi.nlm.nih.gov/32460021/]
- 90. Homology-Directed Repair in Zebrafish: Witchcraft and Wizardry? - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7793982/]
- 91. Standardizing Exposure Media to Enhance the Rigor and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12574817/]
- 92. Standardizing Exposure Media to Enhance the Rigor and ... [https://pubmed.ncbi.nlm.nih.gov/41090622/]
- 93. Optimal tagging strategies for illuminating expression ... [https://www.nature.com/articles/s42003-023-05686-1]
- 94. CRISPR screening redefines therapeutic target ... [https://www.sciencedirect.com/science/article/pii/S2095177925001741]
- 95. Generation of a zebrafish knock-in line expressing MYC ... [https://www.sciencedirect.com/science/article/abs/pii/S0006291X22004429]
- 96. Systematic quantification of HDR and NHEJ reveals effects ... [https://www.nature.com/articles/srep23549]
- 97. Predictable NHEJ Insertion and Assessment of HDR ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9037586/]