Nodal Signaling in Zebrafish Mesendodermal Patterning: From Molecular Mechanisms to Research Applications
This comprehensive review explores the pivotal role of Nodal signaling in zebrafish mesendoderm induction and patterning, providing researchers and drug development professionals with current mechanistic insights and practical methodologies.
Nodal Signaling in Zebrafish Mesendodermal Patterning: From Molecular Mechanisms to Research Applications
Abstract
This comprehensive review explores the pivotal role of Nodal signaling in zebrafish mesendoderm induction and patterning, providing researchers and drug development professionals with current mechanistic insights and practical methodologies. We examine foundational principles including ligand-receptor interactions, feedback regulation, and maternal control of Nodal gene expression. The article details cutting-edge techniques for manipulating and monitoring Nodal signaling, addresses common experimental challenges, and validates zebrafish as a powerful model for studying TGF-β signaling pathways relevant to human development and disease.
Core Mechanisms of Nodal Signaling in Mesendoderm Formation
The Nodal signaling pathway is a cornerstone of vertebrate body plan establishment, with its components orchestrating a complex sequence of patterning events during early embryogenesis. In zebrafish, the Nodal-related ligands Squint (Sqt/Ndr1), Cyclops (Cyc/Ndr2), and Southpaw (Spaw/Ndr3) function through a sophisticated receptor complex to induce and pattern the mesoderm and endoderm. This technical review synthesizes current understanding of their distinct and overlapping functions, quantitative phenotypic outcomes, receptor interactions, and experimental methodologies. Framed within the broader context of mesendodermal patterning research, we provide structured data compilations, signaling pathway visualizations, and essential research tools to facilitate advanced investigation of this critical developmental pathway.
Nodal ligands, belonging to the Transforming Growth Factor-β (TGF-β) superfamily, are secreted cytokines that play indispensable roles in zebrafish embryogenesis. The three zebrafish Nodal ligands—Squint (Sqt), Cyclops (Cyc), and Southpaw (Spaw)—exhibit distinct yet partially redundant functions in establishing the embryonic axes and germ layers [1] [2]. These signals are transduced through a conserved receptor complex comprising Type I (Acvr1) and Type II (Acvr2) Activin receptors, alongside the essential EGF-CFC family co-receptor One-eyed pinhead (Oep) [3] [4]. Signal activation initiates an intracellular phosphorylation cascade culminating in Smad2/3 translocation to the nucleus and regulation of target gene expression. The precise spatiotemporal control of this pathway is critical for normal development; its disruption leads to severe defects including loss of mesendodermal tissues, cyclopia, and impaired left-right asymmetry [5] [6]. This review systematically examines the specialized functions of each ligand, their integrated operation within the signaling network, and the experimental frameworks used for their investigation.
Core Ligands: Functions, Expression, and Phenotypes
Squint (Sqt/Ndr1)
Squint functions as a potent long-range morphogen during early blastula stages. Maternal Sqt transcripts are deposited in the oocyte and are later joined by zygotic expression at the blastula margin, forming a signaling gradient that patterns the mesendoderm [1] [7]. Unlike other Nodal ligands, Sqt exhibits unique long-range signaling activity and contains an evolutionarily conserved 3' untranslated region (UTR) that facilitates dorsal targeting of its mRNA [1]. The penetrance of sqt mutant phenotypes is notably variable and influenced by genetic modifiers and environmental factors such as temperature, suggesting Sqt may provide evolutionary advantage by buffering embryos against genetic and environmental perturbations [1].
Table 1: Zebrafish Nodal Ligands: Functions and Mutant Phenotypes
| Ligand | Primary Functions | Expression Dynamics | Mutant Phenotypes | Genetic Redundancy |
|---|---|---|---|---|
| Squint (Sqt/Ndr1) | Mesendoderm induction, Dorsal organizer formation, Long-range patterning | Maternal and zygotic; blastula margin, dorsal lineage | Variable: delayed organizer formation, cyclopia, loss of ventral brain; viable adults to lethal | Partially redundant with Cyc |
| Cyclops (Cyc/Ndr2) | Mesendoderm formation, Ventral neural tube, Floor plate specification | Zygotic; blastula margin, midline mesendoderm | Cyclopia, loss of ventral diencephalon and floor plate, left-right defects; lethal | Partially redundant with Sqt |
| Southpaw (Spaw/Ndr3) | Left-right asymmetry establishment | Late segmentation; left lateral plate mesoderm (LPM) | Situs inversus, heart looping defects | Specialized function |
Cyclops (Cyc/Ndr2) and Southpaw (Spaw/Ndr3)
Cyclops acts predominantly as a short-range signal with expression initiating in mesendoderm precursor cells during the blastula stage and persisting in midline structures throughout gastrulation [1] [7]. While Cyc and Sqt show functional redundancy in mesendoderm induction, Cyc has unique essential functions in patterning the ventral neural tube and establishing the floor plate [5]. In contrast to the incomplete penetrance of sqt mutants, cyc deficiency produces fully penetrant cyclopia and embryonic lethality [1]. Southpaw functions predominantly after gastrulation, exhibiting left-sided expression in the lateral plate mesoderm where it directs the establishment of left-right asymmetry for visceral organ patterning [2] [6]. Unlike the early widespread functions of Sqt and Cyc, Spaw activity is temporally and spatially restricted to later asymmetry determination.
Quantitative Phenotypic Analysis
Table 2: Quantitative Phenotypes in Nodal Signaling Mutants
| Genotype / Condition | Mesendoderm Defects | Cyclopia Penetrance | Neural Tube Defects | Left-Right Defects |
|---|---|---|---|---|
| sqt (zygotic) | Mild to severe (dose-dependent) | Incomplete (13-34% in adults) | Variable: elongated/divided pineal | Not reported |
| cyc (zygotic) | Moderate (ventral CNS) | Complete (100%) | None (round pineal) | Randomized |
| sqt; cyc double mutant | Complete loss | Complete (100%) | Not specified | Not specified |
| MZoep | Complete loss | Complete (100%) | Severe (widely separated pineal) | Not specified |
| acvr1b-a; acvr1b-b double mutant | Complete loss | Complete (100%) | Not specified | Not specified |
| SB-431542 treatment | Complete loss | Complete (100%) | Not specified | Not specified |
Receptor Complexes and Signaling Mechanisms
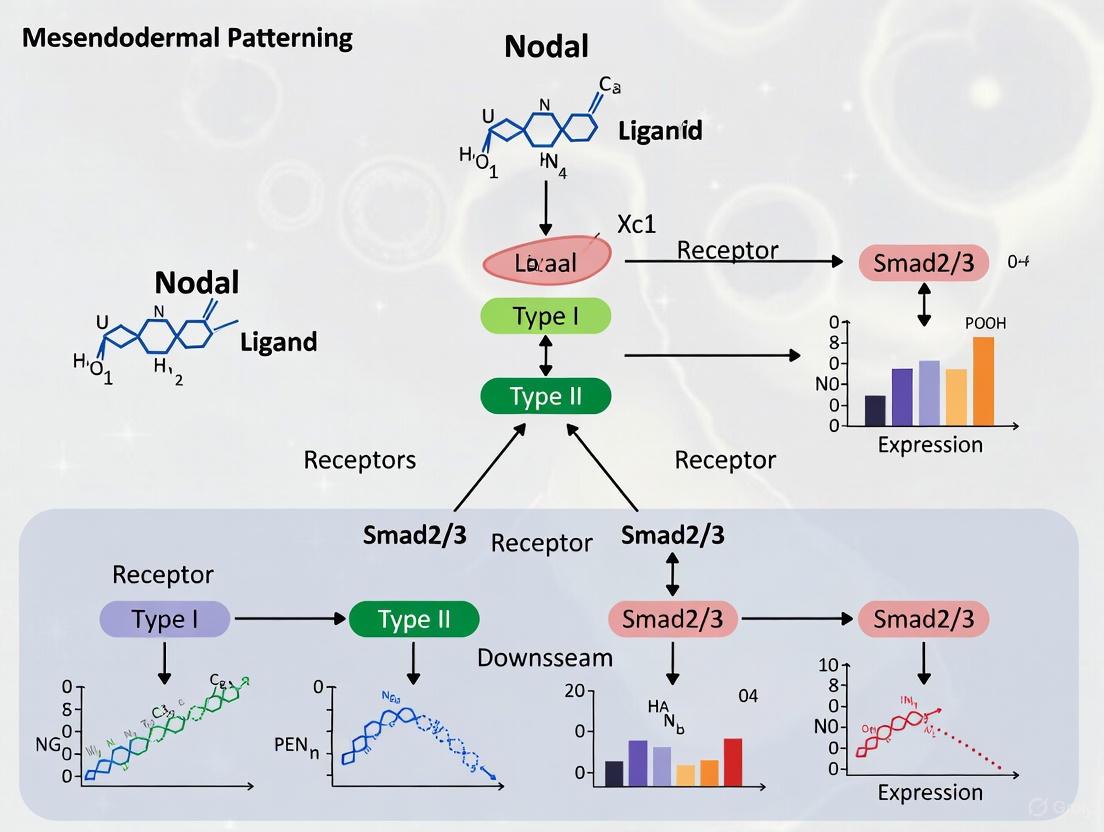
Nodal signal transduction is mediated by a sophisticated cell surface receptor system. The core complex includes Type I receptors (Acvr1b-a, Acvr1b-b), Type II receptors (Acvr2a, Acvr2b-a, Acvr2b-b), and the indispensable EGF-CFC co-receptor Oep [3] [4]. Upon ligand binding, Type II receptors phosphorylate Type I receptors, which subsequently activate the downstream Smad2/3 transcription factors. The co-receptor Oep plays a particularly critical role beyond simple signal facilitation; it regulates the spatial distribution of Nodal ligands by controlling their extracellular diffusion and enhancing cellular sensitivity to the signals [3]. Recent genetic analyses reveal that Type I receptors Acvr1b-a and Acvr1b-b function redundantly as the primary mediators of Nodal signaling, while Type II receptors exhibit both Nodal-dependent and independent functions in embryonic patterning [4].
Nodal Signaling Pathway and Key Components: This diagram illustrates the core Nodal signaling pathway in zebrafish, from ligand-receptor binding to target gene activation, including regulatory feedback mechanisms.
Experimental Approaches and Methodologies
Genetic Loss-of-Function Strategies
Genetic analysis remains fundamental to Nodal pathway investigation. Maternal-zygotic mutants provide the most complete loss-of-function scenarios, as in MZoep mutants which lack both maternally deposited and zygotically expressed gene products, resulting in complete absence of mesendoderm [5]. Compound mutants reveal functional redundancy; while single sqt or cyc mutants show specific defects, sqt;cyc double mutants display complete absence of mesoderm and endoderm, demonstrating their collective essential role [7]. CRISPR/Cas9-generated mutants have enabled precise dissection of receptor functions, revealing that simultaneous knockout of both acvr1b-a and acvr1b-b Type I receptors is required to recapitulate the complete Nodal loss-of-function phenotype [4].
Pharmacological Inhibition
Small molecule inhibitors provide temporal control over Nodal signaling disruption. SB-431542 and SB-505124 specifically target the kinase activity of ALK4/5/7 Type I receptors, completely blocking signal transduction when applied after the mid-blastula transition [7]. This pharmacological approach demonstrated that Nodal signaling is required during a specific competency window from mid-to-late blastula stages (3-5 hours post-fertilization) for sequential specification of mesendodermal derivatives [7]. Treatment with 800 μM SB-431542 produces phenotypes indistinguishable from sqt;cyc double mutants, confirming its efficacy as a Nodal pathway inhibitor [7].
Quantitative Imaging and Gradient Analysis
Advanced imaging techniques have revealed the biophysical properties of Nodal morphogen gradients. Fluorescently tagged ligands (e.g., Sqt-GFP, Cyc-GFP) enable direct visualization of ligand distribution, demonstrating that Sqt has greater effective range than Cyc [3]. These studies revealed that the co-receptor Oep restricts Nodal spread through receptor-mediated capture, shaping the morphogen gradient that patterns the germ layers [3]. Computational modeling combined with live imaging shows that without Oep replenishment, the Nodal gradient transforms into a traveling wave, highlighting the dynamic regulation of signaling distribution [3].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Nodal Signaling Studies
| Reagent / Tool | Type | Primary Application | Key Characteristics / Function |
|---|---|---|---|
| SB-431542 | Small molecule inhibitor | Temporal disruption of Nodal signaling | Inhibits ALK4/5/7 receptors; blocks signaling after MBT |
| Sqt, Cyc, Spaw mutants | Genetic models | Loss-of-function studies | Alleles: sqtcz35, sqthi975 (predicted nulls) |
| Oep (MZoep) mutants | Genetic model | Complete pathway blockade | Lacks maternal and zygotic Oep; no mesendoderm |
| Acvr1b-a; Acvr1b-b double mutants | Genetic model | Receptor function studies | Phenocopies Nodal loss-of-function |
| Sqt/Cyc-GFP fusions | Imaging tools | Ligand distribution studies | Measures diffusion coefficients and gradient formation |
| gdf3 mutants | Genetic model | Co-ligand function analysis | Maternal-zygotic mutants show reduced Nodal signaling |
| Nodal receptor morpholinos | Knockdown tools | Acute protein depletion | Combinatorial F0 knockdown for receptor analysis |
Regulatory Networks and Co-factors
Nodal signaling operates within a sophisticated regulatory network featuring multiple feedback mechanisms. Positive feedback amplifies the initial signal, as Nodal ligands induce their own expression as well as that of their co-factors [3] [2]. Negative feedback occurs through the induction of Lefty antagonists, which diffuse rapidly to restrict signal range and prevent overactivation [3]. The TGF-β family member Gdf3 (Vg1 ortholog) serves as an essential co-ligand, forming heterodimers with Sqt and Cyc to enhance signaling robustness [6]. Maternal depletion of Gdf3 results in severe reduction of Nodal signaling, demonstrating its role as a critical modulator rather than a redundant component [6].
Nodal Gradient Formation and Regulation: This diagram illustrates how the Nodal signaling gradient is established through diffusion from its source and shaped by co-receptor binding and feedback inhibition.
The zebrafish Nodal signaling pathway, with its sophisticated repertoire of ligands, receptors, and regulatory mechanisms, provides a powerful model system for understanding vertebrate embryonic patterning principles. The functional specialization of Squint, Cyclops, and Southpaw—despite their structural similarities—demonstrates how gene duplication and divergence can generate complexity in developmental networks. From a technical perspective, the comprehensive toolkit of genetic mutants, pharmacological inhibitors, and quantitative imaging approaches enables precise dissection of this essential pathway. Understanding these mechanisms has broader implications for regenerative medicine and disease modeling, particularly given the re-emergence of Nodal signaling in tumorigenesis and its role in maintaining stem cell pluripotency. Future research will likely focus on quantitative modeling of signal dynamics, single-cell analysis of cellular responses, and translational applications targeting Nodal signaling in cancer therapeutics.
This whitepaper synthesizes current research on the maternal regulation of Nodal signaling, a pivotal pathway in zebrafish mesendodermal patterning. We elucidate the essential and complementary roles of the maternal T-box transcription factor Eomesodermin A (Eomesa) and Huluwa (Hwa)-activated β-catenin signaling in the spatiotemporal initiation of zygotic ndr1 (squint) and ndr2 (cyclops) expression. While Hwa/β-catenin is the primary activator of ndr1 on the dorsal side, maternal Eomesa is crucial for its expression in the lateroventral margin and is the major regulator of ndr2. This guide provides a detailed dissection of the experimental evidence, including comprehensive quantitative data and methodologies, which together establish a model wherein maternal factors orchestrate a precise Nodal signaling landscape to ensure robust mesendoderm induction.
In vertebrate embryogenesis, the establishment of the body plan hinges on the precise specification of mesodermal and endodermal tissues, a process known as mesendodermal patterning. The Nodal signaling pathway, belonging to the Transforming Growth Factor-β (TGF-β) superfamily, is the principal inducer of this process across species, from sea urchins to mammals [8] [9]. In zebrafish, the Nodal-related genes ndr1/squint and ndr2/cyclops are redundantly required for the formation of most mesodermal and endodermal tissues; simultaneous deficiency of both genes leads to a near-complete loss of these germ layers [10] [8]. The regulation of ndr1 and ndr2 expression is therefore a critical control point in development. Following fertilization, early zebrafish development is directed by maternal gene products deposited into the egg. The transition to zygotic control occurs at the Mid-Blastula Transition (MBT), and it is at this juncture that maternal factors directly activate the expression of zygotic genes, including the nodal genes [10]. This whitepaper focuses on two key maternal determinants—Eomesodermin and the Hwa/β-catenin signaling axis—and their interplay in controlling the Nodal landscape that patterns the zebrafish embryo.
Maternal Regulators of Nodal Expression
Eomesodermin (Eomesa): A Versatile Maternal Determinant
Eomesodermin A (Eomesa) is a maternally expressed T-box transcription factor. Its transcripts are distributed in a vegetal-to-animal gradient during cleavage stages, prefiguring its role in marginal cell fates [10] [11]. Beyond its well-documented synergy with Bon and Gata5 to induce the endoderm marker cas (sox32) [12], Eomesa is a fundamental upstream regulator of Nodal gene expression. Genetic studies using maternal-zygotic eomesa mutants (Meomesa) revealed that Eomesa is indispensable for the initial expression of ndr2 and for the lateroventral expression of ndr1 [10]. This positions Eomesa as a functional counterpart to the Xenopus T-box factor VegT, which is known to activate Nodal-related genes [10].
Hwa/β-catenin Signaling: The Dorsal Initiator
The dorsal-vental axis in zebrafish is established through the activity of the maternal factor Huluwa (Hwa), which encodes a transmembrane protein essential for stabilizing β-catenin on the future dorsal side of the embryo [10]. Mutants lacking maternal hwa (Mhwa) are severely ventralized, phenocopying β-catenin deficient mutants [10]. This Hwa/β-catenin signaling pathway acts as a major dorsal activator of ndr1 expression. The requirement for β-catenin signaling in dorsal Nodal induction demonstrates a conserved mechanism with Xenopus, where nuclear β-catenin synergizes with VegT to enhance Nodal expression [10].
Integrated Model of Regulation
The genetic interaction between these pathways was definitively established using Meomesa;Mhwa double mutants. In these embryos, the expression of both ndr1 and ndr2 is completely abolished, indicating that the functions of maternal Eomesa and Hwa are together essential for the initiation of Nodal signaling [10]. However, these factors contribute differentially to the expression of each Nodal gene and across different regions of the embryo, as detailed in the quantitative analysis below.
Quantitative Analysis of Genetic Interactions
The distinct and overlapping contributions of maternal Eomesa, Hwa/β-catenin, and Nodal autoregulation to ndr1 and ndr2 expression are quantified and summarized in the table below.
Table 1: Relative Contributions of Maternal Factors to ndr1 and ndr2 Expression
| Gene | Dorsal Margin Expression | Lateroventral Margin Expression | Primary Regulator | Secondary Regulator(s) |
|---|---|---|---|---|
ndr1 (squint) |
Severely reduced or absent in Mhwa mutants [10] |
Severely reduced or absent in Meomesa mutants [10] |
Hwa/β-catenin (dorsal); Eomesa (lateroventral) [10] | Nodal autoregulation (ventral expansion) [10] |
ndr2 (cyclops) |
Minor reduction in Mhwa mutants [10] |
Severely reduced or absent in Meomesa mutants [10] |
Maternal Eomesa [10] | Minor contribution from Hwa/β-catenin and Nodal autoregulation [10] |
Table 2: Phenotypic Consequences of Mutations in Maternal and Nodal Pathway Components
| Genotype / Condition | Mesendoderm Phenotype | Key Molecular Deficits |
|---|---|---|
MZoep (No Nodal co-receptor) |
Near-complete loss of mesoderm and endoderm; failure of neural tube closure [5] [9] | Absence of ndr1 and ndr2 signaling [9] |
Meomesa |
Defects in endoderm marker expression; delayed epiboly initiation [11] | Loss of lateroventral ndr1 and most ndr2 expression [10] |
Mhwa |
Severe ventralization (Class I) [10] | Loss of dorsal ndr1 expression [10] |
Meomesa;Mhwa |
Synthetic severe phenotype | Complete abolition of ndr1 and ndr2 expression [10] |
| Nodal Inhibition (SB431542) | Disruption of mesendodermal patterning [10] | Reduced ventral expansion of ndr1 domain [10] |
Experimental Protocols for Key Studies
Genetic Dissection Using Maternal Mutants
Objective: To determine the individual and combined requirements of maternal eomesa and hwa in the spatiotemporal regulation of zygotic ndr1 and ndr2 expression.
Methodology:
- Zebrafish Strains: The following mutant lines were utilized:
- Generation of Maternal-Zygotic Mutants:
- Maternal
eomesa(Meomesa): Homozygouseomesamutant females were generated and their eggs, devoid of functional maternal Eomesa protein, were fertilized with mutant sperm [11]. - Maternal
hwa(Mhwa): A similar strategy was employed usinghwamutant females [10]. - Double Mutants (
Meomesa;Mhwa): Double heterozygotes (eomesa tsu007/+ ;hwa tsu01sm/+) were crossed to generate double homozygous mothers [10].
- Maternal
- Spatiotemporal Expression Analysis:
- Whole-mount In Situ Hybridization (WISH): Embryos were collected at specific developmental stages (e.g., 3.3 hpf to shield stage) and fixed. Digoxigenin-labeled antisense RNA probes for
ndr1andndr2were used to visualize transcript localization [10] [11]. - Pharmacological Inhibition: The small molecule SB431542, a specific inhibitor of the Activin/Nodal type I receptor ALK4, was used to treat wild-type and mutant embryos to assess the contribution of Nodal autoregulation to its own expression domains [10].
- Whole-mount In Situ Hybridization (WISH): Embryos were collected at specific developmental stages (e.g., 3.3 hpf to shield stage) and fixed. Digoxigenin-labeled antisense RNA probes for
Molecular Analysis of the Eomesa Nexus
Objective: To define the molecular mechanism by which Eomesa integrates Nodal signaling and endoderm specification.
Methodology:
- Gel Shift Assays (EMSA): Recombinant Eomesa protein or embryo extracts were incubated with a labeled DNA probe containing a T-box binding site from the
caspromoter. Supershift or competition with unlabeled wild-type/mutant oligonucleotides confirmed specific binding [12]. - Co-Immunoprecipitation (Co-IP): Plasmids expressing Eomesa, Bon, and Gata5 were transfected into cultured cells. Protein complexes were immunoprecipitated using an antibody against one factor (e.g., Eomesa) and the immunoblot was probed for the others (e.g., Bon and Gata5) to demonstrate direct physical interaction [12].
- Promoter-Reporter Assays: Wild-type and mutant versions of the
caspromoter (e.g., with a mutated Eomesa binding site) were cloned upstream of a luciferase reporter gene. These constructs were injected into zebrafish embryos, with or without co-injection ofeomesamRNA, and luciferase activity was measured to assess transcriptional synergy [12].
Signaling Pathway and Regulatory Logic
The following diagram synthesizes the complex regulatory interactions between maternal factors, Nodal genes, and their downstream targets as established in the cited research.
Diagram 1: Maternal and Regulatory Control of Nodal Signaling. This diagram illustrates the primary pathways through which maternal Hwa/β-catenin and maternal Eomesa activate the expression of the Nodal genes ndr1 and ndr2. The resulting Nodal ligand signals through a receptor complex requiring the EGF-CFC co-receptor Oep, leading to the formation of an active Smad/FoxH1 complex. This complex drives the expression of mesendoderm target genes and reinforces Nodal expression through a positive feedback loop.
The Scientist's Toolkit: Key Research Reagents
This table catalogues essential reagents used in the featured studies to facilitate experimental replication and further investigation.
Table 3: Essential Research Reagents for Studying Maternal Control of Nodal Signaling
| Reagent / Tool | Type | Key Function in Research | Example Use Case |
|---|---|---|---|
eomesa tsu007/fh105 |
Mutant Allele | CRISPR/TILLING-generated loss-of-function mutants for dissecting maternal vs. zygotic roles [10] [11] | Generation of maternal-zygotic (Meomesa) mutants to analyze ndr2 expression [10] |
hwa tsu01sm |
Mutant Allele | Loss-of-function mutant to disrupt maternal β-catenin signaling and dorsal axis specification [10] | Generation of Mhwa mutants to assess dorsal-specific ndr1 loss [10] |
| SB431542 | Small Molecule Inhibitor | Selective inhibitor of the TGF-β/Activin/Nodal type I receptor ALK4/5/7 [10] | Inhibition of Nodal autoregulation to study its role in ndr1 expression domain maintenance [10] |
| Anti-Eomesa Antibody | Custom Antibody | Polyclonal antibody for protein detection via Western Blot and whole-mount immunohistochemistry [11] | Confirmation of Eomesa protein loss in mutant embryos and analysis of its expression gradient [11] |
ndr1/sqt & ndr2/cyc RNA Probes |
In Situ Hybridization Probe | Digoxigenin-labeled antisense RNA for spatial visualization of transcript expression [10] [11] | Mapping the dynamic expression domains of ndr1 and ndr2 in wild-type and mutant embryos [10] |
| OptoNodal2 System | Optogenetic Tool | Light-controllable Nodal receptor system for high-resolution spatial/temporal perturbation of signaling [13] | Creating synthetic Nodal signaling patterns to study fate decision-making in live embryos [13] |
The body of research synthesized in this whitepaper firmly establishes that the maternal factors Eomesodermin and Hwa/β-catenin are non-redundant, upstream initiators of the Nodal signaling cascade in zebrafish. They act as regional specialists—Hwa/β-catenin on the dorsal side and Eomesa broadly throughout the margin with a predominant role for ndr2—to combinatorially ensure the precise onset of ndr1 and ndr2 expression, thereby launching the mesendodermal gene regulatory network.
Future research will likely focus on several frontiers. First, identifying the direct transcriptional targets of Eomesa and β-catenin at the ndr1 and ndr2 promoters will provide a more precise molecular understanding of their activation mechanisms. Second, the integration of new technologies, such as the improved OptoNodal2 system [13], will enable researchers to move beyond static loss-of-function studies and actively probe how dynamic Nodal signaling patterns are interpreted by embryonic cells. Finally, investigating the robustness of this network, particularly the role of feedback inhibitors like Lefty within the activator-inhibitor motif [14], will be crucial for understanding how this critical developmental system buffers against genetic and environmental variability. For drug development professionals, the exquisite regulation of Nodal signaling offers a paradigm for understanding how morphogen pathways can be targeted, with implications for regenerative medicine and cancer biology, where Nodal signaling is often reactivated.
The formation of mesoderm and endoderm (mesendoderm) in the early zebrafish embryo is orchestrated by a finely-tuned signaling system centered on Nodal proteins, a subset of the Transforming Growth Factor-β (TGF-β) superfamily [8] [15]. A defining feature of this system is a negative feedback loop wherein Nodal signaling activates the expression of its own inhibitors, the Lefty proteins [16] [8]. This activator-inhibitor relationship is considered a paradigm for how feedback inhibition controls pattern formation in vertebrate development [16]. The core circuitry involves Nodal ligands such as Squint (Sqt) and Cyclops (Cyc) promoting their own transcription (autoregulation) while simultaneously inducing the expression of Lefty1 and Lefty2, which in turn diffuse and inhibit Nodal signaling extracellularly [16] [17]. This review synthesizes current research on the mechanisms and functional significance of this feedback loop, with a specific focus on its critical role in ensuring robust zebrafish mesendodermal patterning.
Core Molecular Mechanisms of the Nodal/Lefty Feedback Loop
Nodal Signaling and Autoregulation
Nodal signaling is initiated when a mature Nodal ligand (e.g., Squint or Cyclops) binds to a cell-surface receptor complex. This complex consists of Type I and Type II Activin serine/threonine kinase receptors and an essential EGF-CFC co-receptor (One-eyed pinhead, Oep, in zebrafish) [8] [18] [15]. Ligand binding leads to the phosphorylation of the intracellular signal transducers Smad2 and Smad3. These phosphorylated Smads form a complex with Smad4, which translocates to the nucleus [8] [18]. Within the nucleus, this Smad complex associates with transcription factors such as FoxH1 to activate the expression of target genes [18] [15]. Critically, these target genes include the squint and cyclops genes themselves, establishing a positive autoregulatory loop that amplifies and sustains Nodal signaling [8]. This same transcriptional complex also activates the expression of the lefty genes, instigating the negative feedback arm of the circuit [16].
Lefty-Mediated Inhibition
The Lefty proteins (Lefty1 and Lefty2) are divergent, secreted members of the TGF-β superfamily that function as potent extracellular antagonists of Nodal signaling [8] [19]. Research has elucidated two distinct mechanistic modes by which Lefty proteins inhibit Nodal activity, providing efficient and stringent regulation [19]:
- Direct Ligand Binding: Lefty can directly interact with the Nodal ligand in the extracellular space, physically preventing Nodal from accessing and binding to its receptor complex [19].
- Co-receptor Interference: Lefty can bind directly to the EGF-CFC co-receptor (Oep/Cripto), inhibiting the ability of this essential cofactor to participate in the formation of an active Nodal receptor complex [19].
Table 1: Modes of Lefty Inhibition of Nodal Signaling
| Mechanism | Molecular Interaction | Functional Consequence |
|---|---|---|
| Direct Ligand Blockade | Lefty binds to Nodal ligand | Prevents Nodal from interacting with its receptors |
| Co-receptor Interference | Lefty binds to EGF-CFC (Oep) | Disrupts formation of the active receptor complex |
Pathway Visualization
The following diagram illustrates the core components and interactions of the Nodal/Lefty feedback loop:
Biophysical Properties Governing Signal Propagation
The different signaling ranges of Nodal and Lefty are not merely a function of production sites but are critically determined by their distinct biophysical properties and interactions with the extracellular environment. Single-molecule tracking studies in live zebrafish embryos have provided direct support for the hindered diffusion model, revealing how tissue geometry and binding interactions shape morphogen gradients [20].
Hindered Diffusion of Nodal and Lefty
In vivo measurements show that while Nodal and Lefty molecules have similar free diffusion coefficients in open extracellular "cavities," their mobility is significantly reduced in regions of close cell-cell contact ("interfaces") [20]. However, Nodal ligands exhibit a much higher "bound fraction" and longer binding times (tens of seconds) on cell surfaces compared to Lefty proteins [20]. This is attributed to Nodal's high-affinity binding to its receptors and co-receptors. This transient trapping on cell surfaces hinders Nodal movement, restricting it to short-range action. In contrast, the more mobile Lefty proteins, which lack this strong receptor binding, can diffuse over longer distances to exert their inhibitory influence [20].
Quantitative Diffusion Data
Table 2: Biophysical Properties of Nodal and Lefty Proteins from Single-Molecule Tracking
| Protein | Relative Local Mobility (in cavities) | Bound Fraction on Cell Surfaces | Typical Binding Time | Resulting Signaling Range |
|---|---|---|---|---|
| Cyclops (Nodal) | Low | High | Tens of seconds | Ultra-short range (few micrometers) |
| Squint (Nodal) | Intermediate | Intermediate | Tens of seconds | Short-to-mid range |
| Lefty1 | High | Low | Shorter | Long range |
| Lefty2 | Very High | Very Low | Shortest | Ultra-long range (near uniform) |
Experimental Evidence from Zebrafish Models
Phenotypic Consequences of Disrupted Feedback
Genetic loss-of-function experiments in zebrafish have been instrumental in deciphering the role of Lefty-mediated feedback. While lefty1 or lefty2 single mutants are viable with mild or no patterning defects, lefty1-/-;lefty2-/- double mutants are embryonic lethal [16]. These double mutants exhibit a dramatic expansion of Nodal signaling domains and a consequent excess of mesendoderm specification, leading to severe morphological defects including the loss of eyes, heart, and tail [16]. This phenotype demonstrates that without Lefty inhibition, Nodal signaling becomes hyperactive and spatially uncontrolled, disrupting normal embryonic patterning.
Key Experimental Protocol: Rescuing Patterning without Feedback
A pivotal experiment by Rogers et al. (2017) tested whether the feedback connection itself, rather than just the presence of inhibitor, is essential for development [16].
- Objective: To determine if Nodal patterning can function without inhibitory feedback by decoupling Lefty production from Nodal signaling.
- Experimental Subjects: lefty1-/-;lefty2-/- double mutant zebrafish embryos.
- Methodology - Two Rescue Conditions:
- Ectopic Lefty Expression: lefty mRNA was expressed ectopically in locations far from its normal expression domain in the mutant embryos.
- Uniform Pharmacological Inhibition: Mutant embryos were bathed in a solution of a Nodal inhibitor drug (e.g., SB505124), providing spatially and temporally uniform inhibition.
- Key Findings: Both rescue methods successfully restored normal mesendoderm patterning and viability to the lefty mutants [16]. This demonstrates that the precise, feedback-coupled expression of Lefty is not absolutely required for successful development. The system can function with uniform, non-feedback inhibition.
- Critical Insight on Robustness: While viable, the pharmacologically-rescued embryos were "fragile." They were less tolerant to mild perturbations in Nodal signaling levels compared to wild-type embryos, which can dynamically adjust Lefty expression to compensate for such fluctuations [16]. This indicates that the primary function of the Nodal-Lefty feedback loop is to ensure robustness and stability of the patterning process against genetic and environmental variations.
The Scientist's Toolkit: Key Research Reagents
Table 3: Essential Reagents for Studying Nodal/Lefty Signaling in Zebrafish
| Reagent / Tool | Function / Description | Key Use Case |
|---|---|---|
| lefty1/2 Mutant Zebrafish | Genetic null alleles (e.g., lefty1a145, lefty2a146) generated via TALENs or CRISPR. | Modeling loss of feedback inhibition; studying phenotypes of expanded Nodal signaling [16]. |
| Nodal Inhibitor Drugs (e.g., SB505124) | Small molecule inhibitors that block Activin/Nodal type I receptors. | Temporally controlled inhibition of pathway; rescuing lefty mutants without feedback [16]. |
| HaloTag-Labeled Morphogens | Nodal and Lefty proteins fused to the HaloTag for covalent, bright dye labeling. | Single-molecule tracking of diffusion and binding in live embryos [20]. |
| memGFP mRNA | mRNA encoding membrane-targeted GFP. | Visualizing cell outlines and defining extracellular spaces for single-molecule analysis [20]. |
| Anti-pSmad2 Antibodies | Antibodies specific to the phosphorylated (active) form of Smad2. | Immunohistochemical readout of active Nodal signaling domains in fixed embryos [16]. |
The Nodal/Lefty feedback loop is a cornerstone of zebrafish mesendodermal patterning. The experimental evidence confirms that the primary role of this intricate circuit is not to initiate patterning per se, but to buffer the system against perturbations, thereby ensuring developmental robustness [16]. The ability to pattern successfully with uniform inhibition, albeit with reduced tolerance to fluctuation, underscores that the spatial coupling of activator and inhibitor through feedback is a key evolutionary adaptation for reliability. Furthermore, disruptions in this finely balanced system have direct clinical relevance, as aberrant Nodal signaling is linked to congenital heart defects and laterality disorders like heterotaxy in humans [18]. A deep understanding of these feedback mechanisms therefore not only illuminates fundamental principles of embryonic patterning but also provides a foundation for exploring therapeutic interventions for birth defects.
The transformation of a fertilized egg into a complex organism requires the precise spatial and temporal coordination of cell fate decisions. A cornerstone of this process in vertebrate embryos is the formation of the three primary germ layers—ectoderm, mesoderm, and endoderm—which give rise to all adult tissues and organs. In zebrafish, as in other vertebrates, the TGF-β signaling molecule Nodal serves as the primary inducer of mesendodermal fates, orchestrating a sophisticated gene regulatory network that patterns the embryonic axes. The mesendoderm represents a common progenitor population that subsequently segregates into definitive mesodermal and endodermal lineages, with Nodal signaling levels acting as a morphogen to determine specific cell fates. Cells exposed to high Nodal concentrations adopt endodermal fates, intermediate levels direct mesodermal differentiation, while low or absent signaling permits ectodermal specification [21].
This whitepaper examines the molecular machinery through which Nodal signaling translates concentration gradients into discrete transcriptional programs and cellular behaviors. We explore the downstream transcription factors that interpret this signaling, the cis-regulatory elements that integrate these signals, and the terminal differentiation genes that ultimately execute germ layer-specific functions. Recent technological advances, including optogenetic perturbation and large-scale transcription factor interaction mapping, have provided unprecedented insight into the logic of this developmental system. Understanding these mechanisms not only illuminates fundamental biological principles but also informs efforts in regenerative medicine, where controlled differentiation of stem cells into specific lineages remains a critical challenge.
The Core Nodal Signaling Pathway and its Regulation
The Nodal signaling pathway comprises a conserved set of components that transmit extracellular signals to the nucleus, culminating in changes in gene expression. Nodal ligands – primarily Cyclops and Squint in zebrafish – function as morphogens that form concentration gradients emanating from the embryonic margin [21]. These ligands signal through a cell surface receptor complex consisting of type I and type II activin receptors along with the EGF-CFC co-receptor One-eyed pinhead (Oep). Genetic studies have demonstrated that Oep is not merely a permissive co-factor but actively shapes the Nodal signaling gradient by regulating ligand spread and cellular sensitivity [21].
Upon receptor activation, intracellular Smad2/3 transcription factors become phosphorylated, form complexes with Smad4, and translocate to the nucleus where they participate in transcriptional regulation. The signaling range and intensity are finely tuned by feedback mechanisms, including positive feedback on Nodal ligand production and negative feedback through induction of Lefty antagonists, which form an activator-inhibitor pair with Nodal ligands [14] [21]. This regulatory architecture enables the system to generate robust patterning outcomes despite potential environmental fluctuations.
Table 1: Core Components of the Nodal Signaling Pathway in Zebrafish
| Component Type | Key Elements | Function in Signaling |
|---|---|---|
| Ligands | Cyclops, Squint | Diffusible morphogens forming concentration gradients; often heterodimers with Vg1 |
| Receptors/Co-receptors | Type I/II Activin Receptors, Oep | Cell surface complex that binds ligands and initiates signaling |
| Intracellular Transducers | Smad2/3, Smad4 | Phosphorylated and form complexes that translocate to nucleus |
| Transcription Factors | FoxH1, Mix-type proteins | Cooperate with Smads to activate target genes |
| Antagonists | Lefty1, Lefty2 | Diffusible inhibitors that restrict signaling range |
Figure 1: Core Nodal Signaling Pathway with Feedback Loops. Nodal ligand binding activates receptor complexes, leading to Smad phosphorylation and nuclear translocation. Target gene activation includes positive feedback on Nodal production and negative feedback through Lefty antagonists.
From Signal to Pattern: Gradient Formation and Interpretation
The establishment of a Nodal signaling gradient is a dynamic process initiated by ligand secretion from the yolk syncytial layer (YSL) beneath the embryonic margin. Research has revealed that diffusive transport of Nodal ligands is sufficient to generate this gradient, contrary to models proposing obligatory feedback-driven relay mechanisms [21]. The shape and range of this gradient are critically determined by the EGF-CFC co-receptor Oep, which regulates ligand capture by target cells. In oep mutants, Nodal activity becomes nearly uniform throughout the embryo, demonstrating Oep's essential role in restricting ligand spread [21].
Cells interpret their position within the Nodal gradient through concentration-dependent activation of target genes. The signaling duration and intensity are translated into distinct transcriptional outputs through the combinatorial action of Smad complexes with various transcription factors. This interpretation mechanism enables cells to adopt specific fates according to their positional coordinates: high signaling levels activate endodermal genes like sox32 and sox17, intermediate levels induce mesodermal regulators such as ntl and gsc, while low or absent signaling permits ectodermal default programs [21].
Recent work has revealed that metabolic cues intersect with Nodal signaling to influence germ layer patterning. Glycolytic activity has been shown to modulate Nodal and Wnt signaling pathways, thereby influencing the proportional allocation of cells to different germ layers [22]. This connection between metabolism and patterning adds an additional layer of regulation that may link developmental programs to nutritional status.
Downstream Transcription Factors and Their Target Genes
The transcriptional response to Nodal signaling is mediated by a hierarchy of transcription factors that directly interpret Smad input and activate cell-type-specific genetic programs. Large-scale interaction mapping using CAP-SELEX has revealed an extensive network of transcription factor (TF) interactions that dramatically expand the gene regulatory code [23]. This methodology, which systematically tests TF-TF-DNA interactions, identified 2,198 interacting TF pairs among 58,000 tested combinations, with 1,329 showing preferred spacing/orientation and 1,131 forming novel composite motifs distinct from individual TF specificities [23].
Key Nodal-responsive transcription factors include FoxH1, which forms complexes with Smad2/3, and members of the Mix-type homeodomain family (e.g., Bon, Mixer, Mezzo). These factors activate cascades of gene expression that progressively specify mesodermal and endodermal sublineages. The discovery that TF-TF interactions frequently cross family boundaries and create novel DNA-binding specificities helps resolve the "hox specificity paradox," wherein TFs with similar binding specificities in vitro achieve distinct functional outcomes in vivo [23].
Table 2: Key Transcription Factor Interactions in Mesendodermal Patterning
| Transcription Factor Pair | Interaction Type | Functional Significance |
|---|---|---|
| FOXI1–ELF2 | Composite motif formation | Creates novel DNA binding specificity distinct from individual factors |
| HOXB13–MEIS1 | Spacing/orientation preference | Cooperative binding with specific distance constraints |
| BACH2–LMX1A | Long-range interaction (8-9bp gap) | Unusual example of cooperation over longer distances |
| GLI2–RFX3 | Cross-family interaction | Links Hedgehog signaling to cilia-related gene regulation |
| POU5F1 (OCT4)–SOX2 | Well-characterized pair | Maintains pluripotency; paradigm for TF cooperation |
The functional significance of these interactions is evident in their enrichment at cell-type-specific regulatory elements, where they integrate positional information to drive appropriate gene expression programs. For instance, TFs that define embryonic axes frequently interact with different partners and bind distinct composite motifs, explaining how similar DNA-binding domains can achieve diverse developmental outcomes [23].
Experimental Approaches and Methodologies
Optogenetic Perturbation of Nodal Signaling
Recent advances in optogenetics have enabled unprecedented spatial and temporal control over Nodal signaling, permitting direct testing of patterning models. An improved optoNodal2 system was developed by fusing Nodal receptors to the light-sensitive heterodimerizing pair Cry2/CIB1N, while sequestering the type II receptor to the cytosol [13]. This system eliminates dark activity and improves response kinetics without sacrificing dynamic range. Researchers adapted an ultra-widefield microscopy platform for parallel light patterning in up to 36 embryos simultaneously, demonstrating precise spatial control over Nodal signaling activity and downstream gene expression [13].
The experimental workflow involves several key steps:
- Embryo preparation: Zebrafish embryos expressing optoNodal2 components are collected and mounted for imaging and illumination.
- Light patterning: Custom illumination patterns are applied to activate Nodal signaling in defined spatial domains.
- Response monitoring: Signaling activity is tracked using live reporters of pathway activity (e.g., Smad localization) and target gene expression.
- Phenotypic analysis: Morphogenetic outcomes such as endodermal precursor internalization are quantified.
This approach has demonstrated that patterned Nodal activation can drive precise internalization of endodermal precursors and rescue developmental defects in Nodal signaling mutants, establishing a toolkit for systematic exploration of Nodal signaling patterns [13].
CAP-SELEX for Mapping Transcription Factor Interactions
The CAP-SELEX (consecutive-affinity-purification systematic evolution of ligands by exponential enrichment) method provides a high-throughput approach for identifying cooperative binding between transcription factors [23]. The adapted 384-well microplate format enables screening of thousands of TF-TF combinations through these key steps:
- TF expression and pairing: Human transcription factors are expressed in E. coli and combined into pairwise combinations (58,754 pairs in the recent study).
- DNA library incubation: TF pairs are incubated with a random oligonucleotide library.
- Consecutive affinity purification: Complexes containing both TFs and their bound DNA sequences are purified through sequential affinity tags.
- Sequencing and analysis: Bound DNA ligands are sequenced and analyzed using specialized algorithms to detect spacing/orientation preferences and novel composite motifs.
Two novel algorithms were developed for data analysis: a mutual information-based approach that identifies TF pairs with preferential binding to specific spacings/orientations, and a k-mer enrichment method that detects composite motifs differing from individual TF specificities [23].
Single-Cell Multiomics for Regulatory Network Inference
Single-cell RNA sequencing combined with single-cell ATAC sequencing enables the reconstruction of enhancer-driven gene regulatory networks with high resolution [24]. This approach involves:
- Cell isolation and processing: Single-cell suspensions are prepared from developing embryos or patterned stem cell models.
- Parallel library preparation: Both transcriptome and chromatin accessibility libraries are generated from the same cells.
- Data integration: Transcriptional and epigenomic profiles are combined to infer regulatory relationships.
- Regulon assembly: Transcription factors and their potential target genes are connected based on correlated activity patterns across cells.
Application of this method to T cell differentiation revealed how transcription factors like BATF and KLF2 govern cell fate decisions in the tumor microenvironment [24], illustrating approaches applicable to mesendodermal patterning studies.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Studying Nodal Signaling and Germ Layer Specification
| Reagent/Tool | Type | Key Features and Applications |
|---|---|---|
| OptoNodal2 System | Optogenetic reagent | Light-controllable Nodal signaling; Cry2/CIB1N heterodimerizing pair; no dark activity; improved kinetics [13] |
| CAP-SELEX Platform | TF interaction mapping | 384-well format; screens >58,000 TF pairs; identifies spacing preferences and composite motifs [23] |
| Ultra-widefield Microscopy | Imaging platform | Parallel light patterning in 36 embryos; precise spatial control of signaling [13] |
| Tg(myl7:EGFP-CAAX) | Transgenic zebrafish line | Membrane-targeted GFP in myocardial cells; enables live imaging of heart morphogenesis [25] |
| scRNA-seq + scATAC-seq | Multiomic profiling | Paired single-cell transcriptome and epigenome; reveals enhancer-driven regulons [24] |
Morphogenetic Outcomes: From Gene Expression to Tissue Architecture
The ultimate readout of germ layer specification is the transformation of transcriptional programs into complex three-dimensional tissues. Nodal signaling not only patterns gene expression but also directs morphogenetic behaviors that shape embryonic structures. In zebrafish heart development, Nodal signaling through the ligand Southpaw regulates asymmetric cellular behaviors that drive clockwise rotation of the heart tube [25].
High-resolution live imaging has revealed that myocardial cells undergo oriented cell rearrangement and cell shape changes that collectively drive convergent extension of the cardiac disc [25]. Interestingly, left-sided cells exhibit more active rearrangement and shape changes compared to right-sided cells, resulting in asymmetric deformation that rotates the heart tube. This left-right asymmetry is abolished when Nodal signaling is disrupted, demonstrating that Nodal directly modulates the cellular behaviors underlying organogenesis [25].
Figure 2: Nodal Regulation of Heart Tube Morphogenesis. Asymmetric Nodal signaling drives left-right differences in cellular behaviors, leading to tissue-scale asymmetric deformation and clockwise heart tube rotation.
The morphogenetic process can be divided into two temporally distinct phases: an early phase dominated by oriented cell rearrangement through intercalation, and a later phase driven primarily by cell shape changes [25]. This sophisticated coordination of cellular behaviors ensures the robust establishment of organ asymmetry essential for proper physiological function.
The journey from Nodal signaling activation to germ layer specification represents a paradigm of embryonic patterning, integrating dynamic signaling gradients, sophisticated transcriptional networks, and physical morphogenetic processes. Recent technological advances—particularly in optogenetics [13], large-scale TF interaction mapping [23], and single-cell multiomics [24]—have provided unprecedented resolution into these processes.
Key emerging principles include the central role of TF-TF composite motifs in expanding the regulatory lexicon [23], the importance of co-receptor expression in shaping morphogen gradients [21], and the direct regulation of cellular morphogenetic behaviors by patterning signals [25]. The integration of metabolic cues with signaling pathways adds another layer of regulation that connects developmental programs to physiological conditions [22].
Future research directions will likely focus on achieving quantitative, predictive models of pattern formation that incorporate the full complexity of TF interactions, feedback regulation, and cellular behaviors. The application of synthetic biology approaches to reconstruct patterning circuits in stem cell models will further test our understanding of these principles. As we deepen our knowledge of how transcription factors guide cells from pluripotency to differentiated states, we advance both fundamental biology and the prospects for controlled tissue engineering and regenerative medicine applications.
Within the broader study of zebrafish mesendodermal patterning research, understanding the cellular behaviors that translate molecular signals into physical form is paramount. The Nodal signaling pathway, a member of the TGF-β superfamily, serves as a master regulator in early vertebrate development. Its function extends beyond simple fate specification to directly orchestrating the complex morphogenetic movements that shape the embryo. This technical guide focuses on two fundamental cellular processes—convergent extension (CE) and other key tissue morphogenesis behaviors—driven by Nodal signaling. We examine how Nodal coordinates these processes through precise control of cell rearrangement, cell shape change, and directed migration, drawing upon recent optogenetic, live-imaging, and explant studies to provide a mechanistic framework for researchers and drug development professionals.
The Nodal Signaling Pathway: Mechanism and Regulation
The core Nodal pathway is an evolutionarily conserved system for controlling cell fate and behavior during embryonic development [8]. Activation begins when a Nodal ligand binds to a complex comprising Activin type I (Acvr1b) and type II serine/threonine kinase receptors at the cell surface. This interaction is dependent on EGF-CFC co-receptors (e.g., One-eyed pinhead/Oep in zebrafish, Cripto in mammals), which are essential for productive signal transduction [8]. Receptor activation leads to the phosphorylation of intracellular Smad2/3 proteins, which then form a complex with Smad4. This Smad complex translocates into the nucleus, where it associates with transcription factors such as FoxH1 to activate the expression of target genes, including Nodal itself (forming a positive feedback loop), Lefty (a secreted antagonist), and Sox32 (a key endodermal determinant) [8] [26].
The pathway is tightly regulated at multiple levels to ensure precise spatiotemporal signaling dynamics. Extracellular antagonists like Lefty and DAN family proteins (e.g., Cerberus) inhibit Nodal signaling by preventing receptor binding [8]. Intracellular negative regulators include Ectodermin, which promotes Smad4 mono-ubiquitination and nuclear export, and PPM1A, a phosphatase that dephosphorylates and inactivates Smad2/3 [8]. Additionally, the microRNA-430/427/302 family post-transcriptionally represses components like Nodal and Lefty, adding another layer of control to fine-tune signaling output [8]. The following diagram illustrates the core pathway and its key regulators.
Diagram of the core Nodal signaling pathway and its key regulators. The pathway is initiated by Nodal binding to its receptors and co-receptor, leading to Smad-dependent transcription of target genes. Multiple negative feedback mechanisms, including extracellular antagonists and intracellular inhibitors, precisely regulate signaling output.
Quantitative Data on Nodal-Driven Cellular Behaviors
Recent research has quantified the profound impact of Nodal signaling on specific cellular behaviors during gastrulation and organogenesis. The data reveal how Nodal governs tissue-level morphogenesis by regulating the magnitude and asymmetry of cellular processes.
Table 1: Quantitative Data on Nodal-Dependent Cellular Behaviors during Zebrafish Development
| Cellular Behavior | Tissue Context | Quantitative Measurement | Effect of Nodal Loss | Citation |
|---|---|---|---|---|
| Cell Rearrangement (Intercalation) | Left Heart Primordium | ~50% reduction in circumferential length in 9 hours (early phase, driver) | Abolished left-right asymmetry in cell behavior | [25] |
| Cell Shape Change (Shortening) | Left Heart Primordium | ~80% extension in perpendicular axis; progressive cell shortening (later phase, driver) | Abolished left-right asymmetry in cell behavior | [25] |
| Tissue Convergence | Left vs. Right Heart Primordia | More rapid convergence of the left primordium | Asymmetric convergence abolished; heart tube rotation fails | [25] |
| Unidirectional Ingression | Ectopic Endodermal Cells | Radial, highly polarized migration to inner layer; not random walk | Ingression requires Nodal signaling and Sox32 | [26] |
| Patterned Internalization | Endodermal Precursors | Precise spatial control driven by optogenetic Nodal patterns | Mutant defects rescued by synthetic Nodal patterns | [13] |
The BMP/Nodal Signaling Ratio as a Morphogenetic Regulator
Beyond absolute levels, the relative concentration of Nodal to other morphogens provides critical patterning information. Research using zebrafish explants has identified a critical developmental window during which the BMP/Nodal ratio dictates the type of convergent extension movements [27]. Precise temporal manipulation of these pathways revealed that:
- A high BMP/Nodal ratio specifically induces and enhances neuroectoderm-driven convergent extension.
- A low BMP/Nodal ratio specifically induces and enhances mesoderm-driven convergent extension. These findings demonstrate that the temporal dynamics of morphogen signaling ratios activate distinct morphogenetic programs within individual tissues [27].
Experimental Protocols for Investigating Nodal-Driven Behaviors
High-Resolution Live Imaging of Heart Tube Morphogenesis
This protocol enables the visualization and quantification of cellular behaviors during heart tube formation in zebrafish.
- Transgenic Line: Generate or use the Tg(myl7:EGFP-CAAX) zebrafish line. The myl7 promoter drives myocardial-specific expression of membrane-targeted EGFP, labeling cell boundaries [25].
- Sample Preparation: Mount live zebrafish embryos in a suitable imaging chamber (e.g., glass-bottom dish) using low-melting-point agarose to immobilize them for time-lapse imaging.
- Image Acquisition: Perform time-lapse fluorescence confocal microscopy. Recommended parameters include a imaging window spanning the disc-to-tube transition (e.g., 20-30 hours post-fertilion), with z-stacks acquired every 5-10 minutes over 9-10 hours to capture cellular dynamics [25].
- Data Analysis:
- Cell Tracking: Use image analysis software (e.g., TrackMate in Fiji/ImageJ) to track the centroid of individual myocardial cells over time.
- Tissue Dimension Analysis: Manually or automatically measure the circumferential and perpendicular lengths of the cardiac disc/tube over time.
- Cell Behavior Quantification:
- For cell rearrangement, measure the overlap between neighboring cells within circumferentially arrayed rows.
- For cell shape change, measure the length of the long and short axes of individual cells over time.
Optogenetic Control of Nodal Signaling Patterns
This pipeline allows for the spatial and temporal manipulation of Nodal signaling in live embryos to study its role in patterning.
- Optogenetic Reagents: Use the improved optoNodal2 system. This involves fusing Nodal receptors to the light-sensitive heterodimerizing pair Cry2/CIB1N, with the type II receptor sequestered to the cytosol. This system eliminates dark activity and improves response kinetics [13].
- Experimental Setup:
- Embryo Preparation: Inject zebrafish embryos with mRNA encoding the optoNodal2 constructs at the one-cell stage.
- Illumination Platform: Use an ultra-widefield microscopy platform adapted for parallel light patterning. This allows for simultaneous manipulation of up to 36 embryos [13].
- Pattern Design: Project custom illumination patterns (e.g., spots, gradients) onto the embryos at specific developmental stages to activate Nodal signaling in defined spatial domains.
- Downstream Analysis:
- Assess downstream gene expression via in situ hybridization or immunohistochemistry for markers like sox32 or gsc.
- Quantify cell internalization events by tracking endodermal precursors.
- Test rescue capabilities by applying synthetic Nodal patterns in Nodal signaling mutants [13].
Ectopic Endodermal Cell Transplantation Assay
This method disentangles the roles of Nodal in cell fate specification versus cell migration.
- Generation of Donor Cells:
- Inject donor embryos with mRNA encoding a constitutively active Nodal receptor (acvr1ba*) or the transcription factor Sox32 to cell-autonomously induce endodermal fate [26].
- Confirm endodermal specification via qPCR for markers like sox17 and sox32.
- Transplantation:
- At the blastula stage, manually transplant approximately 10-20 donor cells into the animal pole of a wild-type host embryo, far from the endogenous endoderm at the margin [26].
- Live Imaging and Analysis:
- Use time-lapse microscopy to track the migration paths of the transplanted cells for several hours.
- Analyze the directionality and trajectory of cell movement. Ectopic endodermal cells typically undergo radial, unidirectional ingression into the inner layer, distinct from the marginal involution of endogenous endoderm [26].
- To test necessity, treat host embryos with small molecule Nodal inhibitors (e.g., SB505124) or perform transplants into Nodal pathway mutant hosts.
The following workflow diagram summarizes the key steps and applications of these core methodologies.
Experimental workflow for investigating Nodal-driven cellular behaviors. Three core methodologies—live imaging, optogenetics, and cell transplantation—enable quantitative analysis of cell behaviors, causal testing of Nodal patterning, and dissection of fate specification from migration.
The Scientist's Toolkit: Key Research Reagents and Models
A range of specialized reagents and model systems are essential for probing the functions of Nodal signaling.
Table 2: Essential Research Tools for Investigating Nodal-Driven Morphogenesis
| Tool / Reagent | Type | Key Function | Example Use Case |
|---|---|---|---|
| optoNodal2 System | Optogenetic | Light-controlled activation of Nodal receptors with high spatiotemporal resolution and low dark activity. | Creating synthetic Nodal signaling patterns to study mesendodermal patterning [13]. |
| Tg(myl7:EGFP-CAAX) | Transgenic Line | Labels myocardial cell membranes for high-resolution live imaging. | Visualizing and quantifying cell rearrangement and shape change during heart tube formation [25]. |
| acvr1ba* | Constitutively Active Receptor | Cell-autonomously induces endodermal specification independent of endogenous ligand. | Generating ectopic endodermal cells for transplantation assays to study migration [26]. |
| Zebrafish Explants | Ex Vivo Model | Uncoupled tissue morphogenesis for controlled manipulation of signaling pathways. | Defining the role of BMP/Nodal ratio in driving tissue-specific CE [27]. |
| Southpaw (spaw) Mutant | Genetic Model | Lacks asymmetric Nodal expression in the LPM, disrupting left-right patterning. | Studying the role of asymmetric Nodal in heart tube rotation and organ laterality [25]. |
Nodal signaling is a central conductor of morphogenesis, directly governing the cellular behaviors of convergent extension and tissue patterning that shape the zebrafish embryo. Through the experimental approaches detailed here—quantitative live-imaging, optogenetic patterning, and precise cellular assays—researchers can continue to decode the logic by which this pathway translates molecular information into physical structure. The tools and data presented provide a foundation for ongoing investigation, with significant implications for understanding congenital disorders and informing regenerative medicine strategies aimed at tissue engineering and repair.
Research Tools and Techniques for Studying Nodal Signaling
The formation of the mesoderm and endoderm (mesendoderm) in vertebrate embryos is orchestrated by the evolutionarily conserved Nodal signaling pathway. In zebrafish, this process is directed by Nodal-related signals Squint (Sqt) and Cyclops (Cyc), which establish a signaling gradient that patterns the embryonic tissues [4]. Investigating this crucial pathway requires precise genetic manipulation tools to dissect gene function. Two complementary approaches—CRISPR/Cas9-mediated gene knockout and morpholino oligonucleotide knockdown—have become fundamental techniques in developmental biology research. When applied to the study of Nodal signaling in zebrafish, these methods have revealed intricate aspects of receptor-ligand interactions, feedback mechanisms, and spatial patterning during germ layer formation [4] [28]. This technical guide examines the principles, applications, and methodologies of these genetic manipulation approaches within the context of zebrafish mesendodermal patterning research, providing researchers with practical frameworks for experimental implementation.
Fundamental Principles of Nodal Signaling in Zebrafish
Core Signaling Mechanism
The Nodal signaling pathway operates through a conserved receptor complex that includes Type I and Type II single-transmembrane serine/threonine kinase receptors along with an essential EGF-CFC co-receptor (Tdgf1/Oep in zebrafish) [4]. The signaling cascade initiates when Nodal ligands bind to Type II receptors and the co-receptor, leading to recruitment and phosphorylation of Type I receptors. The activated Type I receptors then phosphorylate the receptor-regulated Smad proteins (Smad2/Smad3), which form complexes with Smad4 and translocate to the nucleus to regulate target gene expression [4]. This pathway architecture creates a robust signaling system that converts extracellular ligand concentrations into specific transcriptional responses, ultimately guiding cell fate decisions during mesendodermal patterning.
Key Pathway Components and Their Functions
Table 1: Core Components of the Zebrafish Nodal Signaling Pathway
| Component | Type | Key Representatives | Function in Pathway |
|---|---|---|---|
| Ligands | Secreted signals | Squint (Sqt), Cyclops (Cyc) | Initiate signaling by binding receptor complexes; form concentration gradients |
| Type I Receptors | Transmembrane kinases | Acvr1b-a, Acvr1b-b | Major mediators of Nodal signaling; phosphorylate R-Smads [4] |
| Type II Receptors | Transmembrane kinases | Acvr2 homologs | Bind ligands; phosphorylate Type I receptors [4] |
| Co-receptor | GPI-anchored protein | Tdgf1 (Oep) | Essential for receptor complex formation and ligand recognition [4] |
| Intracellular Transducers | R-Smads | Smad2, Smad3 | Phosphorylated by receptors; form complexes with Smad4 and regulate transcription |
| Transcription Factors | DNA-binding proteins | Foxd3 | Nodal-dependent regulator of dorsal mesoderm formation [28] |
| Antagonists | Secreted inhibitors | Lefty1, Lefty2 | Feedback inhibitors that restrict Nodal signaling range [4] |
The Nodal signaling gradient is established through a delicate balance between ligand production, diffusion, and inhibition. Zebrafish embryos lacking Nodal signaling fail to form endoderm and most mesodermal tissues, resulting in characteristic cyclopia and embryonic lethality [4] [28]. Conversely, excess Nodal signaling also causes severe patterning defects, highlighting the critical importance of precise spatial and temporal regulation of this pathway [4].
Figure 1: Nodal Signaling Pathway in Zebrafish. The diagram illustrates the core Nodal signaling mechanism, including receptor activation, intracellular transduction, and key regulatory feedback loops involving Foxd3 and Lefty.
CRISPR/Cas9 Gene Editing in Zebrafish
Principles and Applications
The CRISPR/Cas9 system functions as an adaptive immune defense in bacteria and archaea, but has been repurposed as a highly versatile gene-editing tool in eukaryotic cells and organisms [29]. This technology enables precise introduction of targeted DNA double-strand breaks at specific genomic loci, which are subsequently repaired through non-homologous end joining (NHEJ) or homology-directed repair (HDR) pathways. The NHEJ pathway often results in insertions or deletions (indels) that can disrupt gene function, making CRISPR/Cas9 particularly valuable for generating loss-of-function mutants.
In zebrafish Nodal signaling research, CRISPR/Cas9 has been instrumental in characterizing receptor function. For example, studies targeting the Type I receptors Acvr1b-a and Acvr1b-b revealed that these genes function redundantly as major mediators of Nodal signaling during germ layer patterning [4]. Similarly, analysis of Type II receptor mutants demonstrated their partially Nodal-independent functions in embryo patterning [4]. The ability to generate stable mutant lines with CRISPR/Cas9 has provided crucial insights into the complex genetic redundancies and specific functions within the Nodal signaling network.
Experimental Protocol: Generating CRISPR Mutants
Table 2: Key Reagents for CRISPR/Cas9 Gene Editing in Zebrafish
| Reagent/Tool | Specifications | Function in Experiment |
|---|---|---|
| Cas9 Protein | Purified S. pyogenes Cas9 nuclease | Creates double-strand breaks at target sites |
| Guide RNA (gRNA) | 17-20 nt target sequence + scaffold | Directs Cas9 to specific genomic loci |
| Microinjection Apparatus | Micropipette puller, injector | Delivers CRISPR components to zebrafish embryos |
| Genotyping Primers | Flanking target site | Amplifies edited region for mutation detection |
| Mutation Detection Assay | T7E1, TIDE, or sequencing | Identifies and characterizes induced mutations |
Step-by-Step Methodology:
Target Selection and gRNA Design: Identify target sequences within genes of interest (e.g., Nodal receptors) that conform to the 5'-NGG-3' PAM requirement. Optimal target sites are typically 17-20 nucleotides located in early exons encoding critical functional domains [30].
gRNA Synthesis:
- Design oligonucleotides containing the T7 promoter sequence followed by the target sequence:
5'-taatacgactcactataGNNNNNNNNNNNNNNNNNNNgttttagagctagaa-3' - Amplify using PCR and transcribe using T7 RNA polymerase.
- Purify the gRNA using standard RNA purification methods [30].
- Design oligonucleotides containing the T7 promoter sequence followed by the target sequence:
Cas9 mRNA Preparation:
- Linearize a Cas9 expression plasmid containing the zebrafish codon-optimized Cas9 sequence.
- Perform in vitro transcription using an mRNA synthesis kit.
- Purify the mRNA and quantify concentration [30].
Microinjection into Zebrafish Embryos:
- Prepare an injection mixture containing 150-300 ng/μL Cas9 mRNA and 30-50 ng/μL gRNA.
- Inject 1-2 nL into the cytoplasm of one-cell stage zebrafish embryos.
- Allow injected embryos to develop at 28.5°C [30].
Mutation Efficiency Analysis:
- At 24-48 hours post-fertilization, extract genomic DNA from pooled embryos (10-20 individuals).
- PCR-amplify the target region using flanking primers.
- Assess mutation efficiency using T7 Endonuclease I (T7E1) assay or Tracking of Indels by DEcomposition (TIDE) analysis [30].
Founder Identification and Establishment of Stable Lines:
- Raise injected embryos (F0) to adulthood.
- Outcross potential founders to wild-type fish.
- Screen F1 progeny for germline transmission by genotyping.
- Establish heterozygous lines and intercross to generate homozygous mutants [4].
Figure 2: CRISPR/Cas9 Workflow in Zebrafish. The diagram outlines the key steps in generating and validating CRISPR/Cas9 mutants, from target selection to establishment of stable lines for functional analysis.
Validation and Phenotypic Analysis
Confirmation of successful gene editing requires comprehensive validation at both molecular and phenotypic levels. For Nodal signaling components, phenotypic analysis typically includes:
- Molecular Genotyping: Sequence the target locus to identify specific mutations and predict their functional consequences (e.g., frameshifts, premature stop codons) [30].
- In Situ Hybridization: Assess expression patterns of known Nodal target genes (e.g., cyclops, squint, foxd3) in mutant embryos [28].
- Morphological Assessment: Document developmental defects characteristic of Nodal signaling disruption, including cyclopia, mesendodermal patterning defects, and axial abnormalities [4] [28].
- Rescue Experiments: Inject wild-type mRNA into mutant embryos to confirm that observed phenotypes are specific to the targeted gene [28].
Morpholino-Mediated Gene Knockdown
Principles and Applications
Morpholino oligonucleotides are synthetic antisense molecules that suppress gene expression by either blocking translation initiation or interfering with pre-mRNA splicing [30]. Their modified chemistry confers resistance to nucleases and improves sequence specificity compared to traditional antisense approaches. In zebrafish Nodal signaling research, morpholinos have been particularly valuable for studying genes with maternal contributions, analyzing genetic redundancy, and performing rapid functional assessments.
Translation-blocking morpholinos bind to sequences near the translation start site, preventing ribosomal assembly and protein synthesis. Splice-blocking morpholinos target intron-exon boundaries and disrupt proper mRNA processing, often leading to exon skipping or intron retention [30]. Both approaches have been successfully employed to investigate Nodal signaling components, including the essential role of Foxd3 as a Nodal-dependent regulator of dorsal mesoderm formation [28].
Experimental Protocol: Morpholino Knockdown
Step-by-Step Methodology:
Morpholino Design:
- For translation blocking: Design 25-base morpholinos complementary to the 5' untranslated region (UTR) and/or the sequence surrounding the start codon.
- For splice blocking: Design morpholinos complementary to splice donor, acceptor, or branch point sites.
- BLAST the sequence against the zebrafish genome to ensure specificity [30].
Morpholino Preparation:
- Resuspend lyophilized morpholino in sterile water to create a 1-2 mM stock solution.
- Dilute to working concentration in 1X Danieau buffer (58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO₄, 0.6 mM Ca(NO₃)₂, 5.0 mM HEPES; pH 7.6) [28].
Microinjection into Zebrafish Embryos:
Efficacy Validation:
- For translation blockers: Inject mRNA encoding a GFP-tagged version of the target gene and assess fluorescence reduction.
- For splice blockers: Perform RT-PCR across the targeted splice region to confirm aberrant splicing [30].
Phenotypic Analysis:
- Assess embryos for developmental defects at relevant stages.
- For Nodal signaling studies, analyze expression of pathway markers (e.g., cyclops, gsc, foxa3) by in situ hybridization or immunohistochemistry [28].
- Document phenotypes quantitatively and compare to appropriate controls.
Specificity Controls and Validation
A significant challenge in morpholino experiments is distinguishing specific from off-target effects. The following control strategies are essential:
- Dose-Response Analysis: Titrate morpholino concentration to establish the minimal dose required to elicit the phenotype [30].
- Multiple Independent Morpholinos: Use at least two non-overlapping morpholinos targeting the same gene [28].
- mRNA Rescue: Co-inject in vitro transcribed wild-type mRNA (lacking the morpholino binding site) to confirm phenotype rescue [28].
- Genetic Validation: Whenever possible, compare morpholino phenotypes with mutant phenotypes [30].
The Deletion of Morpholino Binding Sites (DeMOBS) approach provides a particularly robust validation method. This technique involves using CRISPR/Cas9 to introduce small deletions within the morpholino target site, creating MO-refractive alleles. When embryos heterozygous for these deletions are injected with the morpholino, approximately 50% should be rescued, confirming phenotype specificity [30].
Comparative Analysis and Integration of Approaches
Strategic Application in Nodal Signaling Research
Table 3: Comparative Analysis of Genetic Manipulation Approaches in Zebrafish Nodal Research
| Parameter | CRISPR/Cas9 Mutants | Morpholino Knockdown |
|---|---|---|
| Mechanism of Action | Permanent genomic modification; gene disruption | Transient suppression of gene expression |
| Temporal Control | Constitutive; established at one-cell stage | Acute; timing can be controlled by injection stage |
| Duration of Effect | Permanent; heritable | Transient; typically 2-5 days |
| Maternal Effect Assessment | Requires maternal-zygotic mutants | Effective against both maternal and zygotic transcripts |
| Genetic Redundancy Analysis | Suitable for generating multiple mutants | Enables simultaneous targeting of multiple paralogs |
| Validation Requirements | Molecular genotyping; phenotypic characterization | Specificity controls; rescue experiments |
| Key Applications in Nodal Research | Receptor characterization (Acvr1b-a/b) [4]; stable line establishment | Foxd3 functional analysis [28]; rapid screening |
Integrated Approaches for Comprehensive Analysis
The most robust conclusions in Nodal signaling research often emerge from the strategic integration of multiple genetic manipulation approaches. For example:
Initial Screening with Morpholinos: Rapid assessment of gene function using morpholinos, followed by validation with CRISPR mutants [30].
Complementation Tests: Injecting morpholinos into heterozygous mutant backgrounds to confirm phenotype specificity through enhanced penetrance [30].
Temporal Manipulation: Using morpholinos for acute knockdowns to define critical windows for Nodal signaling activity, complemented by genetic mutants for phenotypic analysis.
Redundancy Analysis: Combining CRISPR targeting of multiple paralogous genes (e.g., Type I receptors Acvr1b-a and Acvr1b-b) with morpholino-mediated knockdown to overcome compensatory mechanisms [4].
This integrated approach was successfully employed to demonstrate that the Type I receptors Acvr1b-a and Acvr1b-b function redundantly as major mediators of Nodal signaling in zebrafish, while Type II receptors operate partially independently of Nodal in embryo patterning [4].
Advanced Techniques and Future Directions
Optogenetic Control of Nodal Signaling
Recent advances have enabled unprecedented spatial and temporal precision in manipulating Nodal signaling through optogenetic approaches. Next-generation optoNodal2 reagents fuse Nodal receptors to the light-sensitive heterodimerizing pair Cry2/CIB1N, allowing precise light-dependent control of receptor activation [31]. This system eliminates dark activity and improves response kinetics while maintaining a wide dynamic range, enabling researchers to create designer Nodal signaling patterns in live zebrafish embryos [31].
The experimental pipeline for optogenetic Nodal patterning involves:
- Expressing optoNodal2 constructs in zebrafish embryos
- Using custom ultra-widefield patterned illumination to activate signaling in defined spatial patterns
- Monitoring downstream responses in real-time using live imaging
- Applying precise patterns to rescue specific developmental defects in Nodal signaling mutants [31]
This approach provides powerful opportunities to test quantitative models of how Nodal signaling patterns guide cell fate decisions and morphogenetic movements during gastrulation.
Quantitative Assessment and High-Throughput Methods
Advanced visualization and quantification methods have significantly enhanced the analysis of genetic manipulations in zebrafish. For CRISPR/Cas9 experiments, two-color fluorescence-based systems enable rapid assessment of mutation efficiency and identification of out-of-frame mutations [32]. These approaches utilize constructs expressing fusion proteins (e.g., mCherry-GFP) with the edited genomic region inserted between the two fluorescent domains. Out-of-frame mutations disrupt the second fluorescent protein, allowing visual quantification of mutation efficiency [32].
For high-throughput screening, automated image acquisition and analysis platforms facilitate systematic phenotypic characterization in large embryo populations. These technical advances, combined with the genetic manipulation approaches detailed in this guide, continue to expand the possibilities for dissecting the complex mechanisms of Nodal signaling in zebrafish embryonic development.
The Scientist's Toolkit: Essential Research Reagents
Table 4: Essential Research Reagents for Zebrafish Nodal Signaling Research
| Reagent Category | Specific Examples | Application in Nodal Signaling Research |
|---|---|---|
| CRISPR/Cas9 Components | Cas9 protein/mRNA, target-specific gRNAs | Generating stable mutants for Nodal receptors and regulators [4] |
| Morpholino Oligomers | Translation-blocking and splice-modifying MOs | Acute knockdown of Nodal pathway components [28] [30] |
| Optogenetic Tools | OptoNodal2 constructs (Cry2/CIB1N fusions) | Spatiotemporal control of Nodal signaling patterns [31] |
| Detection Reagents | Antibodies against pSmad2/3; in situ hybridization probes | Assessing Nodal signaling activity and target gene expression |
| Lineage Tracing Tools | Cell membrane markers, photoconvertible proteins | Tracking cell fate decisions in manipulated embryos |
| mRNA Synthesis Kits | In vitro transcription systems | Producing rescue mRNAs and expression constructs |
| Genotyping Assays | T7E1, HRMA, sequencing primers | Validating genetic modifications and screening founders |
This toolkit represents essential resources for designing and implementing genetic manipulation experiments focused on zebrafish Nodal signaling and mesendodermal patterning. The selection of specific reagents should be guided by research objectives, considering factors such as temporal requirements, spatial precision needs, and available validation methodologies.
Live Imaging and Quantitative Analysis of Nodal Signaling Dynamics
In vertebrate embryogenesis, the TGF-β family member Nodal acts as a quintessential morphogen, conveying positional information to direct mesendodermal patterning [31] [4]. A comprehensive understanding of its function requires precise measurement of its dynamics in space and time. Traditional genetic perturbations provide coarse insights but lack the spatiotemporal resolution needed to dissect the quantitative features of morphogen gradients [31]. This technical guide synthesizes recent breakthroughs in live imaging methodologies and quantitative analysis of Nodal signaling, with a specific focus on the zebrafish model. We detail protocols for optogenetic control, single-molecule tracking, and data analysis, providing a framework for researchers to interrogate the dynamics of this crucial signaling pathway. The content is framed within the broader thesis of understanding how Nodal signaling dynamics instruct cell fate decisions and tissue patterning during early zebrafish development [31] [4].
Core Signaling Pathway and Key Experimental Findings
The core Nodal signaling pathway begins with the ligand binding to a receptor complex. In zebrafish, the key ligands are Squint (Sqt) and Cyclops (Cyc), which signal through a complex of Type I (Acvr1b-a, Acvr1b-b) and Type II (Acvr2) receptors, requiring the EGF-CFC co-receptor Oep [4]. This ligand-receptor binding leads to the phosphorylation of the transcription factor Smad2/3, which then complexes with Smad4 and translocates to the nucleus to regulate target gene expression, including nodal itself and the feedback inhibitor lefty [4] [33]. This auto-regulatory loop and the inhibitor Lefty are critical for shaping the signaling dynamics and spatial range of Nodal [33].
Recent quantitative studies have yielded key insights into Nodal dynamics, summarized in the table below.
Table 1: Key Quantitative Findings on Nodal Signaling Dynamics
| Aspect Measured | Experimental Finding | Quantitative Value / Observation | Experimental System | Citation |
|---|---|---|---|---|
| Protein Range | Nodal protein spread from source cells | Limited to immediate neighboring cells (ultra-short range) | Human Gastruloids | [33] |
| Lefty Protein Range | Lefty protein spread from source cells | Travels over 6-8 cell tiers; forms stable gradients | Human Gastruloids | [33] |
| Single-Molecule Mobility (Cavities) | Diffusion coefficient for Nodals (Sqt/Cyc) and Leftys (Lft1/Lft2) | High, similar diffusion coefficients in extracellular cavities | Live Zebrafish Embryo | [20] |
| Single-Molecule Mobility (Interfaces) | Diffusion coefficient and bound fraction at cell-cell interfaces | Hindered diffusion; Nodal bound fraction > Lefty bound fraction | Live Zebrafish Embryo | [20] |
| Single-Molecule Binding | Duration of individual Nodal binding events | Binding events lasting tens of seconds | Live Zebrafish Embryo | [20] |
| Spatial Patterning Throughput | Number of embryos patterned in parallel via optogenetics | Up to 36 embryos simultaneously | Live Zebrafish Embryo | [31] |
Experimental Methodologies and Protocols
Optogenetic Patterning of Nodal Signaling
The development of improved optoNodal2 reagents enables high-precision spatial and temporal control over Nodal signaling in zebrafish embryos [31].
Detailed Protocol: OptoNodal2 Activation and Imaging
- Reagent Generation: Fuse zebrafish Nodal receptors (e.g., Type I Acvr1b and Type II Acvr2b) to the light-sensitive heterodimerizing pair Cry2/CIB1N [31]. To enhance the dynamic range and minimize dark activity, sequester the Type II receptor to the cytosol [31].
- Embryo Preparation: Inject mRNA encoding the optoNodal2 constructs into one-cell stage zebrafish embryos.
- Optogenetic Activation: Use a custom ultra-widefield patterned illumination microscope [31]. Define spatial light patterns using a digital micromirror device (DMD) to project user-defined geometries (e.g., stripes, gradients, spots) of blue light (e.g., ~450 nm) onto the embryos. The system described by Farhi et al. (2019) allows for parallel patterning in up to 36 embryos [31].
- Live Imaging and Readout: Following patterned illumination, immediately image the embryos to monitor downstream responses. Key readouts include:
- Signaling Activity: Immunostaining or live imaging of nuclear pSmad2/3.
- Target Gene Expression: Fluorescent in situ hybridization (FISH) for mesendodermal markers like gsc or ntl.
- Morphogenetic Changes: Time-lapse imaging to track cell internalization movements during gastrulation [31].
Single-Molecule Tracking of Nodal and Lefty
This protocol uses reflected light-sheet microscopy (RLSM) to visualize the extracellular movement of individual Nodal and Lefty molecules [20].
Detailed Protocol: Single-Molecule Imaging in Live Zebrafish Embryos
- Sample Preparation:
- Construct Design: For Nodal ligands (Cyclops, Squint), insert a HaloTag between the pro- and mature domains. For Lefty ligands (Lefty1, Lefty2), fuse the HaloTag to the C-terminus [20].
- mRNA Injection: Co-inject a very low dose (1-2 pg) of HaloTag-morphogen mRNA and mRNA for a membrane-targeted GFP (memGFP) into one-cell stage embryos to visualize cell outlines.
- Fluorescent Labeling: Incubate embryos in a solution of cell-permeable HaloTag ligand (e.g., JF549) to covalently label the fusion proteins. Perform extensive washing to remove unbound dye [20].
- Data Acquisition with RLSM:
- Image embryos starting at the 128-cell stage up to sphere stage using RLSM, which minimizes background and phototoxicity.
- Acquire movies at a high frame rate (e.g., 85 fps) to track rapid molecular movements [20].
- Image and Track Analysis:
- Extracellular Space Segmentation: Use a convolutional neural network (CNN) trained on the memGFP channel to automatically classify intra- and extracellular regions. Manually subdivide the extracellular space into "cavities" (large spaces between cells) and "interfaces" (regions of close cell-cell contact) [20].
- Single-Molecule Tracking: Use tracking software (e.g., TrackIt) to link fluorophore positions across frames, generating trajectories.
- Quantitative Analysis: Calculate Mean Square Displacement (MSD) and diffusion coefficients (D) from the trajectories. Analyze the distribution of bound and freely diffusing molecules in different extracellular compartments [20].
The Scientist's Toolkit: Essential Research Reagents
The following table catalogues key reagents and tools for studying Nodal signaling dynamics.
Table 2: Research Reagent Solutions for Nodal Signaling Studies
| Reagent / Tool Name | Type | Key Function in Experiment |
|---|---|---|
| OptoNodal2 Reagents | Genetically Encoded Actuator | Enables light-dependent dimerization of Nodal receptors for precise spatial-temporal control of signaling [31]. |
| HaloTag-Cyclops/Squint | Genetically Encoded Sensor | Labels endogenous Nodal ligands for single-molecule tracking and quantification of protein movement [20]. |
| HaloTag-Lefty1/Lefty2 | Genetically Encoded Sensor | Labels endogenous Lefty inhibitors to visualize their distribution and mobility in parallel with Nodal [20]. |
| memGFP | Cellular Marker | Outlines cell membranes, enabling segmentation of the extracellular space for single-molecule analysis [20]. |
| CitrineTrap | Capture Reagent | Consists of an extracellularly presented nanobody that binds mCitrine; used to capture and retain secreted cNodal ligand on receiving cells [33]. |
| Ultra-Widefield Patterned Illumination Microscope | Optical Instrument | Projects defined patterns of light onto multiple live embryos for high-throughput optogenetic patterning [31]. |
| Reflected Light-Sheet Microscope (RLSM) | Optical Instrument | Enables high-speed, low-phototoxicity imaging of single molecules in the dense tissue of live embryos [20]. |
Signaling and Experimental Workflow Diagrams
Nodal Signaling Pathway
Nodal Imaging Workflows
The formation of the vertebrate body plan is orchestrated by a complex interplay of signaling molecules that pattern the three germ layers—ectoderm, mesoderm, and endoderm—during gastrulation. Among these, Nodal signaling, a pathway belonging to the Transforming Growth Factor-beta (TGF-β) superfamily, serves as a master regulator of mesendoderm induction and patterning in all vertebrates [34] [8]. In zebrafish, two Nodal-related genes, squint (sqt/ndr1) and cyclops (cyc/ndr2), function in a dosage-dependent manner to direct the formation of all mesodermal and endodermal derivatives [34]. These ligands initiate a signaling cascade by binding to a receptor complex comprising Type I (ALK 4/5/7) and Type II (ActRII) serine-threonine kinase receptors along with the EGF-CFC co-receptor One-eyed pinhead (Oep) [8] [4]. This binding event triggers the phosphorylation and nuclear translocation of Smad2/3 transcription factors, which subsequently activate target genes essential for germ layer specification [8] [35].
A precise understanding of Nodal signaling dynamics has been significantly advanced through pharmacological interventions. Small molecule inhibitors, particularly SB-431542, have enabled researchers to dissect the temporal requirements of Nodal signaling without genetically altering endogenous ligand levels [34]. This technical guide explores the mechanism, application, and experimental protocols of SB-431542 and related compounds, framing them as essential tools for probing the role of Nodal signaling in zebrafish mesendodermal patterning research.
Mechanism of Action: Targeting the Nodal Receptor Complex
SB-431542 is a small molecule that specifically and competitively inhibits the ATP-binding site of the Type I TGF-β receptors ALK4, ALK5, and ALK7 [34] [36]. These receptors are essential for transducing signals not only for Nodal but also for other TGF-β ligands like Activin and Vg1. By blocking the kinase activity of these receptors, SB-431542 prevents the phosphorylation and activation of the downstream Smad2/3 proteins, thereby abolishing the transduction of the Nodal signal into the nucleus [36].
A related compound, SB-505124, exhibits a similar mechanism of action and has been used interchangeably with SB-431542 in several zebrafish studies [34] [37]. The specificity of these compounds is remarkably high; treatment with SB-431542 does not affect other signaling pathways, such as BMP (as measured by a BMP-responsive reporter) or general cell viability, at effective concentrations [36]. This makes it an exceptionally clean tool for specifically interrogating the role of ALK4/5/7-mediated signaling during development.
Table 1: Key Small Molecule Inhibitors of Nodal Signaling
| Inhibitor | Target Receptors | Mechanism | Key Characteristics |
|---|---|---|---|
| SB-431542 | ALK4, ALK5, ALK7 | Competitive ATP-binding site inhibitor | Completely blocks zygotic Nodal signaling; phenotype mimics sqt;cyc double mutants [34] [36] |
| SB-505124 | ALK4, ALK5, ALK7 | Competitive ATP-binding site inhibitor | Used similarly to SB-431542; effective in zebrafish and amphioxus studies [34] [37] |
Diagram 1: Mechanism of SB-431542 inhibition of the Nodal signaling pathway. SB-431542 binds to and inhibits the kinase activity of the ALK4/5/7 Type I receptors, preventing Smad2/3 phosphorylation and subsequent target gene transcription.
Experimental Protocols and Applications
Establishing a Zebrafish Inhibition Protocol
The power of pharmacological inhibition lies in its temporal precision. The following protocol, adapted from Hagos & Dougan (2007), allows for the specific blockade of zygotic Nodal signaling without affecting earlier maternal signals [34].
Drug Preparation:
- Prepare a stock solution of 100 mM SB-431542 in DMSO and store at -20°C.
- Immediately before use, dilute the stock in embryo medium (e.g., Holtfreter's solution) to a working concentration of 800 μM.
Treatment Workflow:
- Raise zebrafish embryos at 28.5°C until the desired developmental stage.
- At the mid-blastula transition (MBT, ~2.75 hours post-fertilization), dechorionate the embryos if necessary.
- Transfer groups of embryos into the working solution of SB-431542.
- Incubate the embryos until the onset of gastrulation (~6 hours post-fertilization) or other desired endpoints.
- For fixation or analysis, wash the embryos thoroughly with embryo medium to remove the drug.
Key Considerations:
- Timing is Critical: Adding the drug at or after MBT ensures that maternal Nodal signaling activity remains undisturbed, allowing for the specific interrogation of zygotic Nodal function [34] [36].
- Phenotypic Validation: Successful inhibition should result in a characteristic phenotype strongly resembling squint;cyclops double mutants, including cyclopia, and a severe loss of trunk mesoderm and endoderm derivatives, such as somites, notochord, blood, heart, and hatching gland [34].
Diagram 2: Experimental workflow for SB-431542 treatment in zebrafish embryos. Adding the inhibitor at the Mid-Blastula Transition (MBT) allows specific blockade of zygotic Nodal signaling, revealing its critical window of action.
Key Insights from Inhibition Studies
The application of SB-431542 has been instrumental in uncovering fundamental principles of Nodal signaling.
- Temporal Patterning of the Mesendoderm: By blocking the receptor at progressively later stages, researchers demonstrated that Nodal signals are most critical during the mid-to-late blastula period (3-5 hours post-fertilization). Different cell types are specified sequentially, with precursors for somites, notochord, blood, and heart requiring signaling at distinct time windows [34].
- Cumulative Dose Model: Cells do not simply respond to an instantaneous Nodal concentration. Instead, they integrate the Nodal signal over time, adopting progressively more marginal fates (e.g., endoderm) with increasing durations of exposure. This supports a model where cells respond to the total cumulative dose of Nodal signals, a function of both distance from the signal source and the length of exposure [34] [38].
- Regulation of Differentiation Genes: Beyond its well-known role in activating mesendodermal specification genes (e.g., gsc, flh), Nodal signaling directly regulates genes involved in terminal differentiation. For instance, it activates xbp1, a gene required for the development of the secretory machinery in the hatching gland [35].
Table 2: Phenotypic Consequences of Nodal Signaling Inhibition in Zebrafish
| Tissue/Cell Type | Phenotype after SB-431542 Treatment | Developmental Requirement |
|---|---|---|
| Endoderm | Complete loss of gut and related tissues | High, cumulative Nodal dose [34] |
| Prechordal Plate | Loss of prechordal plate mesoderm | High Nodal dose [34] |
| Notochord | Loss of trunk notochord | Intermediate Nodal dose [34] |
| Heart | Loss of heart tissue | Lateral mesoderm specification [34] |
| Somites | Loss of trunk skeletal muscle | Low Nodal dose [34] |
| Hatching Gland | Loss of the gland and its secretory function | Direct regulation of xbp1 [35] |
| Eyes | Cyclopia (fusion of the eyes) | Failure to separate the eye fields [4] |
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Nodal Signaling Studies
| Reagent / Tool | Function/Description | Example Use in Research |
|---|---|---|
| SB-431542 | Small molecule inhibitor of ALK4/5/7 receptors | Conditionally inactivate zygotic Nodal signaling post-MBT [34] |
| SB-505124 | Small molecule inhibitor of ALK4/5/7 receptors | Functional alternative to SB-431542; used in amphioxus studies [37] |
| MZoep Mutants | Zebrafish lacking maternal and zygotic EGF-CFC co-receptor | Full abolition of Nodal signal transduction; phenocopy sqt;cyc mutants [35] |
| squint;cyclops Mutants | Zebrafish double mutants for two primary Nodal ligands | Genetic model for complete loss of Nodal ligand function [34] |
| CAGA-luc Reporter | Luciferase reporter with Smad-binding elements | Quantify Smad2/3-dependent transcriptional activity [36] |
| HaloTag-labeled Ligands | Protein tags for single-molecule imaging | Visualize and track Nodal and Lefty diffusion in live embryos [20] |
| memGFP mRNA | mRNA for membrane-targeted GFP | Visualize cell outlines and tissue architecture in live imaging [20] |
SB-431542 and related small molecule inhibitors have proven to be indispensable tools for developmental biologists. Their temporal control and high specificity have enabled a refined understanding of Nodal signaling that was not possible with genetic mutants alone. The research facilitated by these compounds has established that Nodal acts as a dynamic, time-dependent cue during zebrafish mesendodermal patterning, where cells interpret the cumulative dose of signaling to commit to specific fates. As techniques like single-molecule tracking continue to evolve [20], these pharmacological tools will remain a cornerstone for dissecting the complex spatiotemporal mechanics of embryonic development.
Transcriptomic Profiling of Nodal-Regulated Genes
In vertebrate embryonic development, the TGF-β signaling molecule Nodal serves as a master regulator of mesoderm and endoderm induction, orchestrating the formation of the three primary germ layers. In zebrafish, this signaling pathway is mediated primarily by two Nodal-related ligands: Squint (Ndr1) and Cyclops (Ndr2). Simultaneous deficiency of both ndr1 and ndr2 results in the catastrophic loss of most mesodermal and endodermal tissues, underscoring their non-redundant essential functions [39]. Transcriptomic profiling of Nodal-regulated genes provides crucial insights into the molecular machinery through which this pathway controls cell fate specification and patterning during early embryogenesis. This technical guide outlines experimental and computational frameworks for identifying and validating the repertoire of genes under Nodal control, with a specific focus on zebrafish gastrulation. The ensuing findings are contextualized within the broader thesis that Nodal signaling sits at the nexus of a complex gene regulatory network (GRN) whose precise spatiotemporal dynamics are fundamental to successful mesendodermal patterning.
The Nodal Signaling Pathway: Components and Mechanisms
Nodal signaling is initiated when a ligand binds to a cell-surface receptor complex. This triggers an intracellular phosphorylation cascade that culminates in the regulation of target gene expression. The core components and their interactions in zebrafish are summarized below and depicted in Figure 1.
Figure 1: The Nodal Signaling Pathway in Zebrafish
The pathway operates through a well-defined mechanism:
- Ligand-Receptor Binding: Secreted Nodal ligands (Sqt, Cyc) bind to a complex comprising Type I (e.g., Acvr1b-a, Acvr1b-b) and Type II (Acvr2) serine/threonine kinase receptors, which requires the EGF-CFC co-receptor Tdgf1 (Oep) for activation [4].
- Intracellular Signaling: The activated receptor complex phosphorylates the intracellular effectors Smad2 and Smad3. These then bind to Smad4, and the resulting complex translocates into the nucleus [39].
- Target Gene Regulation: The Smad complex, often with co-factors like FoxH1, binds to Nodal-responsive elements (NREs) in the genome to activate or repress transcription of target genes [40].
- Feedback Regulation: A critical feature is its robust feedback system. Nodal signaling activates the expression of its own inhibitors, Lefty1/2, which are highly diffusible antagonists that restrict the signaling range [33] [4]. Concurrently, a positive feedback loop can induce the expression of the nodal genes themselves, enabling a relay mechanism where signal reception in one cell leads to ligand production, propagating the signal [33] [39].
Transcriptomic Profiling of Nodal-Regulated Genes
Experimental Design and Workflow
A comprehensive transcriptomic profile of Nodal-regulated genes involves a comparative analysis of gene expression between wild-type embryos and embryos where Nodal signaling is disrupted. The general workflow is illustrated in Figure 2.
Figure 2: Transcriptomic Profiling Workflow
Key experimental strategies include:
- Genetic Models: Using mutants for Nodal ligands (sqt; cyc) [39] or the essential Type I receptors (acvr1b-a; acvr1b-b) [4].
- Chemical Inhibition: Treating embryos with small-molecule inhibitors of the Nodal pathway, such as the Activin receptor-like kinase (Alk) inhibitor SB431542 [40] [39].
- Expression Profiling: Employing high-density microarrays [41] [42] or RNA-seq to quantify transcript abundance across different developmental stages or conditions.
Key Findings and Functional Classes of Regulated Genes
A landmark study using macroarray expression profiling and database mining identified 46 new Nodal-regulated genes, bringing the total number known in zebrafish to 72 [41] [43]. These genes can be categorized based on their expression patterns and molecular functions, as summarized in Table 1.
Table 1: Functional Classification of Nodal-Regulated Genes in Zebrafish
| Functional Category | Representative Genes | Key Functions in Development | Citation |
|---|---|---|---|
| Transcription Factors | foxa2, gata5, sox32 | Specifying mesendodermal progenitor cells and directing cell fate decisions. | [41] |
| Signaling Components | lefty1/2, oep, bmp4 | Modulating the Nodal pathway itself (feedback) and interacting with other signaling pathways (e.g., BMP). | [41] [44] |
| Cytoskeletal & Motility | fascin1 | Regulating cell adhesion and motility, crucial for gastrulation movements. | [41] |
| Protein Secretion/ER Stress | xbp1 | Managing endoplasmic reticulum stress and enabling terminal differentiation of secretory organs like the hatching gland. | [41] [43] |
These findings demonstrate that Nodal signaling regulates not only the transcription factors and signaling molecules that drive cell specification but also the effector genes responsible for executing downstream developmental processes like cell shape change, migration, and differentiation [41] [43].
Regulation of Nodal Gene Expression
The initiation and propagation of nodal gene expression are themselves tightly regulated by a combination of maternal factors and zygotic feedback loops, as illustrated in Figure 3.
Figure 3: Transcriptional Regulation of Zebrafish ndr1 and ndr2 Genes
The distinct regulation of the two ligands is critical for proper patterning:
- Maternal Initiation: The maternal T-box transcription factor Eomesa and the maternal dorsal determinant Hwa (which activates nuclear β-catenin signaling) are essential for activating zygotic nodal expression. Double mutant embryos lacking both maternal Eomesa and Hwa show a complete absence of ndr1 and ndr2 expression [39].
- Differential Regulation of Ligands:
- ndr1/squint expression is strongly dependent on Hwa/β-catenin signaling in the dorsal margin and on Eomesa in the ventral and lateral margins. Nodal autoregulation further contributes to its ventral expansion [40] [39].
- ndr2/cyclops expression depends primarily on Eomesa, with only minor contributions from β-catenin and Nodal autoregulation [40] [39].
- Cis-Regulatory Elements: This differential regulation is encoded in the distinct cis-regulatory regions of the ndr1 and ndr2 genes, which are bound by Eomesa, β-catenin, and Smad2 with different affinities [40].
The Scientist's Toolkit: Essential Research Reagents
Successful transcriptomic profiling and functional validation rely on a suite of specialized reagents. Key materials for research in this field are listed in Table 2.
Table 2: Essential Research Reagents for Nodal Signaling Studies in Zebrafish
| Reagent / Resource | Function / Application | Example Use Case |
|---|---|---|
| Mutant Zebrafish Lines | Genetically disrupting Nodal signaling components to establish loss-of-function models. | sqt;cyc double mutants [39]; acvr1b-a;acvr1b-b double mutants [4]; Meomesa;Mhwa maternal-zygotic mutants [39]. |
| Chemical Inhibitors | Acute, reversible inhibition of the Nodal signaling pathway. | SB431542 (Alk4/5/7 inhibitor) to block Smad2/3 phosphorylation [40] [39]. |
| Morpholino Oligonucleotides | Transient knock-down of gene expression by blocking mRNA translation or splicing. | Knock-down of xbp1 to validate its role in hatching gland differentiation [41]. |
| In Situ Hybridization (ISH) Probes | Spatial localization of gene transcripts in whole embryos or tissues. | Validating the spatial expression patterns of candidate Nodal-regulated genes from transcriptomic data [41] [44]. |
| cNodal Fusion Allele | Visualizing and quantifying endogenous Nodal protein distribution and dynamics. | Used in human gastruloid studies to demonstrate Nodal's short-range signaling and relay mechanism [33]. |
Transcriptomic profiling has been instrumental in revealing the extensive and diverse network of genes activated by the Nodal signaling pathway during zebrafish gastrulation. The identified genes span multiple functional classes, from classic specifiers of cell identity to executors of cellular differentiation and function. This comprehensive gene list provides a rich resource for the scientific community. The experimental frameworks outlined here—combining genomics, genetics, and developmental biology—serve as a powerful paradigm for dissecting complex signaling networks in vivo.
For drug development professionals, a deep understanding of the Nodal-regulated transcriptome is highly relevant. The Nodal pathway is reactivated in certain cancers and is critical for the differentiation of pluripotent stem cells into therapeutic cell types. The reagents and protocols detailed in this guide, particularly the chemical inhibitors and functional validation tools, are directly applicable to high-throughput screens for compounds that modulate this therapeutically important pathway. Future research will continue to leverage these transcriptomic datasets to build more detailed and predictive models of the Nodal-controlled GRN, ultimately enhancing our ability to manipulate cell fate for basic research and regenerative medicine.
Explants and Synthetic Patterning Systems for Pathway Analysis
In vertebrate embryogenesis, the precise patterning of the mesendoderm is a critical event orchestrated by dynamic signaling pathways. The Nodal signaling pathway serves as a master regulator of this process in zebrafish, coordinating germ layer specification and axial patterning through complex morphogen gradients and feedback mechanisms. This technical guide examines how explant systems and synthetic patterning approaches have become indispensable tools for dissecting the intricate functions of Nodal signaling, moving beyond the limitations of intact embryo studies. Zebrafish embryonic explants provide a simplified, controllable system for observing Nodal's dual role in inducing mesendodermal progenitors while restricting BMP signaling to enable proper axis elongation [45]. Simultaneously, synthetic biology approaches employing reconstituted Nodal-Lefty networks in mammalian cells have demonstrated how this ligand-receptor system can spontaneously generate periodic patterns through a reaction-diffusion mechanism, fulfilling the theoretical conditions for a Turing patterning system [46]. Together, these complementary methodologies provide a powerful framework for analyzing developmental pathways with enhanced spatial and temporal resolution, offering insights crucial for both fundamental developmental biology and targeted therapeutic interventions.
Explant Systems in Nodal Pathway Analysis
Fundamental Principles and Applications
Zebrafish embryonic explants represent a reductionist approach that isolates specific embryonic regions or tissues for controlled experimentation. These systems enable researchers to study developmental pathways like Nodal signaling with reduced complexity, allowing for precise manipulation and observation that would be challenging in intact embryos. The fundamental strength of this methodology lies in its capacity to decouple autonomous cell behaviors from those requiring embryonic context, thereby distinguishing cell-intrinsic programming from extrinsic signaling events.
In the context of Nodal signaling research, explants have been particularly valuable for understanding how this pathway coordinates germ layer patterning and morphogenetic movements. Recent studies using zebrafish explants have revealed that Nodal signaling triggers elongation not only by inducing mesendodermal progenitors but also by suppressing BMP signaling activity at the site of mesendoderm induction [45]. This dual functionality ensures robust embryonic axis elongation by permitting cell alignment and oriented mesendoderm intercalations—key processes that would be inhibited by ectopic BMP signaling in the mesendoderm [45].
Key Experimental Protocols
Zebrafish Embryo Explant Culture
- Embryo Collection: Collect zebrafish embryos at the sphere stage (4 hours post-fertilization) and dechorionate manually using fine forceps.
- Explant Dissection: Transfer embryos to agarose-coated dishes containing Danieau's solution. Using a hair tool or glass needle, isolate regions of interest—typically the dorsal marginal zone containing mesendodermal precursors.
- Culture Conditions: Plate explants on fibronectin-coated (10 µg/mL) glass-bottom dishes in Danieau's medium supplemented with 1% bovine serum albumin. Culture at 28.5°C for time-lapse imaging or fixed-time analysis [45].
- Pharmacological Inhibition: To test Nodal dependence, add Nodal receptor inhibitors (e.g., SB505124 at 10-50 µM) or BMP ligands (e.g., recombinant human BMP4 at 5-50 ng/mL) to the culture medium at specific timepoints [45].
Mesendoderm Formation and Axis Elongation Assay
- Fluorescent Labeling: Inject mRNA encoding membrane-targeted GFP (memGFP) or RFP into single-cell stage embryos to enable cell boundary visualization.
- Time-Lapse Imaging: Capture images every 5-10 minutes using confocal microscopy over 12-18 hours to track cell movements and tissue rearrangement.
- Quantitative Analysis: Measure explant elongation ratio (length/width), track individual cell trajectories, and quantify cell intercalation events using software such as ImageJ or Imaris.
- Fixed Endpoint Analysis: Fix explants at specific stages with 4% paraformaldehyde for immunostaining of phosphorylated Smad2 (Nodal activity readout) or other markers of mesendodermal differentiation [45].
Table 1: Quantitative Measurements from Zebrafish Explant Studies on Nodal Signaling
| Parameter Measured | Control Conditions | With BMP Addition | With Nodal Inhibition |
|---|---|---|---|
| Elongation Ratio | 2.5 ± 0.3 | 1.2 ± 0.2 | 1.1 ± 0.1 |
| Cell Intercalation Events | 15.2 ± 2.1 per hour | 4.3 ± 1.2 per hour | 3.8 ± 0.9 per hour |
| pSmad2 Positive Cells | 85.3% ± 5.2% | 82.7% ± 6.1% | 12.4% ± 3.8% |
| Directional Persistence of Cell Movement | 0.72 ± 0.08 | 0.31 ± 0.05 | 0.28 ± 0.06 |
Data Interpretation and Limitations
When analyzing explant data, it is crucial to recognize that while these systems offer superior experimental control, they lack the full contextual signals of intact embryos. The quantitative data in Table 1 demonstrates Nodal's essential role in promoting explant elongation through facilitating cell intercalation—a process severely disrupted by both BMP addition and Nodal inhibition [45]. However, researchers should validate key findings from explant studies in intact embryos to confirm physiological relevance. For instance, the inhibitory effect of BMP signaling on cell intercalations observed in explants has been confirmed in dorsal embryonic domains, supporting the model that Nodal ensures robust axis elongation by spatially restricting BMP activity [45].
Synthetic Patterning Systems for Nodal Analysis
Theoretical Foundation and System Design
Synthetic patterning represents a complementary approach that reconstructs developmental signaling modules in non-developmental contexts, providing unprecedented control over individual circuit components. This bottom-up methodology enables researchers to test theoretical models of pattern formation, including Turing's reaction-diffusion mechanism, which requires specific interactions between short-range activators and long-range inhibitors [46]. The Nodal-Lefty signaling pair presents an ideal candidate for such reconstruction, as it naturally embodies this activator-inhibitor relationship: Nodal acts as a short-range activator that promotes its own expression while inducing its inhibitor Lefty, which diffuses more extensively to create lateral inhibition [46].
The engineering of synthetic patterning systems follows a modular design principle, where core signaling components are isolated and reconstituted in tractable cellular environments. For mammalian Nodal-Lefty systems, this typically involves engineering HEK293 cells—which lack endogenous Nodal pathway components—with defined genetic modules that recreate the core circuit topology [46]. This approach has demonstrated that the reconstituted Nodal-Lefty network can spontaneously generate stable patterns with a periodicity of approximately 400µm, providing direct experimental validation that this ligand-receptor system possesses inherent patterning capabilities [46].
Implementation of Synthetic Nodal-Lefty Systems
Core Genetic Circuit Construction
- Base Cell Line Preparation: Engineer HEK293 cells to express essential Nodal signaling components they naturally lack—Cryptic co-receptor and FoxH1 transcription factor—creating a responsive cellular background [46].
- Activator Circuit: Introduce (f2)7-Nodal construct, where Nodal expression is driven by a FoxH1-responsive element containing seven repeats of FoxH1 binding sites, creating positive feedback [46].
- Activator-Inhibitor Circuit: Further introduce (f2)7-Lefty2 construct to the activator circuit cells, enabling Nodal-induced expression of its inhibitor Lefty2, completing the Turing-type network topology [46].
- Reporting System: Incorporate (f2)7-luc reporter (Firefly luciferase under FoxH1-responsive elements) for dynamic monitoring of pathway activity, or fluorescent reporters for spatial pattern visualization [46].
Pattern Formation and Analysis Protocol
- Cell Seeding: Plate engineered cells at near-confluent density (≥90% confluence) in 35mm glass-bottom dishes to minimize proliferation effects while allowing cell-cell communication.
- Time-Lapse Monitoring: For luciferase reporters, add D-luciferin (150μg/mL) to culture medium and image bioluminescence every 2-4 hours for 48-72 hours. For fluorescence reporters, use confocal microscopy with environmental control.
- Pattern Quantification: Calculate spatial correlation functions from acquired images to assess pattern periodicity. Measure domain sizes and spatial distribution of signaling activity.
- Perturbation Experiments: Test pattern robustness by adding recombinant Nodal or Lefty proteins, or inhibitors of downstream signaling components.
Table 2: Synthetic Nodal-Lefty Patterning System Parameters and Outcomes
| Circuit Component | Function | Effect on Pattern Formation |
|---|---|---|
| Nodal (Activator) | Short-range activation; induces own expression and Lefty | Necessary for pattern initiation; controls activation strength |
| Lefty (Inhibitor) | Long-range inhibition; suppresses Nodal signaling | Prevents expansion of Nodal domains; stabilizes pattern |
| Cryptic Co-receptor | Essential for Nodal signal reception | Required for circuit function; modulates cellular sensitivity |
| FoxH1 | Nodal-responsive transcription factor | Transduces Nodal signal to transcriptional responses |
| Finger 1 Domain of Nodal | Restricts Nodal diffusion | Critical for differential diffusivity; pattern fails if removed |
Advanced Synthetic Organizer Systems
Recent innovations have extended beyond intracellular circuit engineering to include synthetic organizer cells—engineered cells designed to self-assemble around progenitor cells and provide spatially defined biochemical signals [47]. This approach integrates Spemann and Mangold's organizer concept with synthetic cell adhesion molecules (synCAMs) to create customizable signaling architectures around embryonic stem cells [47]. By programming distinct spatial arrangements of Wnt3A and DKK1 expression, researchers can generate systematically varied morphogen activity gradients that induce different anterior-posterior patterning outcomes [47]. This methodology provides unprecedented control over morphogen presentation, enabling precise dissection of how gradient properties (dynamic range, slope) influence cell fate decisions.
Integrated Signaling Pathways and Experimental Workflows
Nodal Signaling Pathway Architecture
Nodal-Lepty Signaling Circuit
The Nodal signaling pathway functions through a core architecture where extracellular Nodal ligands bind to receptor complexes comprising Type I/II Activin receptors and EGF-CFC family co-receptors (Oep in zebrafish) [21]. This interaction triggers phosphorylation and nuclear translocation of Smad2/3 transcription factors, which complex with Smad4 and partner with FoxH1 to activate transcription of target genes, including nodal genes themselves (positive feedback) and lefty genes (negative feedback) [46] [21]. The EGF-CFC co-receptor Oep plays a surprisingly sophisticated role in this circuit, not only permitting signaling but actively shaping the morphogen gradient by regulating ligand capture rates—a mechanism that controls both signaling range and cellular sensitivity [21]. This pathway architecture explains how the Nodal-Lefty system can generate self-organizing patterns: Nodal acts as short-range activator while the more diffusible Lefty provides long-range inhibition, satisfying the conditions for Turing pattern formation [46].
Integrated Experimental Workflow
Experimental Workflow for Pathway Analysis
The integrated workflow for Nodal pathway analysis combines complementary approaches to establish robust conclusions. Research begins with clearly defined questions about specific aspects of Nodal signaling, followed by selection of the most appropriate experimental system—zebrafish explants for tissue-level morphogenetic events or synthetic systems for circuit-level properties [45] [46]. Experimental design must carefully define both perturbations (genetic, pharmacological, or optogenetic) and quantitative readouts (morphometric, molecular, or dynamic). Data collection employs advanced imaging and quantification methods, while analysis focuses on extracting mechanistic insights from quantitative measurements. Crucially, findings from one system should be validated in alternative contexts—explant observations confirmed in intact embryos or synthetic circuit principles tested in physiological settings—to establish biological relevance and mechanistic generality [45] [46] [21].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagent Solutions for Nodal Signaling Studies
| Reagent/Category | Function/Application | Example Specifics |
|---|---|---|
| Zebrafish Lines | Genetic perturbation of pathway components | oep mutants (show expanded Nodal range); lefty1;lefty2 double mutants |
| Morpholinos | Transient gene knockdown | Nodal ligands (squint, cyclops), receptors, or co-receptors |
| Recombinant Proteins | Pathway activation/inhibition | Recombinant Nodal, BMP4, Lefty proteins for exogenous application |
| Pharmacological Inhibitors | Acute pathway modulation | SB505124 (Nodal receptor inhibitor); Dorsomorphin (BMP inhibitor) |
| Synthetic Biology Tools | Circuit reconstruction | (f2)7-Nodal, (f2)7-Lefty constructs; synthetic receptors; optogenetic tools |
| Cell Adhesion Molecules | Spatial organization in synthetic systems | synCAMs for programming self-assembly architectures [47] |
| Reporting Systems | Monitoring pathway activity | (f2)7-luc reporter; phosphorylated Smad2 antibodies |
| Visualization Tools | Live imaging and quantification | Membrane-targeted GFP/RFP; photoactivatable dyes |
The integration of classical explant systems with modern synthetic patterning approaches provides a powerful multidimensional framework for analyzing Nodal signaling in vertebrate development. Zebrafish embryonic explants offer the advantage of physiological relevance within a controllable environment, enabling researchers to dissect how Nodal coordinates morphogenesis with patterning—particularly its dual role in inducing mesendoderm while restricting BMP signaling to permit axis elongation [45]. Complementarily, synthetic patterning systems provide reductionist platforms for testing fundamental principles of pattern formation, demonstrating that the core Nodal-Lefty circuit possesses inherent self-organizing capabilities that follow theoretical reaction-diffusion principles [46]. The emerging approach of synthetic organizer cells further extends this paradigm by enabling programmable spatial control over morphogen presentation, opening new possibilities for engineering patterned tissues with defined architectures [47]. Together, these methodologies advance our understanding of how embryonic organizers generate positional information during development, with implications for regenerative medicine, disease modeling, and therapeutic development. As these technologies mature, they promise increasingly sophisticated approaches for reconstructing developmental processes and manipulating cell fate decisions in both basic research and clinical applications.
Addressing Experimental Challenges in Nodal Signaling Research
The formation of the vertebrate body plan is orchestrated by precise spatiotemporal gradients of signaling molecules. Among these, the Nodal morphogen plays an indispensable role in mesendoderm patterning, a process fundamental to early embryogenesis. Research in zebrafish has revealed significant functional redundancy among Nodal ligands and their receptors, presenting a challenge for traditional genetic loss-of-function studies. This technical guide synthesizes current methodologies for employing combinatorial mutant strategies to dissect this redundancy, providing a framework for understanding the complex regulatory mechanisms governing Nodal signaling propagation and its role in mesendodermal patterning. The approaches outlined herein have broader implications for investigating redundant signaling systems across developmental biology and disease models.
In zebrafish embryogenesis, the establishment of mesodermal and endodermal tissues—collectively termed mesendoderm—is directed by graded Nodal signaling activity. This signaling system exhibits extensive redundancy at multiple levels, a characteristic that ensures robustness in embryonic patterning but complicates genetic analysis. The core signaling pathway involves Nodal ligands binding to receptor complexes comprising Type I (Acvr1) and Type II (Acvr2) activin receptors along with the EGF-CFC co-receptor Tdgf1 (Oep), leading to phosphorylation of Smad2/3 transcription factors that activate target gene expression [48].
Two principal Nodal ligands—Squint (Sqt) and Cyclops (Cyc)—demonstrate overlapping functions during mesendoderm formation, with single mutants exhibiting partial or no phenotypes despite complete loss of either gene [49]. Similarly, the zebrafish genome encodes multiple Type I and Type II receptor paralogs that are maternally deposited and expressed during early embryogenesis, including three Type I (acvr1b-a, acvr1b-b, acvr1c) and four Type II (acvr2a-a, acvr2a-b, acvr2b-a, acvr2b-b) receptors [48]. This multi-layered redundancy necessitates combinatorial perturbation approaches to uncover the full functional repertoire of these pathway components and their collective impact on the Nodal signaling gradient that patterns the mesendoderm.
Quantitative Profiling of Nodal Signaling Components
Ligand-Receptor Binding Kinetics and Diffusion Properties
Quantitative measurements of ligand-receptor interactions and mobility parameters provide critical insights into the biophysical mechanisms underlying Nodal gradient formation. The data presented in the table below were obtained through advanced techniques including fluorescence correlation spectroscopy (FCS) and single-wavelength fluorescence cross-correlation spectroscopy (SW-FCCS) in live zebrafish embryos [50] [20].
Table 1: Biophysical Properties of Nodal Ligands and Receptors
| Molecule | Diffusion Coefficient (μm²/s) | Receptor Binding Affinity (Kd) | Inhibitor Binding Affinity (Kd to Lefty) | Bound Fraction (%) |
|---|---|---|---|---|
| Cyclops | 5.3 (effective) | 3.7 nM to Acvr2b | 1.8 nM | 45% |
| Squint | 9.8 (effective) | 2.9 nM to Acvr2b | 2.5 nM | 38% |
| Lefty1 | 22.4 (effective) | N/A | N/A | <15% |
| Lefty2 | 27.1 (effective) | N/A | N/A | <10% |
The data reveal crucial differences in mobility and binding characteristics between Nodal ligands and their inhibitors. Nodals exhibit significantly lower effective diffusion coefficients and higher bound fractions compared to Lefty inhibitors, reflecting their stronger interactions with cell-surface receptors [20]. These biophysical differences directly impact their signaling ranges, with Cyclops acting ultra-short range, Squint at short-to-mid range, and Lefty proteins functioning as long-range inhibitors [50].
Phenotypic Consequences of Combinatorial Receptor Loss
Systematic loss-of-function studies have elucidated the functional relationships between different receptor paralogs. The table below summarizes the phenotypic outcomes observed following combinatorial genetic perturbations of Nodal receptors in zebrafish embryos.
Table 2: Phenotypic Spectrum of Combinatorial Receptor Mutants
| Genetic Perturbation | Mesendoderm Patterning Defects | Nodal Signaling Range | Embryonic Lethality | Functional Interpretation |
|---|---|---|---|---|
| Single Type I or Type II receptor mutant | None or mild | Unchanged | No | High functional redundancy |
| acvr1b-a; acvr1b-b double mutant | Severe loss of mesendoderm | Dramatically reduced | Yes | Major mediators of Nodal signaling |
| All Type II receptor compound mutant | Moderate-severe defects | Altered distribution | Partial | Partly Nodal-independent functions |
| acvr1b-a; acvr1b-b; oep compound mutant | Complete loss of mesendoderm | Abolished | Yes | Essential core signaling components |
Strikingly, only the combined loss of both Type I receptors (acvr1b-a and acvr1b-b) recapitulates the classic Nodal loss-of-function phenotype, indicating their non-redundant essential function despite their paralogous relationship [48]. In contrast, Type II receptors exhibit both redundant and Nodal-independent roles, as compound mutants display patterning defects that are only partially attributable to disrupted Nodal signaling.
Experimental Strategies for Combinatorial Perturbation
Genetic Mutant Generation and Validation
Permanent genetic mutants provide the foundation for stable phenotypic analysis. The following protocols outline established methodologies for generating and validating combinatorial Nodal pathway mutants:
Protocol 1: CRISPR-Cas9 F0 Knockout for Multi-Gene Targeting
- Design: Create single-guide RNAs (sgRNAs) targeting exonic regions encoding critical protein domains (e.g., kinase domains of receptors, mature peptide regions of ligands)
- Injection: Co-inject 2-4 sgRNAs (50-100 pg each) with Cas9 protein (300-500 pg) into one-cell stage zebrafish embryos
- Screening: Assess mutagenesis efficiency via T7 endonuclease I assay or restriction fragment length polymorphism (RFLP) analysis at 24 hours post-fertilization (hpf)
- Phenotyping: Analyze F0 embryos for early patterning defects, as this approach effectively circumvents maternal deposition through efficient somatic mutagenesis [48]
Protocol 2: Generation of Stable Compound Mutant Lines
- Sequential Crossing: Cross single mutant lines to generate double and triple heterozygous carriers
- Incrossing: Intercross compound heterozygotes to generate embryos with combinatorial homozygous mutations
- Genotyping: Establish multiplex PCR and sequencing protocols for simultaneous identification of multiple mutant alleles
- Phenotypic Rescue: Validate specificity of genetic interactions through mRNA rescue experiments with wild-type receptor transcripts [48]
Acute Protein Degradation and Knockdown Approaches
For receptors exhibiting maternal transcript deposition, acute protein degradation strategies provide temporal control over gene function:
Protocol 3: Auxin-Inducible Degradation System
- Transgenesis: Generate lines expressing TIR1 F-box protein under ubiquitous or tissue-specific promoters
- Tagging: Use CRISPR-Cas9 to insert minimal AID degron tags into endogenous receptor loci
- Treatment: Apply auxin (indole-3-acetic acid, 500 μM) at specific developmental windows to induce targeted protein degradation
- Control: Include untreated siblings and non-degradable target controls to validate specificity
Protocol 4: Combinatorial Morpholino Knockdown
- Design: Use gene-specific morpholinos targeting translational start sites or splice junctions
- Dosage: Titrate morpholino concentrations (1-8 ng per embryo) to minimize off-target effects while maintaining efficacy
- Validation: Confirm knockdown efficiency via RT-PCR (splice morpholinos) or Western blot (translation blockers)
- Specificity Controls: Include standard control morpholinos and rescue with synthetic mRNA lacking morpholino binding sites [48]
Analytical Methodologies for Assessing Pathway Function
Quantitative Imaging of Signaling Activity and Distribution
Advanced imaging techniques enable direct visualization of Nodal signaling distribution and activity:
Protocol 5: Phospho-Smad2 Immunostaining and Quantification
- Fixation: Collect embryos at 30%-epiboly to shield stages and fix in 4% paraformaldehyde
- Staining: Use anti-phospho-Smad2 primary antibody (1:500) with fluorescent secondary antibodies
- Imaging: Acquire confocal z-stacks of the marginal region with consistent laser power and detection settings
- Quantification: Measure fluorescence intensity as a function of distance from the margin to generate signaling gradient profiles [51]
Protocol 6: Single-Molecule Tracking of Ligand Dynamics
- Sample Preparation: Inject 1-2 pg of HaloTag-tagged Nodal or Lefty mRNA into one-cell stage embryos
- Labeling: Incubate embryos in JF549 dye solution (100 nM) for covalent HaloTag labeling
- Imaging: Use reflected light-sheet microscopy (RLSM) to track individual molecules at 85 frames/second
- Analysis: Classify extracellular space into interfaces and cavities using membrane-GFP marker; calculate diffusion coefficients and bound fractions from trajectory data [20]
Computational Modeling of Gradient Formation
The integration of experimental data with computational models provides a systems-level understanding of how receptor-ligand interactions shape the Nodal gradient:
Protocol 7: Agent-Based Simulation of Morphogen Dispersion
- Framework: Develop a stochastic model representing individual ligand-receptor interactions
- Parameters: Incorporate measured values for diffusion coefficients, binding affinities, and degradation rates
- Spatial Context: Model embryonic geometry based on actual tissue architecture measurements
- Validation: Compare simulation outputs with experimental gradient measurements from phospho-Smad2 staining
- Perturbation Testing: Simulate receptor loss-of-function conditions to predict changes in gradient shape and signaling range [50] [20]
The Scientist's Toolkit: Essential Research Reagents
Table 3: Critical Reagents for Nodal Pathway Combinatorial Studies
| Reagent Category | Specific Examples | Application and Function |
|---|---|---|
| Genetic Tools | acvr1b-a, acvr1b-b mutant lines; squint, cyclops mutant lines; lefty1, lefty2 TALEN mutants | Generation of combinatorial loss-of-function backgrounds; analysis of genetic interactions |
| Visualization Reagents | HaloTag-labeled Squint/Cyclops/Lefty constructs; memGFP membrane marker; phospho-Smad2 antibodies | Live imaging of ligand distribution; cell boundary demarcation; readout of signaling activity |
| Knockdown Reagents | Receptor-specific morpholinos; CRISPR sgRNAs for Acvr1/Acvr2 paralogs | Acute gene function disruption; circumvention of maternal transcript effects |
| Chemical Inhibitors | SB505124 (Alk4/5/7 inhibitor); recombinant Lefty protein | Pharmacological perturbation of Nodal signaling; exogenous inhibition studies |
| Analysis Tools | TrackIt software; custom R/Python scripts for gradient quantification; agent-based modeling frameworks | Single-molecule trajectory analysis; quantitative pattern assessment; computational simulation |
Signaling Pathway and Experimental Strategy Visualization
Nodal Signaling Pathway and Receptor-Ligand Interactions
Diagram 1: Nodal Signaling Pathway with Feedback Loops
Combinatorial Mutant Strategy Workflow
Diagram 2: Combinatorial Mutant Analysis Workflow
Combinatorial mutant strategies have proven essential for unraveling the functional redundancy inherent in the zebrafish Nodal signaling system. The approaches outlined in this technical guide provide a roadmap for systematically investigating how multiple ligand and receptor paralogs cooperate to generate robust patterning signals during mesendoderm formation. The integration of quantitative live imaging, genetic perturbation, and computational modeling represents a powerful framework for understanding how signaling gradients emerge from molecular interactions.
Future advances in genome editing, particularly methods for simultaneous multi-gene targeting and conditional protein degradation, will further enhance our ability to dissect complex genetic networks. The principles established in zebrafish Nodal signaling studies provide a paradigm for investigating redundant developmental systems across model organisms and may inform therapeutic strategies for diseases involving dysregulated TGF-β signaling pathways.
Within the framework of zebrafish mesendodermal patterning research, the Nodal signaling pathway employs sophisticated feedback loops to precisely coordinate germ layer specification and morphogenesis. Disrupting these feedback mechanisms, particularly through the decoupling of inhibitory signals from activation pathways, leads to profound patterning defects and disrupts critical developmental events such as endodermal internalization and cardiac left-right asymmetry. This whitepaper synthesizes current research to provide an in-depth technical guide for studying feedback circuit disruption, offering detailed methodologies for experimental interrogation, quantitative data analysis, and visualization of the core signaling machinery. By establishing standardized approaches for manipulating and monitoring Nodal feedback circuits, this resource aims to equip researchers with the tools necessary to advance both fundamental developmental biology and targeted therapeutic interventions.
The TGF-β superfamily member Nodal serves as a master regulator of vertebrate embryogenesis, directing the formation of mesoderm and endoderm (collectively termed mesendoderm) during gastrulation [4]. In zebrafish, this process is controlled by two Nodal ligands, Squint (Sqt) and Cyclops (Cyc), which initiate a sophisticated signaling cascade upon binding to a receptor complex comprising Type I (Acvr1) and Type II (Acvr2) activin receptors alongside the EGF-CFC co-receptor One-eyed pinhead (Oep/Tdgf1) [4] [52]. This ligand-receptor interaction triggers the phosphorylation and nuclear translocation of Smad2/3 transcription factors, which activate target genes including the key endodermal determinant sox32 and the Nodal genes themselves, establishing a positive feedback loop [26] [4].
A critical feature of this system is its self-regulation through a negative feedback loop mediated by the Lefty protein, itself a Nodal target gene. Lefty acts as a long-range diffusion-prone inhibitor that antagonizes Nodal signaling, creating a dynamic balance that precisely shapes the Nodal morphogen gradient [4]. This balanced feedback circuit is not only essential for correct mesendodermal patterning but also for subsequent morphogenetic processes, including the directed migration of endodermal cells to the embryo's interior and the leftward movement of cardiomyocytes during heart development [26] [52]. Disrupting the delicate equilibrium between Nodal activation and Lefty inhibition—effectively decoupling the inhibitory arm from the signaling arm—leads to severe developmental consequences, ranging from germ layer mispatterning to aberrant organ laterality [4] [52].
Core Signaling Pathway and Feedback Mechanisms
The Nodal signaling pathway operates through a tightly regulated sequence of molecular events that integrate positive and negative feedback to achieve precise spatiotemporal control of gene expression and cell behavior. The core architecture of this pathway can be visualized as a dynamic system with built-in regulatory checks and balances.
Figure 1: The Nodal Signaling Pathway with Integrated Feedback Loops. The core pathway (light gray) shows signal transduction from ligand binding to target gene activation. Positive feedback (red dashed) amplifies signaling through increased Nodal production. Negative feedback (blue dashed) dampens signaling via Lefty inhibition. The output for cell migration (green) requires both sox32 and Nodal reception [26] [4].
Molecular Logic of the Nodal Signaling Cascade
The signaling cascade initiates when Nodal ligands bind to a membrane complex containing Type I receptors (primarily Acvr1b-a and Acvr1b-b in zebrafish), Type II receptors (Acvr2 homologs), and the Oep co-receptor [4]. This assembly activates the kinase domains of the Type II receptors, which subsequently phosphorylate the Type I receptors. The activated Type I receptors then phosphorylate the C-terminal SSXS motif of Smad2 and Smad3 proteins. These phosphorylated Smads form a complex with the co-Smad, Smad4, and translocate to the nucleus where they directly regulate the transcription of target genes [4]. Critical targets include sox32, which drives endodermal specification, and the Nodal genes themselves, creating a positive feedback loop that amplifies the initial signal [26].
Positive and Negative Feedback Regulation
The Nodal pathway employs a self-regulating feedback system where the output of the pathway modulates its own activity:
- Positive Feedback Amplification: Nuclear Smad complexes directly activate transcription of additional Nodal ligands, creating an autocrine circuit that reinforces the initial signal [26]. This autoinduction is crucial for maintaining signaling levels necessary for endodermal specification and for initiating the ingression movements of endodermal cells during gastrulation [26].
- Negative Feedback Restraint: The same nuclear Smad complexes simultaneously activate transcription of Lefty genes, which encode secreted antagonists that bind to Nodal ligands and prevent their interaction with receptors [4]. This inhibitory loop creates a balancing mechanism that limits the spatial range and duration of Nodal signaling.
The functional outcome of this signaling network depends on the successful integration of both specification and migration cues. Research demonstrates that Nodal signaling establishes an "AND" gate where both sox32-dependent endodermal specification and direct Nodal ligand reception are necessary to drive the sorting of endodermal cells to the inner layer of the embryo [26]. Disruption of either component decouples the specification program from the migration program, leading to failed endodermal internalization despite proper fate specification.
Experimental Approaches for Disrupting and Analyzing Feedback Circuits
To dissect the functional consequences of feedback circuit disruption, researchers have developed sophisticated experimental strategies that enable precise manipulation and observation of Nodal signaling components. The following workflow outlines a comprehensive approach for probing feedback mechanisms in zebrafish embryos.
Figure 2: Experimental Workflow for Feedback Circuit Analysis. This pipeline combines genetic perturbation (red), cellular transplantation (green), and quantitative analysis (blue) to dissect Nodal feedback mechanisms [26] [4] [52].
Genetic Perturbation Strategies
A. Combinatorial Receptor Knockdown
- Objective: Determine functional redundancy among Nodal receptors and their role in feedback propagation.
- Protocol:
- Identify target receptors through homology search using human/mouse Acvr1b, Acvr1c, Acvr2a, and Acvr2b protein sequences [4].
- Generate mutant lines using CRISPR-Cas9 targeting:
- Type I receptors: acvr1b-a and acvr1b-b
- Type II receptors: Multiple acvr2 homologs
- Combine mutant alleles through crossing and supplement with morpholino knockdown for complete loss-of-function analysis.
- Assess phenotypic severity relative to Nodal ligand mutants (sqt, cyc) and co-receptor mutants (oep).
- Key Application: This approach revealed that combined loss of acvr1b-a and acvr1b-b phenocopies Nodal loss-of-function, while Type II receptor defects produce partially Nodal-independent phenotypes [4].
B. Late-Zygotic Mutant Analysis
- Objective: Disrupt Nodal signaling specifically during later patterning events without affecting early mesendoderm induction.
- Protocol:
- Utilize maternal-zygotic oep mutants (MZoep) that completely lack Nodal signaling.
- Inject oep mRNA at the 1-cell stage to rescue early mesendoderm induction.
- The rescued embryos (LZoep) specifically lack Nodal signaling during later stages, including left-right patterning.
- Analyze cardiac morphogenesis using transgenic reporters (e.g., cmlc2:gfp) [52].
- Key Application: This temporal dissection revealed that Nodal signaling controls cardiomyocyte migration speed and directionality during heart lateralization without affecting initial heart field specification [52].
Cell Transplantation and Ectopic Patterning Assay
Objective: Decouple cell fate specification from migration by testing the autonomous ability of endodermal cells to internalize without endogenous morphogenetic cues.
Protocol:
- Donor Cell Preparation:
Transplantation Procedure:
- At blastula stages (3-4 hours post-fertilization), isolate donor cells using sharp transplantation pipettes.
- Transplant approximately 10-20 donor cells into the animal pole of wild-type host embryos, far from the endogenous margin where endoderm normally forms.
- Include control transplantations with non-induced cells or lineage tracers.
Time-Lapse Imaging and Analysis:
- Mount embryos in low-melt agarose for imaging.
- Acquire z-stacks every 5-10 minutes for 4-6 hours using confocal microscopy.
- Track individual cell trajectories and quantify migration parameters:
- Speed (μm/min)
- Directionality (net displacement/total path length)
- Final position (inner vs. outer layer)
Interpretation: Wild-type endodermal cells ingress radially into the inner layer, while cells lacking Nodal signaling or sox32 remain on the embryo surface, demonstrating that both specification and Nodal reception are required for internalization [26].
Signaling Distribution and Mobility Assays
Objective: Determine how receptor components influence Nodal ligand distribution and mobility.
Protocol:
- Generate GFP-tagged Nodal ligands (Sqt, Cyc) and Lefty proteins [4].
- Use fluorescence recovery after photobleaching (FRAP) or single-molecule tracking to quantify ligand mobility in wild-type and receptor-deficient backgrounds.
- Measure signaling range using phospho-Smad2/3 immunostaining as a readout of pathway activation.
- Quantify the bound fraction of Nodal ligands versus the freely diffusing fraction.
Key Findings: Nodal ligands exhibit lower effective diffusivity than Lefty inhibitors, largely due to interactions with receptor complexes. In oep co-receptor mutants, Nodal distribution increases dramatically, demonstrating that receptor binding restricts Nodal mobility and shapes the morphogen gradient [4].
Quantitative Analysis of Feedback Disruption Phenotypes
Systematic quantification of phenotypic outcomes following feedback disruption reveals consistent patterns across multiple developmental processes. The following tables summarize key quantitative measurements from representative studies.
Table 1: Quantitative Effects of Nodal Feedback Disruption on Endodermal Cell Behavior [26]
| Experimental Condition | Internalization Efficiency (%) | Migration Speed (μm/min) | Directionality Index | sox32 Expression |
|---|---|---|---|---|
| Wild-type (control) | >95% | 0.8 ± 0.2 | 0.85 ± 0.08 | +++++ |
| acvr1ba* transplantation | 92% ± 5% | 0.7 ± 0.3 | 0.82 ± 0.10 | +++++ |
| sox32 overexpression | 25% ± 10% | 0.3 ± 0.2 | 0.35 ± 0.15 | +++++ |
| Nodal inhibition + acvr1ba* | 15% ± 8% | 0.2 ± 0.1 | 0.28 ± 0.12 | +++++ |
| Nodal inhibition alone | <5% | 0.1 ± 0.1 | 0.15 ± 0.08 | - |
Table 2: Cardiomyocyte Migration Defects in Nodal Signaling Mutants [52]
| Parameter | Wild-type | LZoep Mutant | BMP Mutant | Significance |
|---|---|---|---|---|
| Migration Speed (μm/min) | 0.51 ± 0.09 | 0.32 ± 0.07 | 0.29 ± 0.08 | p < 0.01 |
| Directionality Index | 0.78 ± 0.11 | 0.41 ± 0.13 | 0.38 ± 0.15 | p < 0.001 |
| Leftward Displacement (μm) | 42.5 ± 6.2 | 8.3 ± 4.1 | 7.1 ± 3.8 | p < 0.001 |
| Cardiac Jogging | Normal leftward | Absent/Random | Absent/Random | - |
| Heart Rotation | Clockwise | Absent | Absent | - |
Table 3: Receptor Requirements for Nodal Signaling in Zebrafish [4]
| Receptor Type | Gene | Mutant Phenotype | Combinatorial Phenotype | Role in Feedback |
|---|---|---|---|---|
| Type I | acvr1b-a | Mild or none | Severe mesendoderm defects | Signal transduction |
| Type I | acvr1b-b | Mild or none | (phenocopies Nodal mutants) | Signal transduction |
| Type I | acvr1c | Not detected | - | - |
| Type II | acvr2a | Mild or none | Patterning defects | Ligand binding & distribution |
| Type II | acvr2b-a | Mild or none | (partly Nodal-independent) | Ligand binding & distribution |
| Type II | acvr2b-b | Mild or none | - | - |
| Co-receptor | oep/tdgf1 | Severe Nodal loss-of-function | - | Essential for signal propagation |
The quantitative data reveal several consistent patterns following feedback disruption. First, the migration defects observed in both endodermal and cardiomyocyte populations share remarkable similarity, with significant reductions in speed and directionality representing a common phenotypic signature. Second, the receptor analysis demonstrates substantial functional redundancy within receptor classes, with Type I receptors serving as the primary mediators of Nodal signal transduction while Type II receptors play additional roles in ligand distribution. Finally, the requirement for both sox32 expression and ongoing Nodal reception in endodermal internalization demonstrates that these two outputs of the Nodal pathway cannot be decoupled without disrupting normal morphogenesis.
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Reagents for Studying Nodal Feedback Circuits in Zebrafish
| Reagent Category | Specific Examples | Function/Application | Key References |
|---|---|---|---|
| Genetic Tools | acvr1ba (constitutively active) | Cell-autonomous endoderm induction | [26] |
| sox32 overexpression | Bypass Nodal for endoderm specification | [26] | |
| LZoep mutants | Temporal disruption of Nodal signaling | [52] | |
| cmlc2:gfp transgenic line | Cardiomyocyte visualization and tracking | [52] | |
| Molecular Reagents | Anti-pSmad2/3 antibodies | Readout of Nodal pathway activation | [4] |
| sox17, sox32 RNA probes | Endodermal lineage specification markers | [26] | |
| lft1, lft2 RNA probes | Readout of Nodal feedback inhibition | [4] | |
| Technical Approaches | Cell transplantation | Ectopic patterning assays | [26] |
| Single-molecule tracking | Ligand diffusion and mobility measurements | [4] | |
| 4D confocal time-lapse | Quantitative cell migration analysis | [26] [52] |
The experimental framework presented here provides a comprehensive approach for investigating feedback circuit disruption in Nodal signaling. The methodologies enable researchers to precisely decouple inhibitory mechanisms from activation pathways, revealing how this balance governs both cell fate decisions and morphogenetic behaviors. The consistent finding that Nodal signaling controls migratory parameters—including speed, directionality, and persistence—across multiple cell types suggests a fundamental role for this pathway in coordinating pattern formation with tissue morphogenesis.
Future investigations should leverage emerging technologies to address several key challenges. First, the development of optogenetic tools for Nodal signaling would enable precise spatiotemporal control over feedback loops, allowing researchers to test specific predictions about timing and compartmentalization in feedback regulation. Second, advanced biosensors that simultaneously monitor signaling activity, cell fate, and mechanical forces could reveal how information flows through these circuits to influence cell behavior. Finally, computational modeling approaches that integrate the quantitative data generated by these experimental methods will be essential for predicting system-level behaviors and designing targeted interventions.
The principles governing Nodal feedback circuits extend beyond zebrafish development to mammalian systems and regenerative processes. Furthermore, understanding how to manipulate feedback circuits has implications for biomedical applications, including controlling stem cell differentiation and developing therapeutic strategies for birth defects and cancers driven by dysregulated TGF-β signaling. By establishing standardized approaches for disrupting and analyzing these critical regulatory systems, this technical guide provides a foundation for advancing both basic research and translational applications.
In the intricate landscape of developmental biology, achieving precise spatial and temporal control of signaling molecules represents a fundamental challenge. Morphogen gradients provide crucial positional information during embryogenesis, instructing cells to adopt specific fates based on local signal concentration. The Nodal signaling pathway, a cornerstone of vertebrate development, exemplifies this mechanism, playing an indispensable role in mesendoderm patterning in zebrafish embryos [3] [4]. While traditional models emphasized ligand diffusion and degradation as primary gradient-shaping forces, emerging research reveals that co-receptor expression serves as a potent regulatory layer controlling signal distribution and cellular response.
This technical guide examines the sophisticated mechanisms through which co-receptors, particularly the EGF-CFC family protein One-eyed pinhead (Oep), govern Nodal signaling range and specificity in zebrafish. We explore how manipulation of co-receptor expression and function provides researchers with precise tools to interrogate and control developmental signaling pathways, with implications for both basic developmental biology and therapeutic applications in regenerative medicine.
Biological Foundation: Nodal Signaling and Mesendoderm Patterning
The Nodal Signaling Pathway Core Components
The Nodal signaling cascade constitutes a conserved TGF-β pathway essential for mesendoderm induction and patterning in vertebrate embryos [4]. The core signaling machinery comprises:
- Ligands: Zebrafish employ two primary Nodal ligands - Squint (Sqt/Ndr1) and Cyclops (Cyc/Ndr2) - which function as heterodimers with the TGF-β family member Vg1 [53].
- Receptor Complex: Type I (Acvr1b-a, Acvr1b-b) and Type II (Acvr2) activin receptors form a complex at the cell membrane [4].
- Co-receptor: The EGF-CFC family protein Oep (zebrafish homolog of Tdgf1) is indispensable for signal transduction [3] [4].
- Intracellular Transducers: Phosphorylated Smad2/Smad3 complexes with Smad4 and translocates to the nucleus to activate target gene expression [4].
Table 1: Core Components of the Zebrafish Nodal Signaling Pathway
| Component Type | Elements in Zebrafish | Primary Function |
|---|---|---|
| Ligands | Squint (Sqt/Ndr1), Cyclops (Cyc/Ndr2) | Secreted signals that bind receptor complexes |
| Receptors | Acvr1b-a, Acvr1b-b (Type I), Acvr2 variants (Type II) | Membrane receptors with serine/threonine kinase activity |
| Co-receptor | One-eyed pinhead (Oep) | EGF-CFC protein essential for ligand binding and signaling |
| Intracellular Mediators | Smad2, Smad3, Smad4 | Signal transduction and nuclear gene regulation |
| Extracellular Antagonists | Lefty1, Lefty2 | Feedback inhibitors that restrict signaling range |
Nodal Signaling in Zebrafish Mesendoderm Formation
During zebrafish gastrulation, Nodal ligands secreted from the yolk syncytial layer (YSL) establish a signaling gradient that patterns the embryonic margin [3]. Cells exposed to high Nodal concentrations adopt endodermal fates, while intermediate levels specify mesodermal derivatives, and low levels permit ectodermal differentiation [3]. This concentration-dependent fate specification necessitates exquisite control over the shape, range, and steepness of the Nodal gradient, with co-receptors playing a surprisingly active role in shaping these properties rather than merely facilitating signaling.
Diagram 1: Nodal gradient patterns mesendoderm with Oep regulation
Co-receptor Mechanisms: Controlling Ligand Distribution and Cellular Sensitivity
Oep Regulates Nodal Ligand Capture and Distribution
Contrary to initial models that positioned co-receptors as passive signaling permissors, quantitative studies reveal that Oep actively governs Nodal distribution through ligand capture mechanisms. In wild-type embryos, Nodal activity forms a steep gradient extending approximately 6-8 cell diameters from the margin [3]. Strikingly, in oep mutants, this precise gradient collapses, transforming into a nearly uniform distribution of Nodal activity throughout the embryo [3]. This dramatic expansion demonstrates that Oep-mediated ligand capture restricts Nodal spread, potentially by reducing ligand effective diffusivity through repeated binding and internalization events.
The co-receptor trap model posits that Oep expression creates a "sink" for Nodal ligands, sequestering them and limiting their free diffusion. Supporting this view, single-molecule imaging has confirmed that Nodal ligands spend a substantially higher fraction of time in receptor-bound states compared to the freely-diffusing antagonist Lefty [3]. This differential mobility creates a permeability barrier that confines Nodal signaling to appropriate spatial domains while allowing inhibitors to diffuse more freely and establish signaling boundaries.
Oep Sets Cellular Sensitivity Thresholds
Beyond controlling ligand distribution, Oep expression levels directly modulate cellular sensitivity to Nodal signals. Increasing Oep concentration sensitizes cells to Nodal ligands, lowering the ligand threshold required for pathway activation and target gene induction [3]. This dual functionality—controlling both ligand availability and cellular response—positions co-receptors as central regulators of signaling robustness and precision.
The sensitivity mechanism operates through enhanced formation of active receptor complexes. Oep facilitates the recruitment of Type I receptors into complexes with ligand-bound Type II receptors, increasing signaling efficiency [4]. This amplification enables cells to respond to minute ligand concentrations, a crucial property for interpreting shallow morphogen gradients.
Table 2: Effects of Oep Manipulation on Nodal Signaling Parameters
| Experimental Condition | Signaling Range | Cellular Sensitivity | Gradient Stability | Developmental Outcome |
|---|---|---|---|---|
| Wild-type (Normal Oep) | Normal (6-8 cell diameters) | Normal | Stable gradient | Proper endoderm, mesoderm, ectoderm patterning |
| Oep Loss-of-Function | Expanded (near uniform) | Reduced | Travelling wave | Ectopic mesendoderm, lethal patterning defects |
| Oep Overexpression | Restricted | Enhanced | Sharpened gradient | Limited mesendoderm, potential deficiency |
| Zygotic Oep Mutant (Maternal Oep only) | Progressively expands | Variable | Unstable, transforms to wave | Defective patterning after maternal depletion |
Experimental Approaches: Manipulating Co-receptor Expression
Genetic Loss-of-Function Strategies
Mutant generation provides the most definitive approach for investigating co-receptor function. Zebrafish oep mutants initially identified the essential role of this co-receptor in Nodal signaling [3] [4]. Several methodological considerations enhance the utility of genetic approaches:
- Compound mutants: Since Type I receptors Acvr1b-a and Acvr1b-b function redundantly, only combined disruption fully abrogates Nodal signaling [4].
- Maternal-zygotic mutants: Maternal Oep deposition can mask early zygotic requirements, necessitating complete maternal-zygotic elimination [3].
- Conditional alleles: Tissue-specific or temporally controlled ablation enables stage-specific and region-specific function analysis.
The phenotypic characterization of co-receptor mutants includes:
- Phospho-Smad2 immunostaining to visualize active signaling distribution
- In situ hybridization for direct target genes (e.g., gsc, ntl, sox32)
- Lineage tracing to assess fate specification errors
- Quantitative image analysis to measure gradient properties
Morpholino and CRISPR F0 Knockdown
For rapid assessment of co-receptor function, morpholino antisense oligonucleotides and CRISPR F0 knockout approaches provide valuable alternatives to stable mutant lines [4]. These methods enable:
- Combinatorial targeting of multiple receptor paralogs to overcome redundancy
- Dose-response studies through titrated reagent injection
- Temporal control with inducible or photoactivatable reagents
Technical considerations for these approaches include:
- Morpholino validation using Western blotting or functional assays
- Control for off-target effects with multiple independent reagents
- Phenotypic assessment within appropriate time windows before potential compensation
Overexpression and Misexpression Strategies
Gain-of-function experiments complement loss-of-function studies by testing sufficiency of co-receptors to manipulate signaling range. Common approaches include:
- Synthetic mRNA injection for ubiquitous overexpression
- Plasmid DNA electroporation for regional misexpression
- Inducible expression systems for temporal control
These experiments demonstrate that elevated Oep levels sharpen the Nodal signaling gradient by increasing ligand capture at the source, effectively shortening the signaling range while enhancing local pathway activation [3].
Quantitative Analysis: Measuring Signaling Range and Gradient Properties
Imaging and Quantification Methods
Robust quantification of signaling activity distribution requires specialized imaging and analytical approaches:
- Immunofluorescence for pSmad2: Provides direct readout of pathway activation with cellular resolution
- Fluorescent ligand tagging: GFP-tagged Nodal ligands enable direct visualization of ligand distribution [3]
- Lineage-specific reporters: Transgenic animals with mesendoderm-specific fluorescent reporters facilitate fate mapping
- Computational image analysis: Custom algorithms quantify signaling range, gradient slope, and positional precision
Key quantitative parameters include:
- Signaling range: Distance from source at which activity falls to half-maximal
- Gradient decay length (λ): Characteristic distance over which concentration declines
- Positional error: Precision of boundary placement relative to source
Mathematical Modeling of Gradient Formation
Mathematical models provide a conceptual framework for interpreting experimental manipulations. The co-receptor trap model can be represented through reaction-diffusion equations:
Where [L] represents free ligand concentration, [Oep] is co-receptor density, [L-Oep] is bound complex, D is diffusion coefficient, kon/koff are binding parameters, k_int is internalization rate, and δ is degradation rate.
This model predicts the crucial experimental observation that eliminating co-receptor replenishment (as in zygotic oep mutants) transforms the stable Nodal gradient into a travelling wave [3], demonstrating that continuous co-receptor synthesis is essential for gradient stability.
Diagram 2: Oep replenishment controls gradient stability versus wave formation
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Co-receptor and Nodal Signaling Research
| Reagent Category | Specific Examples | Research Application | Technical Considerations |
|---|---|---|---|
| Genetic Mutants | oep null alleles, acvr1b-a/b double mutants, Maternal-zygotic oep | Loss-of-function analysis, Phenotypic characterization | Redundancy may require multiple mutations; maternal deposition complicates early roles |
| Antisense Oligonucleotides | oep morpholinos, acvr1/2 morpholinos | Acute knockdown, Combinatorial targeting | Dose-dependent effects; potential off-target activity requires controls |
| CRISPR-Cas9 Components | gRNAs targeting receptor genes, Cas9 mRNA/protein | F0 knockout, Stable line generation | Efficient multiplexing enables targeting of redundant family members |
| Expression Constructs | oep mRNA, acvr1/2 expression plasmids, Inducible Oep variants | Overexpression, Misexpression, Structure-function analysis | Titration required to avoid non-physiological effects; inducible systems enable temporal control |
| Signaling Reporters | pSmad2 antibodies, BRE-transgenic lines, Mesendoderm markers | Signaling activity measurement, Fate specification assessment | Multiple reporters confirm specificity; phosphorylation state reflects acute activity |
| Ligand Probes | GFP-tagged Sqt/Cyc, Fluorescent Nodal variants, Cer-S inhibitor | Ligand distribution tracking, Signal inhibition | Tagging may alter ligand properties; functionality must be verified |
Technical Protocols: Key Experimental Methods
Protocol: Quantifying Nodal Signaling Range via pSmad2 Immunostaining
This protocol details the assessment of Nodal signaling distribution in zebrafish embryos through phospho-Smad2 immunofluorescence, applicable for evaluating co-receptor manipulation effects.
Materials:
- Fixed zebrafish embryos (shield stage)
- Anti-phospho-Smad2 (Ser465/467) antibody
- Fluorescent secondary antibody
- Confocal microscopy equipment
- Image analysis software (e.g., Fiji/ImageJ)
Procedure:
- Fixation and Permeabilization: Fix embryos in 4% PFA for 2 hours at room temperature, followed by methanol dehydration and rehydration. Permeabilize with 0.1% Triton X-100 for 30 minutes.
- Immunostaining: Incubate with primary anti-pSmad2 antibody (1:500) overnight at 4°C. After washing, incubate with fluorescent secondary antibody (1:1000) for 2 hours at room temperature.
- Image Acquisition: Capture confocal z-stacks of the marginal region with consistent laser power, gain, and resolution settings across all samples.
- Quantitative Analysis:
- Generate average intensity projections of z-stacks
- Draw a perpendicular line from the margin inward
- Plot fluorescence intensity along this line
- Fit an exponential decay curve: I(x) = I₀e^(-x/λ)
- Calculate decay length (λ) as the signaling range parameter
- Statistical Comparison: Compare λ values between experimental conditions using appropriate statistical tests (e.g., t-test, ANOVA).
Troubleshooting:
- High background: Optimize antibody concentration and blocking conditions
- Uneven staining: Ensure consistent permeabilization across samples
- Signal saturation: Adjust imaging parameters to remain in linear detection range
Protocol: CRISPR-Cas9 Mediated Multiplex Receptor Gene Knockout
This protocol describes simultaneous targeting of multiple Nodal receptor genes to overcome functional redundancy, enabling comprehensive analysis of receptor requirements.
Materials:
- Gene-specific gRNAs for acvr1b-a, acvr1b-b, and acvr2 receptors
- Cas9 protein or mRNA
- Microinjection equipment
- Genotyping primers for each target locus
- T7 endonuclease I or sequencing for mutation detection
Procedure:
- gRNA Design and Synthesis: Design 20-nt guide sequences targeting early exons of each receptor gene. Synthesize gRNAs using T7 polymerase-mediated in vitro transcription.
- Injection Mix Preparation: Combine 50 ng/μL of each gRNA with 300 ng/μL Cas9 protein in nuclease-free water. Include tracer dye (phenol red) for visualization.
- Zebrafish Embryo Injection: Inject 1-2 nL of the mixture into the cell yolk of 1-cell stage embryos.
- Phenotypic Analysis: Score embryos at shield and bud stages for mesendoderm defects, including axial shortening, cyclopia, and reduced mesendodermal markers.
- Mutation Efficiency Assessment: At 24 hpf, extract genomic DNA from pooled injected embryos and assess mutation efficiency using T7 endonuclease I assay or amplicon sequencing.
- Validation: For stable line establishment, raise injected embryos to adulthood and screen for germline transmission.
Technical Notes:
- Include control injections with Cas9 alone or non-targeting gRNA
- Titrate gRNA concentrations to balance efficiency with viability
- For F0 analysis, include uninjected siblings as controls
Applications and Implications: From Basic Research to Therapeutic Potential
The mechanistic insights gleaned from co-receptor manipulation studies extend beyond developmental biology into regenerative medicine and disease modeling. The demonstrated capacity to precisely control morphogen gradient properties through co-receptor expression offers promising approaches for:
- Stem cell engineering: Directing differentiation of pluripotent stem cells toward specific mesendodermal lineages by manipulating co-receptor expression to create defined signaling environments
- Tissue patterning in regenerative contexts: Designing synthetic morphogen systems with engineered co-receptor interactions to pattern tissues in vitro or in vivo
- Therapeutic intervention: Targeting co-receptors in pathological signaling contexts, such as cancer metastasis or fibrosis, where TGF-β signaling plays prominent roles
The zebrafish model continues to provide fundamental insights into these processes, with its external development, genetic tractability, and imaging accessibility enabling detailed analysis of signaling dynamics in vivo. Future research will likely explore the intersection of co-receptor manipulation with additional regulatory mechanisms, including feedback loops, extracellular matrix interactions, and cross-talk with parallel signaling pathways.
Co-receptor expression manipulation represents a powerful strategy for controlling signaling range and specificity in developmental contexts. Research in zebrafish mesendoderm patterning has revealed that the EGF-CFC co-receptor Oep actively shapes the Nodal morphogen gradient through ligand capture and cellular sensitization mechanisms, going far beyond its initial characterization as a passive signaling component. The experimental approaches outlined in this technical guide—from genetic perturbation to quantitative analysis—provide researchers with sophisticated tools to dissect and manipulate signaling gradients with precision. As our understanding of co-receptor biology deepens, so too will our ability to engineer signaling environments for both basic research and therapeutic applications.
Timing and Dosage Considerations for Nodal Pathway Manipulation
In vertebrate embryogenesis, including zebrafish, the establishment of the body plan depends on precisely coordinated signaling events that transform a uniform group of cells into a highly organized, multi-layered structure. Among these events, Nodal signaling stands as a critical regulator responsible for inducing and patterning the mesoderm and endoderm (collectively termed mesendoderm) [4]. This technical guide examines the precise timing and dosage considerations for manipulating the Nodal pathway in zebrafish, framing these concepts within the broader thesis that Nodal functions as a dynamic, dose-dependent signal whose interpretation depends on both concentration and exposure duration. In zebrafish, mesendoderm patterning specifically depends on two secreted Nodal signals: Squint (Sqt) and Cyclops (Cyc) [49] [7]. These ligands act in a dosage-dependent manner to specify different cell fates, with high levels generally promoting more marginal fates (such as endoderm) and lower levels inducing more animal fates (such as somitic mesoderm) [7]. The paradigm of a simple spatial gradient, however, has been complicated by recent findings demonstrating that Nodal activity propagates through a transcriptional relay mechanism rather than passive diffusion, and that cells respond to the cumulative dose of Nodal signals received over time [54] [7].
Molecular Mechanisms of Nodal Signaling
Core Signaling Pathway Components
The Nodal signaling pathway comprises specific molecular components that work in concert to transduce extracellular signals into intracellular responses. Understanding these components is essential for effective pathway manipulation.
Ligands and Receptors: Nodal, a member of the TGF-β superfamily, signals through a receptor complex consisting of Type I and Type II single-transmembrane serine/threonine kinase receptors [4]. Unlike other TGF-β family members, Nodal signaling requires an EGF-CFC co-receptor (Oep in zebrafish) to activate signaling [4]. The ligand binds to Type II receptors and the co-receptor, facilitating recruitment and phosphorylation of Type I receptors.
Signal Transduction: Upon receptor complex oligomerization, Type II receptors phosphorylate Type I receptors in their GS domains, leading to recruitment and phosphorylation of the receptor-regulated Smad proteins Smad2 and Smad3 [4]. The activated pSmad2/pSmad3 proteins associate with the co-factor Smad4 and translocate to the nucleus, where they activate target gene expression, including key developmental regulators and feedback inhibitors [4].
Feedback Regulation: Nodal signaling activates expression of Lefty proteins, which act as long-range feedback inhibitors that antagonize Nodal signaling [54] [4]. This activator-inhibitor relationship is crucial for controlling the spatial and temporal dynamics of Nodal signaling activity.
Visualization of Nodal Signaling Propagation
Nodal signaling pathway and relay mechanism. The pathway involves ligand-receptor binding, intracellular signal transduction, and target gene activation that creates a positive feedback loop for signal propagation.
Critical Timing Windows for Nodal Activity
Developmental Stage-Specific Requirements
Research utilizing pharmacological inhibition of Nodal receptors has revealed precise temporal windows during which Nodal signaling specifies different mesodermal and endodermal cell types. Treatment with SB-431542, which blocks ALK4,5,7 receptors, at different developmental stages demonstrates that Nodal signals are most active during the mid-to-late blastula stages (approximately 3-5 hours post-fertilization) [7]. During this period, cells become progressively committed to different fates in a time-dependent manner.
Table 1: Temporal Sequence of Cell Fate Specification by Nodal Signaling in Zebrafish
| Cell Fate/Tissue Type | Critical Specification Window | Nodal Requirement Level |
|---|---|---|
| Somites | Mid-blastula period | Moderate |
| Notochord | Mid-to-late blastula period | Moderate to High |
| Blood | Late blastula period | High |
| Kupffer's vesicle | Late blastula period | High |
| Hatching gland | Late blastula period | High |
| Heart | Late blastula period | High |
| Endoderm | Mid-to-late blastula period | Very High |
Blocking Nodal signaling at progressively later stages prevents specification of cell types derived from the embryo margin, but not those from more animal regions, suggesting a direct relationship between cell fate and length of exposure to Nodal signals [7]. This timing is crucial because the expression of nodal-related genes and the movement of responding cells are at their most dynamic during these stages.
Experimental Evidence for Timing Dependence
Studies manipulating the timing of Nodal signaling activity provide compelling evidence for time-dependent patterning:
Pharmacological Inhibition: When SB-431542 or SB-505124 is added to embryos after the mid-blastula transition (MBT), it completely blocks the response to zygotic Nodal signals without disturbing earlier signaling activity [7]. Embryos treated with these inhibitors display severe cyclopia and lack all derivatives of mesoderm and endoderm in the head and trunk, closely resembling the phenotype of squint;cyclops double mutants [7].
Mutant Analysis: In squint mutants, cell fate specification is delayed, whereas when Nodal levels are elevated, specification accelerates [7]. This demonstrates that the Nodal dose controls the timing of cell fate specification rather than cells having intrinsically defined periods during which they can adopt particular fates.
Cumulative Dose Response: Cells exposed to a uniform, high dose of Nodal signals adopt progressively more marginal fates with longer exposures [7]. This time-dependent transformation of cell fates indicates that cells respond to the total cumulative dose of Nodal signals to which they are exposed.
Dosage Considerations in Pathway Manipulation
Concentration-Dependent Fate Specification
The concentration of Nodal signaling has a direct impact on the specification of different cell fates, following a dose-response relationship observed across vertebrate species:
High Nodal Levels: Promote the formation of endoderm and prechordal plate [7]. These marginal cell types appear most sensitive to reductions in Nodal levels and require the highest signaling activity for proper specification.
Intermediate Nodal Levels: Induce notochord and heart precursors [7]. These structures require sustained but moderate Nodal signaling during their specification windows.
Low Nodal Levels: Are sufficient for the formation of somitic mesoderm and other more animal derivatives [7]. These tissues are less sensitive to reductions in Nodal activity.
This concentration-dependent effect has been demonstrated in explant experiments, where high doses of Activin-like signals induce marginal cell types, while lower doses induce notochord and muscle [7].
Quantitative Dynamics of Nodal Propagation
Recent research utilizing tagged Nodal proteins has revealed quantitative aspects of Nodal distribution and activity:
Short-Range Signaling: Endogenous Nodal protein is extremely short-range, limited to the immediate neighborhood of source cells [54]. Visualization of fully functional endogenous Nodal protein demonstrates that it fails to spread beyond immediately adjacent cells, contrasting with the longer-range distribution of its inhibitor Lefty.
Relay Mechanism: Nodal activity spreads through a relay mechanism in which Nodal production induces neighboring cells to transcribe Nodal [54]. This propagation mechanism was demonstrated in juxtaposition experiments showing that Nodal signaling requires a functional Nodal gene in receiving cells to propagate beyond immediately adjacent cells.
Receptor-Mediated Distribution: Type I receptor and co-receptor levels can directly influence the diffusion and distribution of Nodals [4]. In the absence of the co-receptor Oep, Nodal's signaling range and distribution dramatically increase, suggesting that receptor binding affects Nodal mobility or stability in the embryo.
Table 2: Nodal Signaling Components and Their Roles in Zebrafish
| Component | Gene Name(s) | Function in Signaling | Effect of Loss-of-Function |
|---|---|---|---|
| Ligands | squint (sqt), cyclops (cyc) | Initiate and maintain Nodal signal | Loss of all mesendoderm in double mutants |
| Type I Receptors | acvr1b-a, acvr1b-b | Major mediators of Nodal signaling | Phenocopy Nodal loss-of-function when both lost |
| Type II Receptors | acvr2a, acvr2b-a, acvr2b-b | Partly redundant, bind ligands | Function partly independently of Nodal |
| Co-receptor | one-eyed pinhead (oep) | Essential for receptor complex formation | Expanded Nodal range, severe patterning defects |
| Inhibitors | lefty1, lefty2 | Long-range feedback inhibition | Excess Nodal signaling, patterning defects |
Experimental Protocols for Nodal Manipulation
Pharmacological Inhibition Approaches
Precise pharmacological inhibition of Nodal signaling allows researchers to target specific temporal windows during development:
Receptor Inhibitor Application: SB-431542 and SB-505124 competitively bind to the ATP binding sites of the ALK 4, 5 and 7 receptors, preventing their kinase activity [7]. These small molecules can be applied to embryos during discrete blastula stages without disrupting signaling at earlier stages or altering endogenous Nodal levels.
Protocol Implementation:
- Prepare stock solutions of SB-431542 in DMSO at concentrations of 20-100 mM.
- Dilute in embryo medium to working concentrations of 100-800 μM.
- Apply to dechorionated zebrafish embryos at specific developmental stages (e.g., 2.75 hours post-fertilization for mid-blastula transition inhibition).
- Maintain embryos in drug solution through the desired developmental window.
- Fix at appropriate stages for molecular analysis or observe phenotypic consequences at later stages.
Validation and Controls: Treated embryos should be compared with DMSO-only controls and assessed for loss of mesendodermal markers such as no-tail (ntl) and floating head (flh) [7]. Properly inhibited embryos display severe cyclopia and lack somites, notochord, blood, heart, and Kupffer's vesicle.
Genetic Manipulation Techniques
Genetic approaches provide complementary methods for manipulating Nodal signaling:
CRISPR/Cas9 Mutagenesis: Generation of mutant lines for Nodal receptors reveals functional redundancy. While single receptor knockouts often show no obvious patterning defects, combined loss of acvr1b-a and acvr1b-b phenocopies Nodal loss-of-function phenotypes [4].
Morpholino Knockdown: Combinatorial morpholino injection can achieve simultaneous knockdown of multiple receptors. For Type II receptors, this approach reveals partial redundancy and Nodal-independent functions [4].
Visualization Tools: Endogenous Nodal protein can be visualized using mCitrine::Nodal fusion alleles, which maintain normal regulation and function while allowing live imaging of protein distribution [54].
Experimental approaches for Nodal pathway manipulation. Both pharmacological and genetic techniques can be applied at specific developmental stages for temporal control of pathway activity.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents for Nodal Pathway Manipulation
| Reagent/Category | Specific Examples | Function/Application | Key Considerations |
|---|---|---|---|
| Pharmacological Inhibitors | SB-431542, SB-505124 | Conditional inhibition of ALK4/5/7 receptors | Enable temporal control without altering ligand levels |
| Genetic Tools | CRISPR/Cas9 mutants (acvr1b-a, acvr1b-b), Morpholinos | Permanent or transient loss-of-function | Functional redundancy requires combinatorial approaches |
| Visualization Reagents | mCitrine::Nodal fusion, Antibodies (pSmad2/3) | Live imaging of protein distribution and signaling activity | Endogenous tagging maintains normal regulation |
| Lineage Markers | no-tail (ntl), floating head (flh) | Assessment of mesendodermal derivatives | Validate loss-of-function phenotypes |
| Receptor Constructs | CitrineTrap, Wild-type receptors | Manipulation and detection of receptor-ligand interactions | Can influence Nodal distribution and range |
Discussion: Integration of Timing and Dosage in Patterning
The emerging model from recent research suggests that Nodal patterning occurs through the integration of both temporal and dosage information. Rather than simply responding to a static concentration gradient, cells appear to monitor and respond to the cumulative dose of Nodal signals received over time [7]. This explains how adjacent cells in the blastula can adopt different fates despite potentially similar instantaneous Nodal concentrations. The dynamic expression of nodal-related genes and the movement of responding cells during the critical mid-to-late blastula period support this time-integrated model of fate specification [7].
The propagation of Nodal signaling through a relay mechanism rather than simple diffusion further complicates the traditional morphogen gradient model [54]. In this relay, Nodal activity induces transcription of Nodal ligand in adjacent cells, creating a wave of signaling that spreads inward from the margin. The speed and extent of this relay are controlled by the feedback inhibitor Lefty, which functions to control the timing of Nodal spread and therefore the proper sequence of mesoderm differentiation [54]. This mechanism represents a sophisticated system for ensuring robust patterning despite potential variations in initial conditions.
The manipulation of Nodal signaling in zebrafish requires careful consideration of both timing and dosage parameters. The critical window for mesendoderm patterning occurs primarily during the mid-to-late blastula stages (3-5 hours post-fertilization), with different tissues requiring different signaling durations and concentrations for proper specification [7]. The traditional view of Nodal as a long-range morphogen has been refined by evidence demonstrating its short-range distribution and relay-dependent propagation [54]. These findings have important implications for experimental design, suggesting that both pharmacological and genetic approaches must account for the temporal dynamics and functional redundancy within the pathway. Future research will continue to elucidate how the quantitative parameters of Nodal signaling are interpreted at the cellular level to generate the diverse array of mesendodermal derivatives in the developing zebrafish embryo.
Validating Tissue-Specific Phenotypes in Left-Right Asymmetry Models
A crucial step in early embryogenesis is the establishment of spatial patterns of signaling activity that break bilateral symmetry and guide the development of left-right asymmetries [13]. The Nodal signaling pathway serves as a fundamental activator-inhibitor pair that regulates mesendodermal patterning and the establishment of laterality in vertebrate embryos [14]. In zebrafish, this pathway governs the internalization of endodermal precursors and controls the precise spatial organization of developing tissues [13]. Understanding how embryonic cells decode these signals to make appropriate fate decisions requires sophisticated tools to perturb morphogen signals with high resolution in space and time. Recent advances in optogenetics and quantitative phenotyping have provided unprecedented capability to systematically explore how Nodal signaling patterns influence tissue-specific phenotypes in left-right asymmetry models [13] [55].
The robustness of Nodal signaling in mesendodermal patterning represents a classic example of how developmental systems buffer against variability while maintaining precision in left-right patterning [14]. Interestingly, this signaling network resembles "antithetic integral feedback" systems in engineering that achieve robust control of cellular averages, though in spatially extended models, additional regulatory mechanisms are required to maintain robust signaling gradients [14]. The functional but fragile nature of mesendodermal patterning in zebrafish without proper inhibitory feedback underscores the critical importance of validated approaches for analyzing tissue-specific phenotypes in asymmetry research.
Core Principles of Phenotype Validation in Asymmetry Research
Defining Validation Metrics and Data Types
Robust validation of tissue-specific phenotypes requires careful distinction between different types of biological measurements and variation. In quantitative cell biology, researchers must differentiate between discrete data (countable, finite values such as the number of cells in an image) and continuous data (measurements that can take any value within a range, such as fluorescence intensity or cell size) [56]. Additionally, qualitative or categorical data represent distinct groups or categories, such as control versus treated conditions [56].
When analyzing left-right asymmetry, several specific metrics are essential for phenotype validation:
- Directional Asymmetry: When the same side is consistently larger than the other across a population [55]
- Absolute Asymmetry: The absolute difference between left and right measurements within an individual [55]
- Among-Individual Variation: The variance of a trait within a population [55]
- Within-Individual Variation: Deviations from anticipated bilateral symmetry, measured through asymmetry metrics [55]
The Quadratic Relationship Between Phenotype Severity and Variation
Recent research has revealed an unexpected universal principle governing the relationship between phenotype severity and variation. Analysis of a zebrafish mef2ca allelic series representing a range of craniofacial phenotype severity demonstrated that severity and variation are positively correlated, but only to a point, with variation collapsing in the most severe conditions [55].
Mathematically, the best fit for this relationship is a quadratic function, where wild-type conditions produce low variation, moderate severity is associated with high variation, and conditions of extreme severity result in low variation [55]. This principle has been observed across both zebrafish craniofacial phenotypes and human genetic disease, suggesting it represents a fundamental biological phenomenon that must be considered when validating tissue-specific phenotypes in asymmetry models.
Table 1: Relationship Between Phenotype Severity and Variation in Zebrafish Models
| Severity Level | Variation Pattern | Developmental Status | Biological Interpretation |
|---|---|---|---|
| Wild-type | Low variation | Canalized | Developmental buffering maintains consistent outcomes |
| Moderate severity | High variation | Decanalized | Perturbed systems exhibit increased variability |
| Extreme severity | Low variation | Neocanalized | Constrained developmental options limit variation |
Experimental Approaches for Validating Tissue-Specific Phenotypes
Optogenetic Control of Nodal Signaling Patterns
The development of improved optogenetic reagents has enabled precise spatial and temporal control over Nodal signaling activity in live zebrafish embryos. The optoNodal2 system utilizes Nodal receptors fused to the light-sensitive heterodimerizing pair Cry2/CIB1N, with the type II receptor sequestered to the cytosol [13]. This system eliminates dark activity and improves response kinetics without sacrificing dynamic range, allowing researchers to create designer Nodal signaling patterns in vivo.
Experimental Protocol: Optogenetic Patterning of Nodal Signaling
- Sample Preparation: Generate zebrafish embryos expressing optoNodal2 constructs under appropriate tissue-specific promoters
- Light Patterning: Adapt an ultra-widefield microscopy platform for parallel light patterning in up to 36 embryos simultaneously
- Spatial Control: Apply precisely defined illumination patterns to activate Nodal signaling in specific tissue regions
- Phenotype Analysis: Monitor downstream effects on mesendodermal patterning and tissue internalization
- Rescue Experiments: Apply patterned illumination to Nodal signaling mutants to assess rescue of developmental defects
This approach allows researchers to test how specific spatiotemporal patterns of Nodal signaling activity influence the establishment of left-right asymmetries in different tissue contexts [13].
Quantitative Analysis of Asymmetry Phenotypes
Validation of tissue-specific phenotypes requires rigorous quantitative approaches that account for both biological and technical variability. Experiments must be conducted with multiple biological repeats, defined as independent experimental replicates performed on different biological samples, to capture biological variation [56]. These should be distinguished from technical repeats, where measurements are taken from the same sample multiple times to assess instrument consistency [56].
Experimental Protocol: Analyzing Craniofacial Asymmetry in Zebrafish
- Sample Collection: Fix zebrafish larvae at consistent developmental stages (e.g., 5-6 days post-fertilization)
- Tissue Staining: Process samples using Alcian Blue and Alizarin Red staining for cartilage and bone, respectively
- Image Acquisition: Capture standardized high-resolution images of craniofacial structures
- Morphometric Analysis: Measure specific linear elements (e.g., symplectic cartilage length) on both left and right sides
- Statistical Analysis: Calculate absolute asymmetry, directional asymmetry, and among-individual variation
This approach has been successfully used to quantify how mutations in mef2ca affect craniofacial asymmetry and how these phenotypes correlate with severity [55].
Table 2: Key Metrics for Validating Asymmetry Phenotypes
| Metric | Calculation Method | Biological Interpretation | Example Application |
|---|---|---|---|
| Absolute Asymmetry | |Left - Right| measurement | Within-individual developmental variation | Zebrafish craniofacial skeletal elements [55] |
| Directional Asymmetry | Mean (Left - Right) across population | Consistent bias toward one side | Drosophila genitalia rotation [57] |
| Among-Individual Variation | Variance of trait measurements within population | Population-level developmental robustness | Mef2ca allelic series analysis [55] |
| Penetrance | Frequency of phenotype associated with a genotype | Reliability of genotype-phenotype relationship | Mutant phenotype analysis [55] |
Visualization and Data Exploration in Asymmetry Research
Effective Data Visualization Strategies
Data visualization is critical for interpreting complex asymmetry phenotypes and communicating findings effectively. The appropriate visualization strategy depends on the specific research question and data type:
- SuperPlots: Combine dot plots and box plots to display individual data points by biological repeat while capturing overall trends [56]
- Box Plots: Show distribution of numerical data values, particularly useful for comparing asymmetry metrics across conditions [58]
- Scatter Plots: Visualize relationships between two numerical variables, such as left versus right measurements [58]
- Heatmaps: Display magnitude of values using different color intensities across a matrix [58]
These visualization approaches enable researchers to identify patterns, detect outliers, and assess biological variability more effectively than numerical data alone [56].
Experimental Workflow for Phenotype Validation
The following diagram illustrates the complete experimental workflow for validating tissue-specific phenotypes in left-right asymmetry models:
Workflow for Validating Tissue-Specific Phenotypes
Nodal Signaling Pathway in Left-Right Asymmetry
The following diagram illustrates the core Nodal signaling pathway and its role in establishing left-right asymmetry:
Nodal Signaling Pathway in Asymmetry
Research Reagent Solutions for Asymmetry Studies
Table 3: Essential Research Reagents for Validating Tissue-Specific Asymmetry Phenotypes
| Reagent/Tool | Function in Validation | Example Application |
|---|---|---|
| OptoNodal2 System | Light-activated control of Nodal signaling patterns | Spatial patterning of mesendodermal precursors in zebrafish [13] |
| fli1:Gal4 Transgene | Neural crest-specific expression; modulates phenotype severity | Decanalization of craniofacial phenotypes in zebrafish mutants [55] |
| Cry2/CIB1N Heterodimerizing Pair | Light-sensitive protein interaction system | Optogenetic control of receptor localization [13] |
| Ultra-Widefield Microscopy | Parallel light patterning in multiple embryos | Simultaneous optogenetic manipulation of up to 36 samples [13] |
| mef2ca Allelic Series | Range of mutation severity for phenotypic analysis | Quadratic relationship between severity and variation [55] |
Interpretation and Integration of Validation Data
Assessing Biological Variability and Reproducibility
Consistently assessing biological variability is crucial for appropriate interpretation of asymmetry phenotypes. Researchers must clearly document both the number of biological repeats and the number of data points per biological repeat to provide a complete picture of reproducibility [56]. The definition of "n" should be explicitly stated in figure legends and methods sections, as confusion about this terminology can lead to misinterpretation of results [56].
The quadratic relationship between phenotype severity and variation provides an important framework for interpreting validation data [55]. In moderate severity conditions, where variation is highest, researchers should expect less consistent phenotypes and plan for correspondingly larger sample sizes. In extreme severity conditions, where variation collapses, smaller sample sizes may be sufficient, but the biological interpretation differs significantly.
Contextualizing Within the Broader Thesis on Nodal Signaling
Validation of tissue-specific phenotypes in left-right asymmetry models contributes significantly to understanding the fundamental principles of Nodal signaling in zebrafish mesendodermal patterning. The functional but fragile nature of this patterning system [14], combined with the recently discovered quadratic relationship between severity and variation [55], suggests that Nodal signaling operates within constrained developmental parameters that balance robustness and flexibility.
The ability to precisely control Nodal signaling patterns with optogenetics [13] provides a powerful approach to test how specific spatiotemporal signaling dynamics influence the consistency of left-right asymmetry outcomes across individuals. This integrated approach—combining precise perturbation tools with rigorous quantitative phenotyping—will continue to advance our understanding of how Nodal signaling patterns are decoded to produce reliable tissue-specific asymmetries during vertebrate development.
Cross-Species Validation and Zebrafish Model Strengths
The Nodal signaling pathway, a pivotal component of the Transforming Growth Factor-beta (TGF-β) superfamily, functions as a master regulatory system governing embryonic patterning across vertebrate species. This morphogen directs fundamental processes including mesendoderm induction, germ layer patterning, and left-right axis establishment through concentration-dependent signaling gradients. The remarkable conservation of Nodal function between zebrafish and mouse provides a powerful comparative framework for unraveling the core principles of embryonic patterning mechanisms while revealing species-specific adaptations. Within the broader context of zebrafish mesendodermal patterning research, this analysis illuminates how a deeply conserved genetic toolkit can be deployed to orchestrate complex tissue organization, offering insights relevant to developmental biology, evolutionary studies, and regenerative medicine applications.
Core Nodal Signaling Pathway: Mechanism and Conservation
The Nodal signaling cascade initiates when ligands bind to cell surface receptor complexes comprising Type I and Type II activin receptors alongside EGF-CFC family co-receptors (Cripto in mouse, One-eyed pinhead/Oep in zebrafish) [8] [21]. This ligand-receptor interaction triggers phosphorylation of cytoplasmic Smad2/3 proteins, which subsequently form complexes with Smad4 and translocate to the nucleus [8]. Within the nucleus, these complexes collaborate with transcription factors such as FoxH1 to activate expression of target genes, including Nodal ligands themselves (creating positive feedback) and extracellular antagonists like Lefty (establishing negative feedback) [8].
Key Pathway Components and Regulatory Mechanisms
The core pathway is regulated through multiple mechanisms that ensure precise spatiotemporal control of signaling activity:
- Ligand Processing: Convertases including Furin and PACE4 cleave immature Nodal precursors to generate mature, active ligands [8].
- Extracellular Antagonism: Lefty proteins act as competitive inhibitors of Nodal signaling, while DAN family proteins (e.g., Cerberus) directly bind Nodal ligands to prevent receptor activation [8].
- Intracellular Modulation: Proteins like Dapper2 promote lysosomal degradation of active receptors, and phosphatases such as PPM1A dephosphorylate Smad2/3 to terminate signaling [8].
- Transcriptional Control: Negative co-regulators including Tgif1/2 compete with Smad2 for binding sites, while microRNAs (miR-430 in zebrafish, miR-302 in mammals) post-transcriptionally regulate pathway components [8].
Table 1: Core Components of the Nodal Signaling Pathway
| Component Type | Zebrafish | Mouse | Function |
|---|---|---|---|
| Ligands | Squint, Cyclops | Nodal | Secreted signaling molecules forming concentration gradients |
| Co-receptors | One-eyed pinhead (Oep) | Cripto | Essential membrane-bound co-factors for receptor complex assembly |
| Extracellular Antagonists | Lefty1, Lefty2 | Lefty1, Lefty2 | Feedback inhibitors that restrict Nodal spread and activity |
| Intracellular Transducers | Smad2, Smad3, Smad4 | Smad2, Smad3, Smad4 | Phosphorylation-dependent transcription factors |
| Nuclear Transcription Factors | FoxH1 | FoxH1 | DNA-binding partners for activated Smad complexes |
Figure 1: Core Nodal Signaling Pathway. The pathway illustrates ligand binding, receptor activation, Smad-mediated signal transduction, and feedback regulation mechanisms conserved between zebrafish and mouse.
Zebrafish-Mouse Comparative Analysis: Developmental Functions
Mesendodermal Patterning and Germ Layer Specification
In both zebrafish and mouse embryos, Nodal signaling plays an indispensable role in mesendoderm induction, the process through which mesodermal and endodermal tissues are specified from naive embryonic cells. Zebrafish with mutations in both squint and cyclops genes (Nodal homologs) fail to develop notochord, heart, kidneys, or blood, demonstrating the essential nature of this pathway for mesendodermal derivatives [8]. Mouse Nodal mutants similarly exhibit severe defects in mesoderm formation and fail to gastrulate properly [8]. The concentration-dependent response to Nodal gradients follows a conserved pattern across both species: high Nodal signaling promotes endodermal fates, intermediate levels specify mesodermal derivatives, and low levels permit ectodermal differentiation [21].
Recent optogenetic studies in zebrafish have enabled unprecedented precision in manipulating Nodal signaling patterns, revealing how embryonic cells decode morphogen concentrations to make appropriate fate decisions [13]. The development of optoNodal2 reagents—which eliminate dark activity while improving response kinetics without sacrificing dynamic range—has allowed researchers to create designer Nodal signaling patterns in live embryos [13]. When coupled with ultra-widefield microscopy platforms enabling parallel light patterning in up to 36 embryos, this system demonstrates precise spatial control over Nodal signaling activity and downstream gene expression, providing powerful experimental leverage for analyzing mesendodermal patterning events [13].
Left-Right Axis Patterning
The conservation of Nodal function in left-right axis establishment represents a paradigm of evolutionary constraint on developmental mechanisms. In both zebrafish and mouse, Nodal signaling operates within a conserved genetic circuit to break bilateral symmetry and establish consistent left-right asymmetry of internal organs [59].
Table 2: Left-Right Patterning Components in Zebrafish and Mouse
| Component | Zebrafish | Mouse | Conserved Function |
|---|---|---|---|
| Nodal Ligand | Southpaw (Spaw) | Nodal | Major effector of left-right asymmetry |
| Gdf1 Partner | Vg1/Gdf3 | Gdf1 | Forms heterodimers with Nodal ligands |
| Asymmetric Inhibitor | Dand5 | Cerl2/Dand5 | Restricts Nodal activity to the left side |
| Midline Barrier | Lefty1 | Lefty1 | Prevents contralateral spreading of Nodal |
| Transcriptional Effector | Pitx2 | Pitx2 | Downstream mediator of asymmetric morphogenesis |
Genetic evidence confirming this conservation comes from zebrafish null mutants for spaw (the Nodal ortholog), dand5, and lefty1, which phenocopy their mouse counterparts with remarkable fidelity [59]. spaw mutants fail to initiate spaw expression in the lateral plate mesoderm and display absent heart looping, mirroring mouse Nodal mutants [59]. Similarly, dand5 and lefty1 mutants develop bilateral spaw expression, analogous to mouse Cerl2 and Lefty1 mutants [59]. Biochemical analyses further demonstrate that zebrafish Spaw and Vg1 form functional heterodimers, comparable to mouse Nodal-Gdf1 complexes [59].
Recent research has identified additional regulators that fine-tune Nodal signaling during left-right patterning. Myosin1G (Myo1G) in zebrafish promotes Nodal signal propagation by associating with SARA-positive endosomes containing activin receptors, thereby enhancing cellular responsiveness to Nodal ligands [60]. Myo1G mutants exhibit delayed left-sided Nodal propagation and tissue-specific laterality defects, particularly in organs most distant from the left-right organizer [60].
Ectoderm Patterning and Placental Development
Beyond mesendodermal patterning, Nodal signaling demonstrates conserved functions in ectoderm patterning and extraembryonic development. In mouse placental development, Nodal and its co-receptor Cripto-1 play distinct but essential roles in trophoblast specification [61] [62]. Trophoblast-specific deletion of Nodal results in decreased implantation site size and placental thickness, primarily due to a smaller labyrinth area alongside expansion of trophoblast giant cells [61] [62]. Conversely, Cripto-1 deletion leads to a smaller junctional zone with reduced spongiotrophoblast cells and disorganized labyrinth structure [62]. These findings highlight both conserved and specialized functions of Nodal pathway components in extraembryonic development.
Experimental Approaches and Methodologies
Genetic Manipulation Strategies
The comparative analysis of Nodal function relies on sophisticated genetic manipulation techniques tailored to zebrafish and mouse model systems:
Zebrafish Mutant Generation: CRISPR-Cas9 has been successfully employed to generate frameshifting mutants for key Nodal pathway components including spaw and dand5 [59]. Maternal-zygotic (MZ) mutants provide particularly robust phenotypes by eliminating both maternally and zygotically contributed gene products [60]. For example, MZ myo1g mutants reveal tissue-specific left-right asymmetry defects with impaired cardiac jogging and abnormal asymmetric gene expression in the dorsal epithalamus [60].
Mouse Conditional Knockouts: Tissue-specific deletion of Nodal pathway components using Cre-lox technology enables analysis of gene function in particular cell lineages. The trophoblast-specific deletion model employing Tat-Cre recombinant protein induces deletion of floxed Nodal or Cripto-1 genes exclusively in the trophectoderm at the blastocyst stage [61] [62]. Treated embryos are transferred into pseudopregnant mice, with implantation sites examined at gestational days 8.5 and 10.5 for placental morphology and trophoblast population analysis [61].
Optogenetic Control of Nodal Signaling
Recent advances in optogenetics have revolutionized the manipulation of Nodal signaling patterns with high spatiotemporal precision in zebrafish embryos [13]. The experimental workflow involves:
Reagent Design: Nodal receptors are fused to the light-sensitive heterodimerizing pair Cry2/CIB1N, with the type II receptor sequestered to the cytosol in the dark state [13].
Optogenetic Stimulation: The improved optoNodal2 system eliminates dark activity while maintaining dynamic range and improving response kinetics [13].
Parallel Processing: An ultra-widefield microscopy platform enables simultaneous light patterning in up to 36 embryos, allowing high-throughput analysis of Nodal-induced phenotypes [13].
Phenotypic Analysis: Patterned Nodal activation drives precisely controlled internalization of endodermal precursors and can rescue developmental defects in Nodal signaling mutants [13].
This approach enables the creation of synthetic Nodal signaling gradients that can test theoretical models of morphogen function and tissue patterning.
Biochemical and Molecular Analyses
Biochemical assays have been instrumental in characterizing protein interactions within the Nodal pathway. Blastoderm assays in zebrafish demonstrate that Vg1 and Spaw are interdependent for target gene induction and form functional heterodimers, contrary to earlier reports [59]. Endosome analysis reveals that Myo1G associates with SARA-positive activin receptor endosomes, with myo1g mutants showing reduced numbers of these signaling compartments and diminished responsiveness to Nodal ligands [60].
Mathematical modeling of Nodal gradient formation, validated through in vivo experimentation, demonstrates that the EGF-CFC co-receptor Oep regulates ligand spread by setting the rate of capture by target cells [21]. Surprisingly, when Oep replenishment is prevented in zygotic oep mutants, the Nodal signaling gradient transforms into a traveling wave, revealing the dynamic regulation of morphogen distribution [21].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Nodal Signaling Studies
| Reagent/Category | Examples/Specific Instances | Function/Application | Species |
|---|---|---|---|
| Optogenetic Tools | optoNodal2 (Cry2/CIB1N receptor fusions) | Precise spatiotemporal control of Nodal signaling with light | Zebrafish |
| Genetic Mutants | spaw¯, dand5¯, lefty1¯, MZ myo1g¯, oep¯ | Loss-of-function analysis of pathway components | Zebrafish |
| Conditional KO Models | Tat-Cre; Nodalfl/fl, Tat-Cre; Cripto-1fl/fl | Tissue-specific deletion of Nodal genes | Mouse |
| Signaling Reporters | Phospho-Smad2/3 immunostaining, BRE-transgenic lines | Readout of pathway activity and response | Both |
| Biochemical Tools | SARA-endosome markers, heterodimerization assays | Protein interaction and trafficking studies | Both |
| Computational Models | Oep-dependent ligand capture models | Predict gradient formation and dynamics | Both |
The comparative analysis of Nodal signaling in zebrafish and mouse reveals a remarkable conservation of core pathway components and regulatory logic alongside species-specific implementations. From mesendoderm induction to left-right patterning, the Nodal pathway exemplifies how a fundamental developmental toolkit can be adapted to shape diverse embryonic structures while maintaining essential functions. The emerging toolkit for manipulating and monitoring Nodal signaling—including optogenetic controls, conditional mutants, and mathematical models—provides unprecedented resolution for probing the mechanisms of morphogen-mediated patterning. These advances not only illuminate fundamental principles of embryonic development but also offer insights relevant to congenital disorders and regenerative medicine strategies that aim to recapitulate developmental programs for therapeutic purposes.
Receptor Homology and Functional Redundancy Across Vertebrates
The formation of the vertebrate body plan is orchestrated by a limited set of evolutionarily conserved signaling pathways. Among these, the Nodal signaling pathway functions as a master regulator of mesendodermal patterning—the process by which embryos form mesoderm and endoderm, the precursors to tissues including muscle, bone, the heart, and the gut [4] [7]. Nodal, a member of the Transforming Growth Factor-β (TGF-β) superfamily, exerts its effects through a receptor complex comprising Type I and Type II activin receptors (Acvr) and an EGF-CFC family co-receptor [4] [3]. A key feature of this system, observed across multiple vertebrate species, is the presence of multiple receptor paralogs, which often exhibit functional redundancy, wherein the loss of a single gene can be compensated for by its paralog, thereby ensuring developmental robustness.
This guide examines the homology and functional redundancy of Nodal receptors within the context of zebrafish mesendodermal patterning research. We synthesize recent genetic evidence, detail key experimental protocols, and provide resources to equip researchers in dissecting the components of this critical developmental pathway.
Nodal Receptor Homology and Evolution
The core Nodal signaling apparatus is highly conserved across vertebrates. The pathway is triggered when a Nodal ligand, which typically forms a heterodimer with a GDF1/3 protein, binds to a receptor complex at the cell surface [63]. This complex consists of a Type I receptor (Acvr1b or Acvr1c), a Type II receptor (Acvr2a or Acvr2b), and an EGF-CFC co-receptor (such as One-eyed pinhead, Oep, in zebrafish) [4] [3]. Receptor complex formation activates an intracellular cascade leading to the phosphorylation and nuclear translocation of Smad2/3 transcription factors, which regulate target gene expression [4].
Genomic analyses reveal that the gene lineages for Nodal and its receptors can be traced back to the ancestor of jawed vertebrates. Following gene duplication events, lineages such as Nodal/Nodal-related and Acvr1b/Acvr1c underwent processes of differential retention and, in some cases, lineage-specific expansion [63]. This evolutionary history has resulted in a variable number of receptor paralogs in different vertebrate species.
Table: Nodal Receptor Homologs in Vertebrates
| Receptor Type | Human/Mouse | Zebrafish | Functional Role |
|---|---|---|---|
| Type I | Acvr1b (Alk4) | Acvr1b-a, Acvr1b-b | Major mediators of Nodal signaling; redundant in zebrafish [4] |
| Type I | Acvr1c (Alk7) | Acvr1c | Role in early zebrafish development is less prominent [4] |
| Type II | Acvr2a | Acvr2a | Partly redundant function in embryo patterning, partially Nodal-independent [4] |
| Type II | Acvr2b | Acvr2b-a, Acvr2b-b | Partly redundant function; Acvr2b-a shown to bind Nodal ligands strongly [4] |
| Co-receptor | Cripto/Tdgf1 | One-eyed pinhead (Oep) | Essential for ligand binding and signal propagation; regulates ligand distribution [4] [3] |
In zebrafish, homology searches have identified three Type I (acvr1b-a, acvr1b-b, acvr1c) and four Type II (acvr2a, acvr2b-a, acvr2b-b, acvr2c) receptor homologs [4]. Except for acvr1c, the transcripts for these receptors are maternally deposited and present during early embryogenesis, positioning them to mediate the earliest Nodal signaling events [4].
Figure 1: Evolutionary history of Nodal signaling components. The core pathway originated in jawed vertebrates, with gene duplication and lineage-specific evolution leading to the receptor complements found in modern species like zebrafish and human/mouse.
Functional Redundancy in Nodal Receptors
Functional redundancy among receptor paralogs is a defining characteristic of the Nodal pathway, providing robustness to the developmental system. This redundancy is, however, partial and hierarchical, with different receptor classes playing distinct roles.
Genetic Evidence from Loss-of-Function Studies
Initial characterizations of single receptor mutants in zebrafish revealed surprisingly mild phenotypes, in stark contrast to the severe defects observed in Nodal ligand (squint; cyclops) or co-receptor (oep) mutants [4] [5]. This suggested that related paralogs might compensate for individual losses.
- Type I Receptors: Only the combined loss of acvr1b-a and acvr1b-b recapitulates the classic Nodal loss-of-function phenotype—a complete absence of mesendoderm derivatives, cyclopia, and embryonic lethality [4]. This demonstrates that these two receptors act redundantly as the primary Type I mediators for Nodal signaling during germ layer patterning.
- Type II Receptors: Combinatorial loss of Type II receptors (e.g., acvr2a, acvr2b-a, acvr2b-b) also causes severe patterning defects and lethality [4]. Intriguingly, these phenotypes are at least partly independent of Nodal signaling, suggesting that Acvr2 homologs may have additional roles beyond transducing Nodal signals [4].
This genetic redundancy extends to the transcriptional effectors downstream of the receptors. The Nodal-dependent specification of mesendoderm requires the combinatorial activities of transcription factors FoxH1 and Eomesodermin (Eomes) [64]. While FoxH1 is essential for axial mesoderm (notochord) formation, Eomes can specify endoderm and nonaxial mesoderm (e.g., paraxial mesoderm, blood) in the absence of FoxH1 activity [64].
Table: Functional Redundancy in Zebrafish Nodal Pathway Components
| Genetic Manipulation | Phenotype | Interpretation |
|---|---|---|
| Single acvr1b-a or acvr1b-b mutant | No obvious patterning defects | Functional redundancy between Type I paralogs |
| Double acvr1b-a; acvr1b-b mutant | Lacks mesendoderm, cyclopic, lethal | These are the major Nodal Type I receptors |
| Compound Type II receptor mutant | Severe patterning defects, lethal | Partly redundant function; partially Nodal-independent |
| FoxH1 mutant (midway) | Lacks notochord, but retains other mesoderm and endoderm | Redundancy with Eomesodermin |
| FoxH1 + Eomesodermin inhibition | Lacks most mesendoderm, phenocopies Nodal loss | Combinatorial activity specifies mesendoderm |
Core Experimental Protocols for Zebrafish Research
This section details key methodologies used to interrogate Nodal receptor function in zebrafish models.
Combinatorial CRISPR-Cas9 and Morpholino Knockdown
To overcome functional redundancy and assess compound loss-of-function phenotypes, researchers employ combinatorial genetic disruption [4].
Protocol:
- Mutant Generation: Generate stable mutant lines for individual receptor genes (acvr1b-a, acvr1b-b, acvr2a, etc.) using CRISPR-Cas9 genome editing. Identify frameshift mutations in the F0 generation and establish homozygous lines.
- Genetic Crosses: Cross single mutants to create double or triple mutant combinations.
- Morpholino Injection: For genes where stable mutants are not available, or to achieve higher-order combinatorial knockdown, inject gene-specific antisense morpholinos into wild-type or mutant embryos at the 1-4 cell stage. Morpholinos are designed to block mRNA translation or splicing.
- Phenotypic Analysis: Assess embryos for mesendodermal patterning defects using morphological criteria and molecular markers (e.g., ntl, flh for mesoderm; sox32 for endoderm) via in situ hybridization or immunofluorescence.
Key Consideration: The use of CRISPR F0 knockout (KO)—injecting Cas9/gRNAs targeting multiple receptors into wild-type embryos—allows for rapid assessment of compound mutant phenotypes without the need for extensive breeding [4].
Pharmacological Inhibition of Nodal Receptors
Small-molecule inhibitors provide a temporal and dose-dependent method to block Nodal signaling [7].
Protocol:
- Compound Selection: Use SB-431542 or SB-505124, which competitively inhibit the ATP-binding site of the ALK4/5/7 Type I receptors.
- Treatment Window: To specifically block zygotic Nodal signaling without affecting maternal contributions, add the drug (e.g., 800 µM SB-431542) to the embryo medium after the mid-blastula transition (MBT, ~3 hours post-fertilization).
- Phenotypic Analysis: Fix embryos at tailbud or later stages and analyze for defects. Treatment results in a complete loss of mesendoderm derivatives, phenocopying squint;cyclops double mutants [7].
Advantage: This method allows researchers to precisely define the temporal requirements for Nodal signaling, for instance, showing that signaling between the mid- and late-blastula stages is critical for sequential specification of somites, notochord, blood, and endoderm [7].
Quantitative Analysis of Nodal Distribution and Signaling
To understand how receptors influence ligand distribution, quantitative imaging techniques are employed.
Protocol:
- Live Imaging: Generate embryos expressing GFP-tagged Nodal ligands (e.g., Squint-GFP, Cyclops-GFP).
- Receptor Perturbation: Image ligand distribution in wild-type versus receptor/co-receptor mutant backgrounds (e.g., oep mutants).
- Fluorescence Recovery After Photobleaching (FRAP): Use FRAP on defined regions of the embryo to measure the effective diffusivity of the GFP-tagged ligands in different genetic contexts.
- Signal Measurement: Quantify the nuclear levels of phosphorylated Smad2 (pSmad2) via immunofluorescence as a direct readout of pathway activity across the embryo.
Application: This approach revealed that loss of the co-receptor Oep dramatically expands the range of Nodal ligands, and that Type I receptor levels can directly modulate Nodal mobility, providing a mechanism for spatial restriction of the signal [4] [3].
Figure 2: Experimental workflow for analyzing Nodal receptor function. Genetic, pharmacological, and imaging approaches provide complementary insights into receptor redundancy and mechanism.
The Scientist's Toolkit: Key Research Reagents
Table: Essential Reagents for Zebrafish Nodal Signaling Research
| Reagent / Tool | Type | Key Function in Research | Example Use Case |
|---|---|---|---|
| SB-431542 | Small-molecule inhibitor | Inhibits ALK4/5/7 Type I receptors | Temporally controlled blockade of zygotic Nodal signaling [7] |
| CRISPR-Cas9 | Genome editing system | Generates stable loss-of-function mutants | Creating single and compound receptor mutant lines [4] |
| Gene-specific Morpholinos | Antisense oligonucleotides | Transient knockdown of target mRNA | Combinatorial F0 knockdown of redundant receptors [4] |
| sqt-GFP / cyc-GFP | GFP-tagged ligand | Visualizes ligand distribution in live embryos | Quantitative measurement of Nodal range in receptor mutants [3] |
| Anti-pSmad2 Antibody | Antibody | Detects activated (phosphorylated) Smad2 | Readout of Nodal signaling activity via immunofluorescence [4] |
| FoxH1 mutant (midway) | Zebrafish mutant line | Lacks functional FoxH1 transcription factor | Studying redundancy with Eomesodermin in mesendoderm specification [64] |
| MZoep mutant | Zebrafish mutant line | Lacks maternal and zygotic EGF-CFC co-receptor | Studying the role of the co-receptor in ligand capture and distribution [4] [3] [5] |
The Nodal signaling pathway relies on a deeply conserved yet flexible receptor system characterized by extensive homology and functional redundancy across vertebrates. In zebrafish, the redundant functions of Acvr1b-a and Acvr1b-b as the primary Type I receptors, alongside the partially Nodal-independent roles of Type II receptors, ensure the robust patterning of the mesendoderm. This redundancy is evident not only at the receptor level but also among downstream transcription factors like FoxH1 and Eomesodermin.
The experimental frameworks of combinatorial genetics, pharmacological inhibition, and quantitative imaging have been instrumental in unraveling these complex functional relationships. Furthermore, the discovery that receptors and co-receptors themselves act as potent regulators of Nodal ligand distribution adds a critical layer to our understanding of morphogen gradient formation. It is now clear that the receptors are not merely passive signal transducers but active participants in shaping the morphogen landscape that patterns the embryo. Continued research into the homology and redundancy of these components will be vital for fully understanding vertebrate development and the evolutionary history of body plan formation.
Zebrafish as a Model for Human TGF-β Signaling Disorders
The Transforming Growth Factor-β (TGF-β) signaling pathway is a master regulator of crucial cellular processes, and its dysregulation underpins a wide spectrum of human disorders. This technical guide elucidates the established utility of zebrafish (Danio rerio) as a powerful in vivo model for deciphering the complexities of TGF-β signaling pathologies. Framed within the broader context of Nodal signaling research in mesendodermal patterning, we detail how zebrafish provide unique insights into the conserved functions of TGF-β ligands, including Nodal and TGF-β3, in development and disease. The document presents consolidated quantitative data from key studies, detailed experimental methodologies for core investigations, and essential research tools, serving as a comprehensive resource for researchers and drug development professionals in the field.
The TGF-β signaling pathway is an evolutionarily conserved system that governs a vast array of cellular functions, including proliferation, differentiation, migration, and apoptosis. In humans, dysregulation of this pathway is implicated in diverse conditions, from craniofacial disorders and fibrosis to cancer [65] [66] [67]. The zebrafish model has emerged as a preeminent system for modeling these human diseases due to its high genetic and functional homology—approximately 70% of human disease genes have a zebrafish counterpart [68]. Key advantages include rapid external development, optical transparency of embryos, ease of genetic manipulation, and high fecundity, facilitating large-scale studies.
This guide places the role of TGF-β signaling within the well-characterized framework of Nodal signaling in zebrafish mesendodermal patterning. Nodal, a member of the TGF-β superfamily, acts as a pivotal morphogen during early embryogenesis. Recent research has moved beyond deterministic gradient models, demonstrating that sustained Nodal signaling establishes a bipotential progenitor state for mesendoderm. Cell fate decisions from this state are modulated by Fgf signaling, revealing a complex, stochastic process that is buffered at later stages to ensure robust embryonic development [69]. Understanding these fundamental mechanisms provides the foundational context for investigating TGF-β-related pathologies in zebrafish.
Core Signaling Pathways: From Nodal to TGF-β
The TGF-β superfamily ligands, including Nodal and TGF-β3, signal through a canonical pathway involving receptor serine/threonine kinases and Smad transcription factors. The specific components and downstream targets dictate the biological outcome.
The Nodal Signaling Pathway in Mesendodermal Patterning
Nodal ligands (e.g., Ndr1/2 in zebrafish) signal through a receptor complex involving the co-receptor Tdgf1 (Oep). This leads to the phosphorylation of Smad2, which complexes with Smad4 and translocates to the nucleus to regulate target gene expression [69]. This pathway is critical for establishing the mesendodermal progenitor state.
Diagram 1: Nodal signaling establishes a bipotential state for stochastic cell fate decisions.
The TGF-β/Smad Pathway in Disease Modeling
TGF-β ligands (e.g., TGF-β3) signal through TβRII and TβRI receptors, leading to the phosphorylation of Smad2/3. The complex with Smad4 regulates genes responsible for extracellular matrix (ECM) production, fibrosis, and immune responses. Inhibitory Smads (Smad6/7) provide negative feedback. Dysregulation of this pathway is a hallmark of fibrotic disorders and craniofacial malformations [65] [66] [67].
Diagram 2: The core TGF-β/Smad pathway drives fibrotic disease processes.
Quantitative Data from Key Zebrafish Studies
Table 1: Quantitative Findings from tgfb3 Knockdown Studies in Zebrafish [65]
| Phenotypic/ Molecular Readout | Experimental Method | Control Group Result | tgfb3 Knockdown Result | Statistical Significance (P-value) |
|---|---|---|---|---|
| tgfb3 mRNA Expression | qPCR (48 hpf) | Normalized to 1 | Significant reduction | < 0.01 |
| tgfb3 Protein Expression | Western Blot (48 hpf) | Normalized to 1 | Significant reduction | < 0.01 |
| Embryos with Abnormal Morphology | In vivo imaging (5 dpf) | -- | 76.67% exhibited defects | -- |
| Key Morphological Defects | In vivo imaging | Normal development | Reduced tail fin, axial curvature, smaller head/eye, enlarged heart | -- |
| Cartilage Development | Alcian Blue Staining (5 dpf) | Normal cartilage structure | Severe malformations | -- |
| Neural Crest Cell Migration | In situ Hybridization | Normal migration | Reduced expression of migration markers | -- |
| NCSC Apoptosis | Flow Cytometry (Annexin V/PI) | Baseline apoptosis | Increased apoptosis | < 0.05 |
| NCSC Migration | Scratch Assay (24h) | Baseline migration rate | Reduced migration rate | < 0.05 |
| Osteogenic Differentiation | Western Blot (Runx2, OSX, ALP) | Normal protein levels | Reduced expression | Varying degrees |
Table 2: Nodal Signaling Dynamics in Early Zebrafish Embryos [69]
| Parameter | Finding | Experimental Method |
|---|---|---|
| Nodal Signaling Range | Extends ~5 cell tiers from margin | Quantitative imaging of P-Smad2 |
| Temporal Window for Specification | ~2 hours (4-6 hpf) | Pharmacological inhibition & transcriptomics |
| Endoderm Progenitor Specification | Requires sustained Nodal signaling | scRNA-seq & genetic analysis |
| Spatial Distribution of sox32+ cells | "Salt and pepper" pattern within first 2 cell tiers | RNAscope in situ hybridization & cell tracking |
| Role of Fgf/Erk | Inhibits endoderm induction; promotes mesoderm | Pharmacological inhibition of Mek1/2 |
| Role of Dusp4 | Localized suppressor of Erk in marginal cells | Mutant analysis |
| Model Conclusion | Nodal establishes competency for stochastic switching, not deterministic fate assignment | Integrated pharmacology, imaging, and transcriptomics |
Detailed Experimental Protocols
Protocol 1: tgfb3 Knockdown and Phenotypic Analysis in Zebrafish
This protocol outlines the method for investigating the role of TGF-β3 in craniofacial development [65].
1. tgfb3 Knockdown via Morpholino Injection
- Materials: Wild-type TU zebrafish strain, Ctrl MO (5′-CCTCTTACCTCAGTTACAATTTAT-3′), tgfb3-targeting DMOs (e1i1-MO + i1e2-MO).
- Procedure: Microinject one-cell stage embryos with a total of 8 ng of morpholinos. The control group receives 8 ng of Ctrl MO, while the experimental group receives 4 ng of each tgfb3-targeting MO (total 8 ng).
- Validation: At 48 hours post-fertilization (hpf), confirm knockdown efficiency by quantifying tgfb3 mRNA levels using qPCR and protein levels using Western blot with an anti-TGFB3 antibody.
2. Analysis of Cartilage and Bone Formation
- Alcian Blue Staining for Cartilage (5 dpf):
- Fix larvae in 4% Paraformaldehyde (PFA) at 4°C overnight.
- Stain with 0.2% Alcian Blue solution at room temperature for 17 hours.
- Bleach in 6% H₂O₂/1% KOH for 1 hour to remove pigment.
- Clear through graded glycerol/KOH solutions (25% → 50%) and image in glycerol.
- Alizarin Red Staining for Bone (5 dpf):
- Fix larvae in 4% PFA overnight.
- Process using a commercial Zebrafish Bone Staining Kit.
- Stain for 1.5 hours, preserve in storage solution, and photograph.
- Alcian Blue Staining for Cartilage (5 dpf):
3. Gene Expression Analysis
- In Situ Hybridization (ISH): Use digoxigenin-labeled probes for neural crest cell formation, migration, and differentiation markers (e.g., sox32, tbxta, tbx16). After hybridization and washing, develop staining using BM Purple AP Substrate and image specimens in glycerol.
- RNA-Seq: Extract total RNA from 48 hpf zebrafish embryo heads. Sequence on an Illumina NovaSeq 6000 platform. Perform differential expression and Gene Ontology (GO) enrichment analysis using tools like DESeq2 and ClusterProfiler.
Protocol 2: Investigating Nodal-Driven Mesendodermal Patterning
This protocol focuses on analyzing the stochastic cell fate decisions governed by Nodal [69].
1. Temporal Dynamics of Marker Expression
- Materials: Zebrafish embryos, RNAscope probes for sox32 (endoderm), tbxta, and tbx16 (mesoderm).
- Procedure: Collect embryos hourly from 4-8 hpf. Perform multiplexed RNAscope in situ hybridization. Develop an in toto quantitative imaging pipeline to segment all nuclei and quantify staining intensity for all three markers simultaneously.
2. Perturbation of Signaling Pathways
- Pharmacological Inhibition: Treat embryos during the specification window (4-6 hpf) with small-molecule inhibitors.
- Nodal Inhibition: Use TGF-β type I receptor kinase inhibitors (e.g., SB431542).
- Fgf/Erk Inhibition: Use Mek1/2 inhibitors (e.g., U0126) to promote endoderm formation.
- Analysis: Quantify the subsequent changes in the proportion of sox32+ endodermal progenitors using the RNAscope and imaging pipeline described above.
- Pharmacological Inhibition: Treat embryos during the specification window (4-6 hpf) with small-molecule inhibitors.
3. Single-Cell Transcriptomics
- Procedure: Perform single-cell RNA sequencing (scRNA-seq) on cells from the marginal zone of wild-type and signaling-perturbed embryos during the specification window.
- Analysis: Use computational tools to reconstruct developmental trajectories and identify the bipotential progenitor state and the gene regulatory networks underlying the stochastic switch to endoderm or mesoderm fates.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagent Solutions for Zebrafish TGF-β/Nodal Signaling Research
| Reagent / Tool | Function / Application | Example / Specification |
|---|---|---|
| Morpholino Oligos (MOs) | Transient knockdown of specific gene expression. | Gene Tools; e.g., tgfb3 DMOs (e1i1-MO + i1e2-MO) [65]. |
| CRISPR/Cas9 | Generation of stable mutant lines. | Used for targeted gene disruption (e.g., sox32/casanova) [69]. |
| Small Molecule Inhibitors | Acute, temporal inhibition of signaling pathways. | Nodal (SB431542), Fgf/Erk (U0126) [69]. |
| RNAscope Probes | High-sensitivity, multiplexed RNA in situ hybridization. | Quantify mRNA for sox32, tbxta, tbx16 in whole embryos [69]. |
| Antibodies for Western Blot | Protein-level detection and quantification. | Anti-TGFB3, pSmad2, Smad2/3, Smad4, Runx2, OSX, ALP, Col1 [65]. |
| Alcian Blue & Alizarin Red | Histological staining for cartilage and bone, respectively. | Critical for skeletal phenotyping in craniofacial studies [65]. |
| scRNA-seq Platforms | Unbiased profiling of cell states and trajectories. | Illumina NovaSeq; analysis via Seurat, SCANPY, or custom pipelines [69] [70]. |
| Flow Cytometry Reagents | Quantification of cell death and apoptosis. | Annexin V-FITC and Propidium Iodide (PI) staining [65]. |
Zebrafish have unequivocally proven their merit as a bona fide model for human TGF-β signaling disorders [71]. The ability to meticulously dissect the roles of specific ligands, such as TGF-β3 in craniofacial development and Nodal in mesendodermal patterning, provides unparalleled mechanistic insights into human disease etiologies. The integration of traditional embryological techniques with cutting-edge tools like single-cell transcriptomics and computational modeling is pushing the boundaries of our understanding, moving the field from deterministic models towards a more nuanced view involving stochasticity and dynamic regulation.
Future research will likely focus on leveraging these sophisticated zebrafish models for in silico drug discovery and therapeutic screening. Advanced computational approaches, like the deep generative model UNAGI applied to single-cell data from human fibrotic diseases, demonstrate the potential for identifying novel drug candidates [70]. The high-throughput and physiological relevance of zebrafish systems make them an ideal platform for validating such computational predictions, ultimately accelerating the development of targeted therapies for TGF-β-related disorders, from fibrosis to congenital craniofacial anomalies.
Quantitative Validation of Nodal Signaling Gradient Formation
The Nodal signaling pathway functions as a quintessential morphogen gradient to orchestrate mesendodermal patterning in vertebrate embryos. This whitepaper synthesizes current research to provide a technical guide for the quantitative validation of this gradient in zebrafish. We detail methodologies for measuring signaling dynamics, outline core quantitative parameters, and present a structured framework for analyzing how the Nodal gradient translates into distinct cell fates. The content is framed within the broader thesis that Nodal signaling establishes a competency window for stochastic cell fate switching rather than acting as a purely deterministic morphogen, a paradigm shift supported by recent single-cell transcriptomic and quantitative imaging data [69].
In zebrafish, the Nodal-related ligands Squint (Sqt/Ndr1) and Cyclops (Cyc/Ndr2) are secreted from the yolk syncytial layer (YSL) at the embryonic margin beginning at the blastula stage [69] [4]. These ligands, as heterodimers with Vg1, diffuse to form a concentration gradient that patterns the mesoderm and endoderm (mesendoderm) [21] [31]. The classical morphogen model posits that cells adopt fates in a concentration-dependent manner: high Nodal signaling induces endoderm, intermediate levels induce mesoderm, and low or absent signaling permits ectodermal fates [21]. However, recent quantitative studies have revealed a more complex reality. Cells in the most marginal tiers, despite experiencing similar high levels of Nodal signaling, stochastically adopt either endodermal or mesodermal fates in a "salt and pepper" pattern [69]. This observation challenges deterministic models and suggests that the gradient does not directly determine fate, but instead establishes a temporal competency window within which bipotential progenitors can undergo a stochastic switch to an endodermal fate, a process modulated by Fgf/Erk signaling [69]. Quantitative validation of the Nodal gradient is therefore essential not merely to confirm its existence, but to understand the dynamic and probabilistic mechanisms of cell fate decision-making.
Quantitative Foundations of the Gradient
The formation, range, and intensity of the Nodal gradient are controlled by a core signaling system and a set of modulatory interactions. The pathway is initiated when Nodal ligands bind to a cell surface complex comprising Type I (e.g., Acvr1b-a, Acvr1b-b) and Type II (e.g., Acvr2b-a) Activin receptors and the EGF-CFC co-receptor One-eyed pinhead (Oep) [4]. This binding triggers the phosphorylation and nuclear translocation of Smad2/3 transcription factors, which activate target gene expression [4].
Table 1: Core Quantitative Parameters of the Nodal Signaling Gradient in Zebrafish
| Parameter | Quantitative Measurement | Biological/Role | Key Regulator(s) |
|---|---|---|---|
| Signaling Range | ~6-8 cell tiers from the margin at shield stage [21] | Defines the spatial domain of mesendodermal patterning. | Ligand diffusion, Oep co-receptor, Lefty inhibitors [21] |
| Signaling Duration | ~2 hours (from ~4 hpf to ~6 hpf) [69] | Creates a temporal window for fate specification; sustained signaling is required for endoderm. | miR-430 (delays Lefty translation), Lefty1/2 [69] |
| Ligand Mobility | Effective diffusivity lower than its inhibitor, Lefty [4] | Determines the rate and shape of gradient formation. | Receptor/co-receptor binding (e.g., Oep) [21] [4] |
| Nuclear pSmad2 | Gradient of intensity, highest at the margin [69] | Direct readout of pathway activity; correlates with signaling level. | Nodal ligand concentration, receptor complex activity |
| Fate Switch Probability | Modulated by Fgf/Erk signaling; lower Erk favors endodermal switch [69] | Determines the likelihood of a bipotential progenitor becoming endoderm. | Dusp4 (Nodal-induced Erk phosphatase) [69] |
A critical quantitative finding is the role of the Oep co-receptor in shaping the gradient. Contrary to being a simple permissive factor, Oep sets the range of Nodal activity by regulating the rate of ligand capture by target cells. In oep mutants, the Nodal signaling range expands dramatically, forming a nearly uniform signaling distribution across the embryo. Furthermore, the failure to replenish Oep via zygotic expression transforms the steady-state Nodal gradient into a traveling wave [21]. This identifies ligand-receptor binding kinetics as a central quantitative parameter controlling gradient shape.
The following diagram illustrates the core Nodal signaling pathway and its key regulators, which establish the quantitative parameters described above.
Diagram 1: The core Nodal signaling pathway and its key regulatory feedback loops. Positive feedback (dashed line) on ligand expression and negative feedback via Lefty inhibitors shape the gradient's dynamics. Cross-talk with the Fgf/Erk pathway, modulated by the Nodal target Dusp4, provides a mechanism for fate modulation.
Methodologies for Quantitative Validation
Validating the Nodal gradient requires a multi-faceted approach that combines precise perturbation, high-resolution imaging, and single-cell analysis.
Experimental Protocols for Measuring Gradient Dynamics
1. Quantitative Immunofluorescence and Image Analysis
- Objective: To quantify the spatial distribution and intensity of active Nodal signaling.
- Protocol:
- Fixation: Collect zebrafish embryos at desired stages (e.g., 40%-epiboly) and fix in 4% paraformaldehyde.
- Staining: Perform standard immunofluorescence using a primary antibody against phosphorylated Smad2 (pSmad2) and a nuclear marker (e.g., DAPI). Include a negative control (e.g., Nodal inhibitor SB505124-treated embryo) to define background [69].
- Imaging: Acquire high-resolution z-stacks of the entire embryo margin using a confocal microscope. Maintain identical laser power, gain, and exposure settings across all samples.
- Quantification: Use image analysis software (e.g., ImageJ, Amira) to segment all nuclei. Measure the mean pSmad2 intensity in each nucleus. Plot the pSmad2 intensity as a function of distance (in cell tiers) from the YSL margin. This generates a quantitative profile of the Nodal signaling gradient [69].
2. Single-Cell RNA Sequencing (scRNA-seq) for Fate Mapping
- Objective: To correlate Nodal signaling levels with transcriptional outputs and cell fate decisions at single-cell resolution.
- Protocol:
- Dissociation: Dissociate cells from the marginal region of embryos at 6-8 hpf into a single-cell suspension.
- Library Preparation: Use a standard scRNA-seq platform (e.g., 10x Genomics) to prepare sequencing libraries.
- Bioinformatic Analysis: Process data to obtain a gene expression matrix. Perform clustering to identify cell populations (e.g., endoderm, mesoderm). Reconstruct developmental trajectories using pseudotime analysis. Overlay expression of Nodal target genes (e.g., sox32, tbxta) and signaling components onto the clusters to map the relationship between signaling and fate [69].
3. Optogenetic Patterning for Functional Validation
- Objective: To test the sufficiency of specific Nodal signaling patterns to induce fate changes.
- Protocol:
- Reagents: Use transgenic embryos expressing optoNodal2 reagents—Nodal receptors fused to the light-sensitive Cry2/CIB1N system [31].
- Light Patterning: Place embryos in a custom ultra-widefield illumination microscope. Project defined light patterns (e.g., spots, gradients) onto the embryos to activate Nodal signaling with high spatiotemporal precision.
- Validation: Fix embryos after illumination and process for in situ hybridization or immunofluorescence for markers like Sox32 (endoderm) or Tbxta (mesoderm). This directly tests if a synthetic Nodal gradient can instruct normal patterning and internalization movements [31].
The workflow for a comprehensive quantitative analysis integrating these methods is summarized below.
Diagram 2: An integrated experimental workflow for the quantitative validation of the Nodal signaling gradient, combining perturbation, multi-modal data collection, and analysis.
Key Experimental Approaches and Findings
The following table summarizes critical experiments that have provided quantitative insights into Nodal gradient function and how they were conducted.
Table 2: Key Experimental Approaches for Validating the Nodal Gradient
| Experimental Approach | Key Findings | Technical Insight |
|---|---|---|
| Pharmacological Inhibition (e.g., SB505124) [69] | Confirmed Nodal necessity; defined the 2-hour signaling window for endoderm specification. | Temporal inhibition reveals requirements for sustained signaling, not just a threshold level. |
| Quantitative pSmad2 Imaging [69] | Established a nuclear pSmad2 gradient; revealed similar high signaling in adjacent cells that adopt different fates. | Provides a direct, quantifiable readout of pathway activity at single-cell resolution. |
| Single-Cell RNA Sequencing [69] | Identified a bipotential mesendoderm progenitor state; revealed stochastic expression of sox32 upon Nodal induction. | Moves beyond population averages to uncover cell-to-cell heterogeneity and fate biases. |
| Genetic Mutant Analysis (e.g., oep, lefty1/2) [21] | Oep restricts ligand spread; loss of Lefty causes gradient expansion and patterning defects. | Demonstrates that gradient shape is set by a balance between ligand dispersal and inhibition/receptor capture. |
| Optogenetic Patterning [31] | Synthetic Nodal patterns can spatially control target genes and cell internalization during gastrulation. | Enables direct functional testing of the instructive capacity of specific signaling patterns. |
The Scientist's Toolkit: Essential Research Reagents
The following table catalogs crucial reagents and tools for designing experiments aimed at quantifying Nodal signaling.
Table 3: Research Reagent Solutions for Nodal Signaling Studies
| Reagent / Tool | Function / Application | Example Use Case |
|---|---|---|
| SB505124 (Small molecule inhibitor) | Inhibits the kinase activity of the Type I Nodal receptor Acvr1b. | Defining the temporal requirements of Nodal signaling via timed application [69]. |
| Anti-pSmad2 Antibody | Recognizes the active, phosphorylated form of Smad2. | Quantitative imaging of the Nodal signaling gradient by immunofluorescence [69]. |
| optoNodal2 System (Cry2/CIB1N-fused receptors) | Light-activatable Nodal receptor system for high spatiotemporal control of signaling. | Generating synthetic Nodal signaling patterns to test fate instruction and scaling [31]. |
| RNAscope Multiplex Assay | High-sensitivity in situ hybridization for detecting mRNA in whole embryos. | Quantifying the spatial expression of Nodal target genes (e.g., sox32, tbxta) with single-cell resolution [69]. |
| oep Mutant Alleles (e.g., oep^m134 ) | Loss-of-function mutations in the essential EGF-CFC co-receptor. | Studying how ligand-receptor binding kinetics shapes gradient range and shape [21]. |
| Acvr1b-a / Acvr1b-b Double Mutants | Compound mutants lacking the major Type I Nodal receptors. | Achieving a near-complete loss of Nodal signaling to study phenotypic consequences [4]. |
The quantitative validation of the Nodal signaling gradient has evolved from simply confirming its existence to understanding the dynamic and probabilistic principles that govern its function. The integration of quantitative imaging, single-cell transcriptomics, and precise optogenetic perturbation has revealed that the gradient establishes a bipotential progenitor state and a temporal competency window, with fate decisions being refined by interactions with pathways like Fgf/Erk [69]. The role of co-receptors like Oep in shaping the gradient through ligand capture highlights the critical importance of receptor-ligand kinetics [21]. Future research, empowered by the toolkit and methodologies outlined herein, will continue to dissect how this robust and adaptable patterning system translates a quantitative chemical signal into the precise arrangement of tissues in the developing embryo.
The establishment of left-right (LR) asymmetry is a fundamental process in embryonic development, governed by a complex interplay of signaling pathways and cytoskeletal regulators. This whitepaper synthesizes current research demonstrating how Myosin1D (Myo1D) and Myosin1G (Myo1G)—two unconventional type I myosins—function as essential, yet mechanistically distinct, regulators of the Nodal signaling pathway in zebrafish. While Myo1D governs the initial symmetry-breaking event at the Left-Right Organizer (LRO), Myo1G potentiates Nodal signal propagation through endosomal trafficking. The evolutionary conservation of these myosins, from Drosophila to zebrafish, highlights their fundamental role in patterning the mesendoderm and other lateralized structures. This review provides a comprehensive analysis of their mechanisms, presents key quantitative data, and offers detailed methodologies and reagents to facilitate further research and therapeutic targeting of this critical pathway.
The TGF-β-related Nodal signaling pathway is a master regulator of vertebrate embryonic patterning. It is indispensable for the induction and patterning of the mesoderm and endoderm (collectively, the mesendoderm) during gastrulation, and later, for the establishment of LR asymmetry [15] [7]. In zebrafish, the Nodal-related ligands Cyclops (Cyc/Ndr2) and Squint (Sqt/Ndr1) act in a dosage-dependent manner to specify all derivatives of the mesoderm and endoderm [72] [7]. Loss of both ligands results in the complete absence of these germ layers [72].
Nodal signaling is mediated by a receptor complex comprising type I (ALK4/5/7) and type II (ActRIIB) serine-threonine kinase receptors, along with the EGF-CFC co-receptor Cripto/Oep [15] [7]. Ligand binding leads to the phosphorylation and nuclear translocation of SMAD2/3 transcription factors, which activate target genes such as pitx2 and lefty [15]. A key feature of the pathway is its auto-regulatory positive feedback loop, enabling the propagation of the Nodal signal across tissues [60]. A critical aspect of this signaling occurs within a specialized population of SARA-positive endosomes, which promote signal transduction by concentrating receptors with their downstream SMAD effectors [60].
Myosin1D: A Conserved Regulator of Symmetry Breaking
Evolutionary Conservation and Genetic Interaction
Myo1D has emerged as an evolutionarily conserved regulator of LR asymmetry from insects to vertebrates [73]. In Drosophila, Myo1D controls the dextral rotation of genitalia and the looping of visceral organs [73]. Strikingly, zebrafish myo1d mRNA can fully rescue the LR defects in Drosophila myo1d mutants, demonstrating deep functional conservation despite the vast phylogenetic distance [73]. The zebrafish genome encodes a paralog, myo1g, and an antagonist, myo1Cb, forming a conserved genetic module for laterality control [73].
Mechanism of Action in the Left-Right Organizer (LRO)
Myo1D's primary role in zebrafish LR asymmetry is to ensure the formation of a functional Left-Right Organizer (LRO), known as Kupffer's Vesicle (KV) [73]. The KV is a transient, ciliated organ where a directional, counter-clockwise fluid flow breaks embryonic symmetry. Myo1D is essential for this process through two key, interrelated functions:
- Cilia Orientation: Myo1D interacts functionally with the Planar Cell Polarity (PCP) component Vangl2 to control the spatial orientation of motile cilia within the KV. This polarized orientation is prerequisite for generating a unified, directional fluid flow [73].
- LRO Morphogenesis: myo1d mutants exhibit a reduction in KV size and the total number of cilia, further impairing flow generation [73].
The dysfunctional LRO flow in myo1d mutants leads to a failure in establishing asymmetric gene expression, including the lateralized degradation of dand5 and the left-sided expression of southpaw (spaw) and pitx2 [73]. Consequently, maternal-zygotic (MZ) myo1d mutants display randomized laterality of the heart, viscera, and brain [73].
Table 1: Phenotypic Consequences of myo1d and myo1g Mutations in Zebrafish
| Gene | LRO Flow | Cardiac Jogging | Brain Laterality | Visceral Laterality | Primary Role |
|---|---|---|---|---|---|
| Myo1D | Severely disrupted [73] | Randomized [73] | Randomized [73] | Randomized [73] | LRO morphogenesis and cilia orientation [73] |
| Myo1G | Normal [60] | Mild defects; enhanced in myo1d;myo1g double mutants [60] | Significant defects (e.g., asymmetric pitx2 expression) [60] | Normal [60] | Potentiating Nodal signal transduction and propagation [60] |
Myosin1G: A Novel Potentiator of Nodal Signal Transduction
In contrast to Myo1D, Myo1G regulates LR asymmetry through a mechanism that is entirely independent of the LRO flow [60]. Instead, Myo1G acts as a context-dependent positive regulator of the Nodal signaling pathway itself, essential for the transfer of laterality information from the LRO to target tissues.
Cellular Mechanism: Association with SARA-Positive Endosomes
Myo1G protein is associated with endosomes positive for SARA (Smad Anchor for Receptor Activation) [60]. These endosomes are known platforms for efficient TGF-β/Nodal signal transduction [60] [15]. In myo1g mutants, the number of SARA-positive Activin receptor endosomes is reduced, leading to a diminished cellular responsiveness to Nodal ligands [60]. This results in a delayed propagation of the left-sided Nodal cascade and tissue-specific laterality defects, particularly in organs most distant from the LRO, such as the brain [60].
Phenotypic Manifestations of myo1g Loss-of-Function
The phenotype of MZ myo1g mutants is distinct from that of MZ myo1d:
- Brain Laterality: The most pronounced defects are seen in the dorsal epithalamus, where asymmetric expression of cyc, lefty1, and pitx2 is frequently lost or randomized [60].
- Cardiac Jogging: Mutants exhibit mild defects in the leftward jogging of the heart, a phenotype that is significantly enhanced in MZ myo1d; MZ myo1g double mutants, indicating genetic interaction [60].
- Visceral Laterality: The liver, pancreas, and gut laterality remains normal, demonstrating the tissue-specific nature of Myo1G function [60].
Table 2: Genetic Interactions between Myosin1 and Nodal Signaling Components
| Genetic Condition | Key Phenotypic Outcome | Functional Interpretation |
|---|---|---|
| myo1g single mutant | Tissue-specific laterality defects (brain), delayed Nodal propagation [60] | Compromised Nodal signal transduction in specific tissues. |
| myo1d; myo1g double mutant | Enhanced cardiac jogging defects compared to either single mutant [60] | Myo1D and Myo1G have distinct, synergistic roles in LR patterning. |
| dnaaf1; myo1g double mutant | Loss of asymmetric pitx2 expression in most embryos [60] | Myo1G acts independently of the ciliary LRO flow; its function is critical when flow is absent. |
| Myo1Cb overexpression | Causes cardiac laterality defects, mimicking myo1d loss-of-function [73] | Myo1Cb acts as a biochemical antagonist of the Myo1D/Myo1G pathway. |
Experimental Protocols for Key Findings
Protocol 1: Analyzing Nodal Signaling Propagation via In Situ Hybridization
This protocol is used to assess the spatial and temporal dynamics of Nodal target gene expression, as shown in studies of myo1g mutants [60].
- Embryo Collection: Collect wild-type and mutant zebrafish embryos at desired developmental stages (e.g., 8-somite to 22-somite stages).
- Fixation: Fix embryos in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) overnight at 4°C.
- Hybridization: Depigment embryos if necessary. Permeabilize embryos with proteinase K. Hybridize with digoxigenin (DIG)-labeled RNA antisense probes for Nodal pathway genes (e.g., dand5, southpaw, lefty2, pitx2).
- Detection: Incubate with anti-DIG antibody conjugated to alkaline phosphatase. Develop color reaction using NBT/BCIP substrate.
- Analysis: Score embryos for symmetric, asymmetric, or absent expression patterns under a dissecting microscope. Statistical analysis of phenotypic penetrance is required.
Protocol 2: Pharmacological Inhibition of Nodal Signaling
This method allows for the temporal control of Nodal pathway activity, crucial for defining critical signaling windows [7].
- Solution Preparation: Prepare stock solutions of small-molecule ALK4/5/7 inhibitors (e.g., SB-431542 or SB-505124) in DMSO.
- Embryo Treatment: At specified stages post-fertilization (e.g., mid-blastula transition), dechorionate embryos and transfer to embryo medium containing the inhibitor (e.g., 800 μM SB-431542). Use DMSO-only treatment as a control.
- Phenotypic Assessment: Culture embryos until desired stages (e.g., 24 hours post-fertilization). Analyze for mesendodermal and LR defects, such as cyclopia, absent somites, notochord, and heart, via microscopy or in situ hybridization.
Protocol 3: Functional Analysis via mRNA Rescue
This genetic rescue experiment confirms the specific requirement for a gene of interest, as performed for myo1d [73].
- Template and RNA Synthesis: Clone the wild-type coding sequence of the gene (e.g., myo1d) into an expression vector. Transcribe capped mRNA in vitro.
- Microinjection: Inject a low dose (e.g., 50-100 pg) of synthesized mRNA into one-cell stage embryos derived from mutant females (for maternal-zygotic mutants).
- Scoring: Raise injected embryos and score for the rescue of the mutant phenotype (e.g., restoration of normal heart jogging and pitx2 expression) compared to uninjected mutants and wild-type controls.
The Scientist's Toolkit: Key Research Reagents
Table 3: Essential Reagents for Studying Myosin and Nodal Signaling
| Reagent / Tool | Type | Key Function / Application | Example Use Case |
|---|---|---|---|
| SB-431542 | Small molecule inhibitor | Selective ATP-competitive inhibitor of ALK4, ALK5, and ALK7 receptors [7]. | Temporally controlled inhibition of zygotic Nodal signaling to define critical windows [7]. |
| SU5402 | Small molecule inhibitor | Pan-inhibitor of FGFR tyrosine kinases [74]. | Studying interaction between FGF and Nodal pathways during cardiac jogging [74]. |
| Tg(hsp70l:dnfgfr1-EGFP) | Transgenic line | Heat-shock inducible dominant-negative FGF receptor [74]. | Spatially and temporally controlled inhibition of FGF signaling. |
| Anti-DIG-AP Antibody | Antibody | Immunological detection of DIG-labeled RNA probes. | Visualizing gene expression patterns via whole-mount in situ hybridization [60] [73]. |
| Maternal-Zygotic (MZ) Mutants | Genetic model | Lack both maternally deposited and zygotically transcribed gene product. | Revealing the full phenotypic severity for genes with maternal contribution (e.g., MZ myo1d) [60] [73]. |
| SARA-GFP Fusion Construct | Molecular construct | Labels endosomes that are platforms for Nodal signal transduction [60]. | Visualizing and quantifying SARA-positive endosomes in live or fixed cells. |
Visualizing the Myosin-Nodal Regulatory Network
The following diagram illustrates the distinct mechanistic roles of Myo1D and Myo1G in regulating Nodal signaling and left-right asymmetry.
Myosin Regulation of Nodal Signaling Pathway
The delineation of Myo1D and Myo1G functions reveals a sophisticated, multi-tiered system for regulating Nodal signaling during zebrafish development. Myo1D acts at the apex of the pathway to break symmetry via the LRO, while Myo1G operates downstream to amplify and ensure the fidelity of the Nodal signal during its propagation. Their synergistic relationship, combined with the antagonistic activity of Myo1Cb, forms a robust regulatory network controlling LR patterning.
Future research should focus on:
- Structural Analysis: Determining the atomic structures of these myosins to understand their specific cargo-binding and actin-interaction properties.
- Human Disease Modeling: Investigating whether mutations in MYO1D or MYO1G are linked to human laterality disorders or congenital heart defects.
- Therapeutic Screening: Exploiting the knowledge of this pathway to develop high-throughput screens for small molecules that can modulate Nodal signaling, with potential applications in regenerative medicine and cancer biology, given Nodal's role in tumorigenesis [60].
This mechanistic understanding, grounded in evolutionary conservation, provides a powerful framework for dissecting the complex interplay between the cytoskeleton and key developmental signaling pathways.
Conclusion
Zebrafish research has established Nodal signaling as a master regulator of mesendodermal patterning, operating through conserved ligand-receptor interactions, sophisticated feedback mechanisms, and maternal-zygotic coordination. The development of advanced genetic tools, live imaging techniques, and computational models has transformed our understanding of how Nodal gradients form and function. Future directions include exploiting zebrafish for high-throughput screening of Nodal pathway modulators with therapeutic potential, engineering synthetic Nodal signaling circuits for tissue engineering, and elucidating how pathway dysregulation contributes to congenital disorders and cancer. The zebrafish model continues to offer unparalleled opportunities for visualizing and manipulating fundamental developmental signaling pathways with direct relevance to human health and disease.
References
- 1. Environmental and Genetic Modifiers of squint Penetrance ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC1994576/]
- 2. Nodal signalling in embryogenesis and tumourigenesis [https://www.sciencedirect.com/science/article/abs/pii/S1357272512004177]
- 3. The pattern of nodal morphogen signaling is shaped by co ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8266389/]
- 4. Regulation of Nodal signaling propagation by receptor ... [https://elifesciences.org/articles/66397]
- 5. Nodal signaling is required for closure of the anterior neural ... [https://bmcdevbiol.biomedcentral.com/articles/10.1186/1471-213X-7-126]
- 6. Gdf3 is required for robust Nodal signaling during germ ... [https://elifesciences.org/articles/28635]
- 7. Time-dependent patterning of the mesoderm and endoderm ... [https://bmcdevbiol.biomedcentral.com/articles/10.1186/1471-213X-7-22]
- 8. Nodal signaling pathway [https://en.wikipedia.org/wiki/Nodal_signaling_pathway]
- 9. Nodal Signaling - an overview [https://www.sciencedirect.com/topics/neuroscience/nodal-signaling]
- 10. Maternal Factors and Nodal Autoregulation Orchestrate ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9178097/]
- 11. Differential regulation of epiboly initiation and progression by ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3259739/]
- 12. Eomesodermin Is a Localized Maternal Determinant ... [https://www.sciencedirect.com/science/article/pii/S1534580705003047]
- 13. Optogenetic control of Nodal signaling patterns [https://pubmed.ncbi.nlm.nih.gov/40145591/]
- 14. Understanding the robustness of Nodal signalling in ... [https://archive.aps.org/smt/2025/mar-g71/10/]
- 15. Nodal Signaling - an overview [https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/nodal-signaling]
- 16. Nodal patterning without Lefty inhibitory feedback is functional ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5720593/]
- 17. Lefty Proteins Are Long-Range Inhibitors of Squint ... [https://www.sciencedirect.com/science/article/pii/S0960982202013623]
- 18. Nodal Signaling and Congenital Heart Defects - NCBI - NIH [https://www.ncbi.nlm.nih.gov/books/NBK500272/]
- 19. Two Modes by which Lefty Proteins Inhibit Nodal Signaling [https://www.sciencedirect.com/science/article/pii/S0960982204001307]
- 20. Single-molecule tracking of Nodal and Lefty in live ... [https://www.nature.com/articles/s41467-022-33704-z]
- 21. The pattern of nodal morphogen signaling is shaped by co- ... [https://elifesciences.org/articles/54894]
- 22. Glycolytic activity instructs germ layer proportions through ... [https://www.sciencedirect.com/science/article/pii/S1934590925001018]
- 23. DNA-guided transcription factor interactions extend human ... [https://www.nature.com/articles/s41586-025-08844-z]
- 24. Enhancer-driven gene regulatory networks reveal ... - PubMed [https://pubmed.ncbi.nlm.nih.gov/40425012/]
- 25. Nodal signaling regulates asymmetric cellular behaviors ... [https://www.nature.com/articles/s42003-022-03826-7]
- 26. Nodal signaling has dual roles in fate specification and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6141772/]
- 27. Temporal dynamics of BMP/Nodal ratio drive tissue-specific ... [https://pubmed.ncbi.nlm.nih.gov/38370754/]
- 28. Foxd3 is an Essential Nodal-Dependent Regulator ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2866760/]
- 29. Visualization analysis of CRISPR/Cas9 gene editing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5064173/]
- 30. Deletion of morpholino binding sites (DeMOBS) to assess ... [https://www.nature.com/articles/s41598-020-71708-1]
- 31. Optogenetic control of Nodal signaling patterns - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12070070/]
- 32. A simple and efficient method to visualize and quantify the ... [https://www.nature.com/articles/srep35488]
- 33. Nodal is a short-range morphogen with activity that ... [https://www.nature.com/articles/s41467-022-28149-3]
- 34. Time-dependent patterning of the mesoderm and endoderm ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC1851950/]
- 35. Nodal Signaling Activates Differentiation Genes During ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC1885460/]
- 36. Activation and roles of ALK4/ALK7-mediated maternal ... [https://www.sciencedirect.com/science/article/abs/pii/S0006291X06009764]
- 37. The Nodal signaling pathway controls left-right asymmetric ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4423147/]
- 38. Time-dependent patterning of the mesoderm and ... [https://pubmed.ncbi.nlm.nih.gov/17391517/]
- 39. Maternal Factors and Nodal Autoregulation Orchestrate ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2022.887987/full]
- 40. Regulatory factor identification for nodal genes in zebrafish ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2022.1047363/full]
- 41. Nodal signaling activates differentiation genes ... - PubMed - NIH [https://pubmed.ncbi.nlm.nih.gov/17306247/]
- 42. Transcriptome Analysis of Zebrafish Embryogenesis Using ... [https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.0010029]
- 43. Nodal signaling activates differentiation genes during ... [https://www.sciencedirect.com/science/article/pii/S0012160607000292]
- 44. Nodal signaling is required for mesodermal and ventral but ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4571091/]
- 45. Robust axis elongation by Nodal-dependent restriction of ... [https://pubmed.ncbi.nlm.nih.gov/38372390/]
- 46. Synthetic mammalian pattern formation driven by ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6303393/]
- 47. Synthetic organizer cells guide development via spatial ... [https://www.sciencedirect.com/science/article/abs/pii/S0092867424013230]
- 48. Regulation of Nodal signaling propagation by receptor ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9612913/]
- 49. The role of the zebrafish nodal-related genes squint ... - PubMed [https://pubmed.ncbi.nlm.nih.gov/12642489/]
- 50. Extracellular interactions and ligand degradation shape the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4887204/]
- 51. Nodal patterning without Lefty inhibitory feedback is ... [https://elifesciences.org/articles/28785]
- 52. Nodal signaling promotes the speed and directional ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2632806/]
- 53. Vg1-Nodal heterodimers are the endogenous inducers of ... [https://elifesciences.org/articles/28183]
- 54. Nodal is a short-range morphogen with activity that spreads ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8789905/]
- 55. A quadratic paradigm describes the relationship between ... [https://www.nature.com/articles/s41467-025-63316-2]
- 56. Practical considerations for data exploration in quantitative ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12045597/]
- 57. Evolution of a novel left-right asymmetry in organ size by co ... [https://pubmed.ncbi.nlm.nih.gov/40600793/]
- 58. Effective Data Visualization in Life Sciences: Tools, charts ... [https://lifesciences.enago.com/blogs/effective-data-visualization-in-life-sciences-tools-charts-and-best-practices]
- 59. Conserved regulation of Nodal-mediated left-right ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6307886/]
- 60. Myosin1G promotes Nodal signaling to control zebrafish ... [https://www.nature.com/articles/s41467-024-50868-y]
- 61. Nodal and cripto-1: distinct functions regulate trophoblast ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1608976/full]
- 62. Nodal and cripto-1: distinct functions regulate trophoblast ... [https://pubmed.ncbi.nlm.nih.gov/40458123/]
- 63. Evolution of nodal and nodal-related genes and the ... [https://pubmed.ncbi.nlm.nih.gov/31210006/]
- 64. Nodal-Dependent Mesendoderm Specification Requires the ... [https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1002072]
- 65. Role of TGF-β3 in Regulating Neural Crest Cell Fate and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12410403/]
- 66. The Function of the TGFβ Signaling Pathway in Connective ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12560638/]
- 67. Role of the TGF‑β/Smad signaling pathway in the transition ... [https://www.spandidos-publications.com/10.3892/ijmm.2025.5603]
- 68. Zebrafish as a model for human epithelial pathology [https://labanimres.biomedcentral.com/articles/10.1186/s42826-025-00238-6]
- 69. Nodal signaling establishes a competency window for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7615190/]
- 70. A deep generative model for deciphering cellular dynamics ... [https://www.nature.com/articles/s41551-025-01423-7]
- 71. Editorial: Zebrafish: bona fide pathophysiological models of ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1624614/full]
- 72. Nodal-related signals establish mesendodermal fate and trunk ... [https://pubmed.ncbi.nlm.nih.gov/10801442/]
- 73. Myosin1D is an evolutionarily conserved regulator of ... [https://www.nature.com/articles/s41467-018-04284-8]
- 74. Synergistic and independent roles for Nodal and FGF in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12539208/]