RNAscope vs. IHC: A Comprehensive Analysis of Sensitivity, Specificity, and Clinical Utility
This article provides a critical comparison between RNAscope in situ hybridization and immunohistochemistry (IHC) for researchers and drug development professionals.
RNAscope vs. IHC: A Comprehensive Analysis of Sensitivity, Specificity, and Clinical Utility
Abstract
This article provides a critical comparison between RNAscope in situ hybridization and immunohistochemistry (IHC) for researchers and drug development professionals. It explores the foundational principles of both technologies, detailing RNAscope's novel double Z-probe design for single-molecule RNA detection and IHC's established protein targeting. The content covers methodological workflows, best practices for assay optimization, and troubleshooting common challenges. A systematic review of validation studies compares the sensitivity and specificity of both techniques across various biomarkers and cancer types, highlighting their complementary roles in clinical diagnostics and biomedical research.
Understanding the Core Technologies: From Protein Detection to RNA Visualization
Immunohistochemistry (IHC) is a foundational technique in pathology and research that utilizes antibody-antigen interactions to visualize the distribution and localization of specific proteins within tissue sections. For decades, IHC has served as the gold standard for identifying cell types, characterizing biological processes, and diagnosing diseases based on protein expression patterns. The technique provides critical spatial context that molecular methods like PCR cannot offer, making it indispensable for understanding cellular environments in complex tissues. This guide explores the established principles of IHC and objectively compares its performance with emerging RNA in situ hybridization techniques, particularly RNAscope, through experimental data and validated methodologies.
Core Principles and Methodological Framework
IHC operates on the principle of specific antibody binding to target antigens within tissue sections, typically formalin-fixed paraffin-embedded (FFPE) samples. The process involves multiple steps: tissue preparation, antigen retrieval, blocking, primary antibody incubation, signal amplification and detection, and counterstaining. The resulting chromogenic or fluorescent signals allow pathologists to identify protein presence, abundance, and cellular localization.
A key strength of IHC lies in its direct detection of functional gene products—proteins—rather than intermediary RNA molecules. This provides more direct insight into biological function, as transcript levels do not always correlate with functional protein expression due to post-transcriptional regulation and protein turnover. The established nature of IHC means extensive validation data exists for numerous biomarkers across various tissue types, creating a robust diagnostic framework.
Table 1: Essential Research Reagents for IHC Workflows
| Reagent Category | Specific Examples | Primary Function |
|---|---|---|
| Primary Antibodies | INSM1 (SP493), UPK2 (BC21), Synaptophysin (SP11), Chromogranin A (LK2H10) | Specifically binds to target protein antigen |
| Detection Systems | OptiView DAB IHC Detection Kit, ultraView Universal DAB Detection Kit | Amplifies signal and enables visualization |
| Counterstains | Hematoxylin II, Bluing Reagent | Provides contrasting nuclear or cellular staining |
| Control Tissues | Tissue microarrays (TMAs) with known expression patterns | Validates assay performance and specificity |
RNAscope as an Emerging Alternative
RNAscope represents a significant advancement in RNA in situ hybridization (ISH) technology. Unlike traditional ISH methods, RNAscope employs a proprietary double Z probe design that enables single-molecule visualization while preserving tissue morphology. This technology provides exceptional sensitivity and specificity for detecting RNA transcripts within their native cellular context [1].
The fundamental distinction between IHC and RNAscope lies in their detection targets: IHC identifies proteins, while RNAscope detects RNA transcripts. This difference has important implications for assay development, validation, and interpretation. RNAscope's probe design requires two adjacent Z probes to hybridize to the target RNA before signal amplification can occur, dramatically reducing background noise and false positives [2]. Each detected dot represents an individual RNA molecule, enabling semi-quantitative analysis directly in tissue sections.
Head-to-Head Performance Comparison
Recent studies have directly compared IHC and RNAscope for detecting various biomarkers across different cancer types. The results demonstrate a complex relationship between these techniques, with performance varying by specific target and tissue context.
In urothelial carcinoma (UC), UPK2 detection showed moderately higher sensitivity with RNAscope (68.0%) compared to IHC (62.6%), though this difference did not reach statistical significance (P = 0.141) [3] [4]. The technologies demonstrated a moderate positive correlation (P < 0.001, R = 0.441), suggesting complementary rather than redundant information. For variant bladder urothelial carcinomas, RNAscope showed a promising trend toward higher detection rates (53.3% vs. 35.6% for IHC, P = 0.057) [4].
A systematic review of 27 studies found that RNAscope has high concordance with PCR-based methods (81.8-100%) but variable concordance with IHC (58.7-95.3%) [1]. This discrepancy reflects the biological reality that mRNA and protein levels don't always correlate due to post-transcriptional regulation, highlighting the complementary nature of these techniques.
Table 2: Quantitative Comparison of IHC vs. RNAscope Performance
| Performance Metric | IHC | RNAscope | Comparative Evidence |
|---|---|---|---|
| Sensitivity for UPK2 in UC | 62.6% | 68.0% | P = 0.141, not statistically significant [3] |
| Sensitivity for UPK2 in Variant UC | 35.6% | 53.3% | Trend toward improvement (P = 0.057) [4] |
| Specificity for UPK2 | Excellent | Excellent | Both maintain high specificity for urothelial tissues [3] |
| Concordance with PCR | N/A | 81.8-100% | High agreement with quantitative methods [1] |
| Detection Target | Proteins | RNA transcripts | Fundamental technological difference [1] |
Experimental Protocols and Workflows
Standard IHC Protocol for Biomarker Detection
The following protocol represents a standardized approach for IHC analysis, as used in comparative studies with RNAscope [3] [4]:
- Tissue Preparation: Cut 3-μm sections from FFPE tissue microarrays (TMAs) and mount on slides.
- Deparaffinization and Rehydration: Process slides through xylene and graded ethanol series.
- Antigen Retrieval: Use automated staining system (e.g., BenchMark ULTRA) with appropriate retrieval solutions.
- Primary Antibody Incubation: Apply target-specific antibodies (e.g., UPK2 at 1:100 dilution; INSM1 at 5 μg/ml) under optimized conditions [3] [5].
- Detection: Employ enzyme-conjugated secondary antibodies with chromogenic substrates (DAB).
- Counterstaining and Analysis: Use hematoxylin for nuclear staining, then evaluate by pathologists.
RNAscope Experimental Workflow
The standardized RNAscope protocol for comparative studies includes [3] [4]:
- Slide Preparation: Deparaffinize TMA sections and sequentially subject to pretreatments.
- Probe Hybridization: Incubate with target-specific probes (e.g., UPK2, NM_006760.4) in HybEZ oven at 40°C for 2 hours.
- Signal Amplification: Use RNAscope 2.0 HD Reagent Kit for signal development.
- Analysis: Score as positive if cytoplasmic staining present in target cells; each dot represents an individual RNA molecule.
Integrated Applications and Complementary Use
The most powerful applications emerge when IHC and RNAscope are used complementarily rather than competitively. Combined protocols enable simultaneous detection of protein and RNA in the same tissue section, providing comprehensive molecular profiling [2] [6].
In neuroscience research, combining RNAscope with IHC has enabled cell-type-specific quantification of inflammatory gene expression. This approach identified that increased IL-1β and NLRP3 mRNA in spinal cord after nerve injury occurs primarily in microglia rather than neurons [2]. Such precise cellular localization of gene expression changes would be impossible with either technique alone.
For diagnostic applications, this integration helps resolve ambiguous cases. When IHC results are equivocal or negative despite strong clinical suspicion, RNAscope can provide confirmatory evidence of gene expression. This is particularly valuable for targets with limited antibody specificity or in poorly differentiated tumors where protein expression may be lost while RNA remains detectable.
Immunohistochemistry remains the established gold standard for protein detection in tissue contexts, with extensive validation across countless biomarkers and disease states. Its direct detection of functional gene products, well-characterized protocols, and integration into diagnostic pathways ensure its continued relevance. RNAscope emerges as a powerful complementary technology with exceptional sensitivity and specificity for RNA detection, particularly valuable when suitable antibodies are unavailable or when transcript-level information is clinically relevant. The most sophisticated applications leverage both technologies in tandem, recognizing that mRNA and protein provide different but complementary biological information. As both technologies evolve, their synergistic use will continue to advance research discovery and diagnostic precision.
For researchers and drug development professionals, accurate in-situ RNA analysis is paramount yet challenging. Traditional methods like immunohistochemistry (IHC) detect proteins but lack direct RNA visualization, while conventional RNA in situ hybridization (ISH) often struggles with sensitivity and specificity issues. The emergence of RNAscope technology represents a significant advancement in molecular pathology, addressing critical limitations through its innovative double Z-probe design. This platform enables single-molecule RNA visualization while preserving tissue morphology in formalin-fixed, paraffin-embedded (FFPE) tissue specimens [7]. As the field increasingly recognizes the importance of spatial context in gene expression, this technology provides researchers with a powerful tool for validating RNA biomarkers within their native tissue microenvironment, offering a complementary approach to traditional protein-based detection methods.
The Double Z-Probe Technology: A Technical Breakdown
Fundamental Design Principles
The RNAscope platform employs a proprietary "double Z" probe design that fundamentally differs from traditional single-probe ISH approaches. This unique architecture enables simultaneous background suppression and signal amplification through a strategic two-step targeting system [8]. Each probe pair consists of two distinct segments (ZZ probes) that must bind adjacent to each other on the target RNA molecule to initiate signal amplification [9]. This adjacent binding requirement ensures that only specifically bound probes generate signal, as single probes or mismatched probes cannot initiate the amplification cascade. The system is engineered to visualize individual RNA molecules as discrete punctate dots under a standard microscope, providing both qualitative localization and quantitative potential through dot counting [7].
Mechanism of Action Workflow
This diagram illustrates the core mechanism of RNAscope's double Z-probe technology. The system requires two independent Z probes binding to adjacent regions of the target RNA to form a complete binding site for the preamplifier molecule. This mandatory cooperative binding provides the foundation for the technology's exceptional specificity. Mismatched or non-specifically bound single probes (shown in the gray pathway) cannot initiate the amplification cascade, thereby suppressing background noise. Once the preamplifier binds, it recruits multiple amplifier molecules, each capable of binding numerous label probes that generate detectable signals [8] [9]. This multi-stage amplification creates a powerful signal from each target RNA molecule while maintaining low background through the initial double Z-probe requirement.
Performance Comparison: RNAscope versus IHC
Quantitative Detection Comparison in Urothelial Carcinoma
Table 1: UPK2 Detection in Urothelial Carcinoma: RNAscope vs. IHC [4]
| Tissue Type | Number of Cases | RNAscope Positive (%) | IHC Positive (%) | P-value |
|---|---|---|---|---|
| Conventional Bladder UC | 127 | 72.4% | 68.5% | P = 0.511 |
| Variant Bladder UC | 45 | 53.3% | 35.6% | P = 0.057 |
| Upper Tract UC | 24 | 70.8% | 70.8% | P = 1.000 |
| Metastatic UC | 23 | 60.9% | 60.9% | P = 1.000 |
| Overall UC | 219 | 68.0% | 62.6% | P = 0.141 |
A comprehensive study evaluating UPK2 expression in urothelial carcinoma (UC) provides direct comparative data between RNAscope and IHC methodologies. The research analyzed 219 samples across various UC subtypes using both techniques [4]. While the overall difference in positivity rates between RNAscope (68.0%) and IHC (62.6%) was not statistically significant (P = 0.141), RNAscope demonstrated a notable trend toward higher sensitivity in detecting UPK2 in variant bladder UCs, with a 17.7% absolute increase in detection rate compared to IHC [4]. Correlation analysis revealed a moderate positive correlation between the two methods (P < 0.001, R = 0.441), suggesting they provide complementary but not identical information [4].
Advantages in Challenging Detection Scenarios
Table 2: Methodological Comparison of RNAscope and IHC
| Parameter | RNAscope | Traditional IHC |
|---|---|---|
| Target Molecule | RNA | Protein |
| Sensitivity | Single-molecule detection [9] | Limited by antibody affinity |
| Specificity Mechanism | Double Z-probe design [8] | Antibody-epitope binding |
| Morphology Preservation | Excellent in FFPE [7] | Excellent in FFPE |
| Multiplexing Capacity | Up to 12 targets with HiPlex [10] | Typically 1-3 targets |
| Quantification Potential | Digital dot counting | Semi-quantitative scoring |
| Background Suppression | Built-in to probe design [9] | Variable by antibody |
Beyond direct detection comparisons, RNAscope offers particular advantages in challenging scenarios where protein expression may not correlate directly with mRNA levels, or when analyzing low-abundance targets. The technology has demonstrated enhanced performance in other cancer types as well. In hepatocellular carcinoma, RNAscope improved Glypican3 (GPC3) and glutamine synthetase (GS) detection sensitivity by 20-30% compared to IHC [4]. Similarly, in lung adenocarcinoma, RNAscope proved more sensitive than IHC for detecting thyroid transcription factor 1 (TTF-1) and Napsin A expression, suggesting its utility for cases where IHC results are negative despite clinical suspicion [4].
Experimental Protocols and Workflows
RNAscope Assay Procedure
The RNAscope assay follows a standardized workflow that can be completed within one day. For FFPE tissues, the process begins with baking slides at 60°C for 1 hour, followed by deparaffinization in xylene and ethanol rehydration [4]. Subsequent steps include:
Pretreatment Series:
- Pretreatment 1: 10 minutes at room temperature
- Pretreatment 2: 20 minutes of boiling
- Pretreatment 3: 30 minutes at 40°C [4]
Probe Hybridization: Target probes are applied and incubated in a HybEZ oven for 2 hours at 40°C [4].
Signal Amplification: Using the RNAscope 2.0 HD Reagent Kit, signals are amplified through a series of 6 amplification steps that build upon the initial probe binding [4].
Signal Detection: Either chromogenic dyes for bright-field microscopy or fluorescent dyes for multiplex analysis are applied [7].
Counterstaining and Mounting: Appropriate counterstains (e.g., hematoxylin for chromogenic, DAPI for fluorescent) are applied before mounting.
Throughout this process, appropriate positive and negative control probes should be included to validate assay performance. The automated version of this protocol can be performed on the Leica BOND RX platform, enhancing reproducibility and throughput for research and clinical applications [11] [9].
Integrated Workflow for Spatial Transcriptomics
For comprehensive biomarker discovery and validation, researchers are increasingly implementing integrated workflows that combine multiple technologies. As illustrated in the diagram, single-cell RNA sequencing (scRNA-seq) and spatial transcriptomics (ST-seq) serve as discovery tools at genome-wide scale, generating hypotheses about potential ligand-receptor interactions and biomarkers [10]. These discoveries can then be validated using targeted approaches like RNAscope (measuring 4-12 genes) and multiplex protein staining (4-9 proteins) [10]. This integrated framework leverages the strengths of each technology: the breadth of scRNA-seq, the spatial context of ST-seq, and the sensitivity and single-cell resolution of RNAscope for final validation. Computational methods like STRISH can then analyze the imaging data to automatically scan whole tissue sections for local co-expression patterns, recapitulating interaction landscapes across entire tissues [10].
Essential Research Reagent Solutions
Table 3: Research Reagent Solutions for RNAscope Implementation
| Reagent/Equipment | Function | Application Notes |
|---|---|---|
| RNAscope Probe Sets | Target-specific ZZ probes | Design requires ~50 bp target region; species-specific available |
| RNAscope 2.0 HD Reagent Kit | Signal amplification and detection | Compatible with chromogenic and fluorescent detection |
| HybEZ Oven | Temperature-controlled hybridization | Essential for proper probe hybridization [4] |
| FFPE Tissue Sections | Standard sample format | 3-5 μm thickness optimal for preservation and penetration |
| Positive Control Probes | Assay validation | Housekeeping genes like PPIB or POLR2A |
| Negative Control Probes | Background assessment | Bacterial genes like DapB |
| BOND RX Platform | Automated processing | Enables high-throughput, standardized staining [9] |
| HALO Image Analysis Software | Quantitative analysis | Enables dot counting and spatial analysis [12] |
Successful implementation of RNAscope technology requires specific reagents and equipment. The core of the system is the target-specific probe sets designed against regions of approximately 50 base pairs of the target RNA [7]. The RNAscope 2.0 HD Reagent Kit provides all necessary components for the amplification and detection steps in a standardized format suitable for both manual and automated protocols [4]. For consistent results, the HybEZ oven provides precise temperature control during the critical hybridization step [4]. Implementation in a research setting should always include appropriate positive and negative control probes to validate each run. For quantitative analysis, platforms like HALO software enable automated dot counting and spatial analysis, providing researchers with objective, reproducible data from their experiments [12].
Discussion and Research Implications
The development of RNAscope technology represents a significant advancement in spatial biology, addressing critical gaps in the researcher's toolkit between genome-wide discovery methods and targeted protein detection. While IHC remains the gold standard for protein detection in clinical settings, RNAscope provides complementary molecular information at the RNA level with exceptional sensitivity and specificity [4] [7]. The technology's double Z-probe design fundamentally solves the longstanding challenges of background noise and insufficient sensitivity that plagued traditional ISH methods [7].
For research and drug development applications, RNAscope offers particular value in several scenarios: validating findings from transcriptomic studies, detecting low-abundance transcripts, analyzing FFPE archival tissues, and providing spatial context for gene expression patterns. The technology's ability to work reliably with FFPE tissues makes it exceptionally valuable for translational research utilizing extensive clinical archives [7]. Furthermore, the growing capabilities for multiplex analysis (up to 12 targets with HiPlex) position RNAscope as a powerful tool for exploring complex cellular interactions and signaling pathways within intact tissue architecture [10].
As spatial biology continues to evolve, integration of RNAscope with other modalities—including IHC, scRNA-seq, and spatial transcriptomics—creates a powerful multidimensional framework for understanding disease mechanisms [10]. This integrated approach enables researchers to move from correlation to causation in their studies of gene expression and protein function, ultimately accelerating biomarker discovery and therapeutic development.
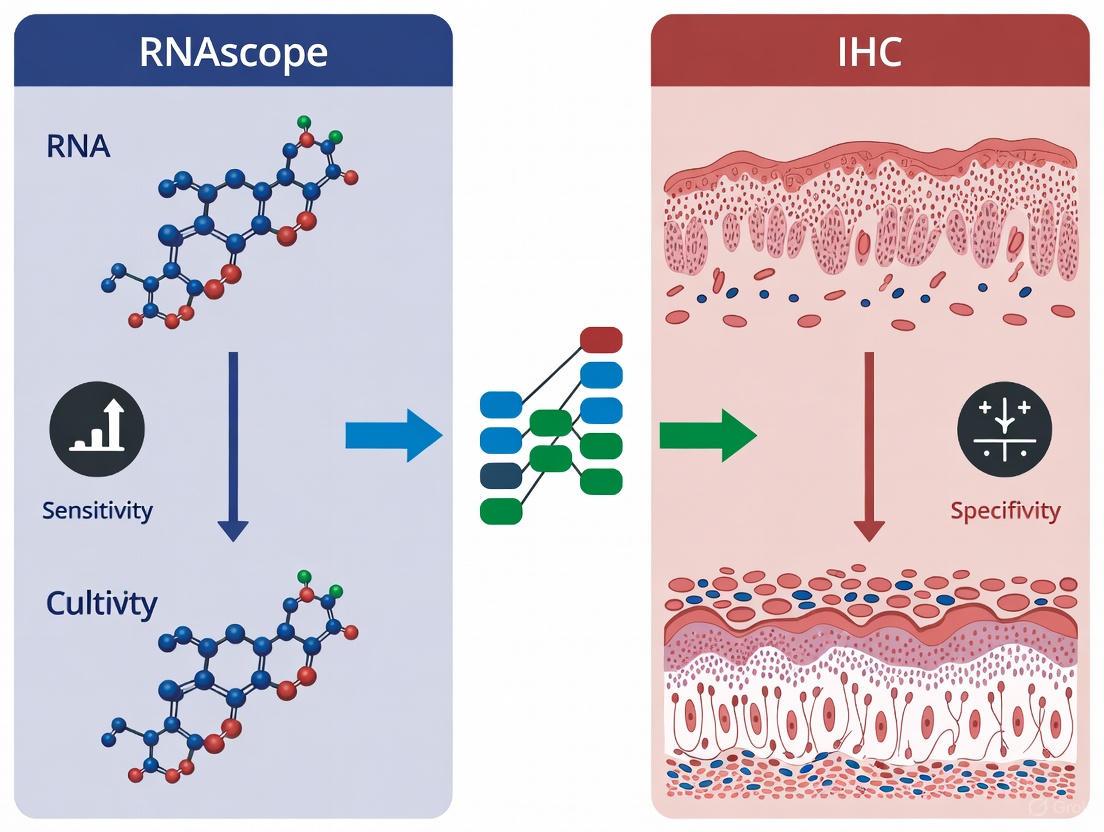
In the pursuit of accurate biomarkers for research and diagnostics, scientists can choose to detect the final functional agents—proteins—or the genetic blueprints that guide their synthesis—RNA. This guide provides an objective comparison of these two fundamental approaches, Immunohistochemistry (IHC) for protein detection and RNA in situ hybridization (ISH), specifically the RNAscope assay, for direct RNA visualization. Framed within the broader thesis of comparing RNAscope and IHC, the data summarized herein reveal a clear trend: direct RNA detection methods, particularly RNAscope, offer superior sensitivity and specificity for identifying gene expression in formalin-fixed paraffin-embedded (FFPE) tissues. This performance advantage is crucial for biomarker discovery and validation in both basic research and drug development.
Methodological Fundamentals at a Glance
The core difference between these techniques lies in their target molecules and detection mechanisms.
- Protein Detection (e.g., IHC): Relies on antibody-antigen interactions to visualize the final functional products of gene expression within their morphological context. While highly specific when antibodies are well-validated, it can be limited by antibody availability, quality, and the stability of the protein epitope, especially in degraded FFPE samples.
- Direct RNA Detection (e.g., RNAscope): Uses nucleic acid probes to hybridize directly to target RNA sequences. The proprietary RNAscope technology employs a "double Z" probe design and signal amplification that allows for highly specific and sensitive detection of RNA transcripts at the single-molecule level, with each dot representing a single RNA molecule [13] [14].
The table below summarizes the key characteristics of these two approaches.
Table 1: Core Characteristics of Protein vs. Direct RNA Detection
| Feature | Protein Detection (IHC) | Direct RNA Detection (RNAscope) |
|---|---|---|
| Target Molecule | Proteins (post-translational products) | RNA (messenger RNA transcripts) |
| Primary Reagent | Antibody | Nucleic Acid Probe |
| Key Mechanism | Antigen-Antibody Binding | Probe-Target RNA Hybridization |
| Signal Readout | Chromogenic or fluorescent stain | Discrete dots (each a single RNA molecule) |
| Sensitivity | Limited by antibody affinity and protein integrity | Single-molecule sensitivity [14] |
| Specificity | Can be compromised by cross-reactivity | High; proprietary probe design minimizes off-target binding [13] [14] |
| Compatibility | Standard FFPE sections | FFPE sections, including partially degraded samples [14] |
Performance Comparison: Sensitivity and Specificity
Head-to-head comparisons in validation studies demonstrate the technical advantages of direct RNA detection with RNAscope.
Experimental Protocol for Validation
A CLIA-guided validation study for a DKK1 RNAscope assay provides a robust framework for comparison [14]. The key methodological steps were:
- Cell Line and Tissue Preparation: Formalin-fixation and paraffin-embedding of cell pellets and gastric/GEJ adenocarcinoma tumor resections.
- Probe Hybridization: Application of the target-specific RNAscope probes and subsequent signal amplification.
- Controls: Each run included positive control probes (e.g., PPIB) to verify RNA integrity and negative control probes (e.g., bacterial dapB) to confirm the absence of background signal.
- Quantification: Both manual pathologist scoring and digital image analysis using algorithms (e.g., QuPath) to generate an H-score, which quantifies the percentage of tumor cells expressing low, medium, and high levels of the target RNA.
- Comparison to IHC: Parallel staining of the same samples with a DKK1 IHC assay for direct comparison.
Key Comparative Data
The validation study yielded quantitative data highlighting the performance differences.
Table 2: Experimental Performance Data: RNAscope vs. IHC
| Assay Parameter | DKK1 RNAscope Assay | DKK1 IHC Assay | Key Finding |
|---|---|---|---|
| Sensitivity | Detected RNA in HeLa cell pellet | No protein signal detected in HeLa cell pellet | RNAscope is more sensitive for low-abundance targets [14] |
| Specificity | No cross-reactivity with other Dickkopf family members (DKK2, DKK3, DKK4, DKKL1) | Information not provided in source | High specificity of designed probes [14] |
| Correlation with Orthogonal Data | Significant correlation with RNA-Seq data (Spearman’s rho = 0.86, p < 0.0001) | Consistent with RNAscope in high and null expressors | RNAscope results are highly accurate and concordant with other mRNA-measuring platforms [14] |
Workflow and Practical Considerations
The experimental workflow and associated tools differ significantly between the two methods. The following diagram illustrates the core technology behind the RNAscope assay.
Research Reagent Solutions
To execute a RNAscope experiment, the following key materials are required:
Table 3: Essential Research Reagents for RNAscope
| Item | Function in the Experiment |
|---|---|
| Target-Specific Probe Set | A pool of oligonucleotide probes designed to bind adjacent segments of the target mRNA; the foundation of assay specificity [14]. |
| Positive Control Probe (e.g., PPIB) | A probe for a constitutively expressed gene used to verify sample RNA integrity [14]. |
| Negative Control Probe (e.g., dapB) | A probe with no target in the sample to assess background noise and non-specific signal [14]. |
| Signal Amplification Reagents | The proprietary preamplifier, amplifier, and enzyme-linked labels that build upon the "ZZ" probes to achieve single-molecule sensitivity [13]. |
| Chromogenic or Fluorescent Substrate | The final reagent that is converted by the enzyme into a stable, visible dot at the site of probe hybridization [13] [15]. |
The objective data from validation studies lead to a clear conclusion: direct RNA detection via the RNAscope assay provides significantly higher sensitivity and specificity compared to IHC for detecting gene expression in FFPE tissues. Its ability to detect low-abundance transcripts and fragmented RNA, coupled with a robust and quantifiable signal output, makes it an superior tool for biomarker research [14].
For researchers and drug development professionals, this has critical implications:
- Biomarker Discovery and Validation: RNAscope increases confidence in biomarker identification, especially for targets where high-quality antibodies are unavailable.
- Clinical Diagnostic Development: The high specificity and sensitivity, combined with compatibility with digital image analysis, make RNAscope a strong candidate for companion diagnostic development [14].
- Retrospective Studies: The ability to work reliably on archived FFPE samples unlocks potential for analyzing vast biobanks.
While IHC remains a valuable and established technique for protein localization, the evidence supports the adoption of direct RNA detection methods like RNAscope for applications where the utmost sensitivity, specificity, and quantitative accuracy are required.
The Significance of Single-Molecule Sensitivity and Cellular Context
In the fields of molecular pathology, drug development, and basic research, the accurate measurement of gene expression is fundamental to understanding disease mechanisms and developing targeted therapies. However, traditional "grind-and-bind" methods like quantitative PCR (qPCR) require RNA extraction, a process that destroys the tissue architecture and loses all spatial information about which cells are expressing the gene of interest [1] [16]. This is a significant limitation because cellular context is often critical to understanding gene function, especially in complex tissues like tumors or the brain, where cell-to-cell heterogeneity plays a major role in disease progression and treatment response [17].
This article objectively compares two powerful techniques that preserve this vital spatial context: Immunohistochemistry (IHC) and RNAscope in situ hybridization (ISH). IHC has long been the clinical workhorse for detecting protein biomarkers in tissue sections. In contrast, RNAscope is a novel RNA ISH technology that enables the direct visualization of RNA molecules within individual cells in their native tissue environment. The core of this comparison hinges on their differing approaches to achieving sensitivity and specificity, and the profound implications this has for single-molecule detection and the preservation of cellular context.
Core Technologies: Principles and Mechanisms
Immunohistochemistry (IHC)
IHC is a well-established technique that leverages antibody-antigen interactions to detect specific proteins within tissue samples.
- Principle: The technique uses primary antibodies that bind specifically to a target protein (antigen) in a prepared tissue section. This binding is then visualized through a detection system, typically involving a secondary antibody conjugated to an enzyme like horseradish peroxidase (HRP). The enzyme catalyzes a chromogenic reaction (e.g., with DAB), producing a colored precipitate that can be seen under a standard bright-field microscope [18].
- What it Measures: IHC detects protein abundance and localization. This is a direct measure of the functional end-product of gene expression.
- Key Considerations: The reliability of IHC is highly dependent on the specificity and quality of the antibodies used. Non-specific binding can lead to false-positive results, and the technique can be difficult to quantify objectively [18].
RNAscope In Situ Hybridization
RNAscope is a groundbreaking RNA ISH method that was developed to overcome the limitations of traditional ISH, namely poor sensitivity and high background noise [16].
- Principle: The technology's foundation is its proprietary double-Z (ZZ) probe design [1] [19] [16]. Each target RNA is detected by multiple pairs of probes. A single probe pair (a "ZZ" pair) must bind adjacent to each other on the target RNA to create a binding site for the pre-amplifier molecule. This requirement is the key to the technology's high specificity, as it is statistically unlikely for two independent probes to bind nonspecifically to a non-target sequence in the correct orientation and proximity.
- Signal Amplification: Once the ZZ probe pair is bound, a multi-step amplification cascade is initiated. The pre-amplifier binds to the probe pair, and it in turn provides binding sites for multiple amplifiers. Finally, each amplifier binds numerous enzyme-linked label probes, resulting in a powerful amplification of the signal—up to 8,000-fold per RNA molecule [1]. This process allows for single-molecule sensitivity, with each individual RNA transcript appearing as a distinct, punctate dot under the microscope [19] [16].
- What it Measures: RNAscope directly detects and visualizes RNA transcripts. This provides a direct readout of gene expression at the transcriptional level.
The following diagram illustrates the unique probe design and signal amplification mechanism of RNAscope.
Performance Comparison: Sensitivity, Specificity, and Concordance
A systematic review evaluating RNAscope in clinical diagnostics provides robust data for a direct comparison with gold standard techniques. The review, which analyzed 27 studies, found that RNAscope is a highly sensitive and specific method [1] [20].
Table 1: Concordance Rates Between RNAscope and Gold Standard Techniques
| Comparison Method | Concordance Rate (CR) Range | Basis of Comparison |
|---|---|---|
| IHC | 58.7% – 95.3% [1] [20] | RNA (RNAscope) vs. Protein (IHC) |
| qPCR / qRT-PCR | 81.8% – 100% [1] [20] | RNA in situ vs. Extracted RNA |
| DNA ISH | 81.8% – 100% [1] [20] | RNA in situ vs. DNA in situ |
The data reveals a key insight: RNAscope has excellent concordance with other nucleic acid-based techniques (qPCR and DNA ISH). However, its concordance with IHC is more variable. This is not necessarily an indication of poor performance by either technique, but rather a reflection that they measure different biomolecules (RNA vs. protein). Discrepancies can arise from post-transcriptional regulation, differences in protein and RNA turnover rates, or issues with antibody specificity in IHC [1].
Case Study: HPV Detection in Oropharyngeal Cancer
A clear example of the practical impact of these technological differences is in detecting high-risk Human Papillomavirus (HPV) in oropharyngeal squamous cell carcinoma (OPSCC). The surrogate IHC marker for HPV is p16, a protein that is overexpressed in HPV-related tumors. However, p16 overexpression can also occur in some HPV-negative tumors, leading to false positives.
RNAscope probes directly target the E6/E7 viral oncogene mRNA, providing direct evidence of active viral infection. Studies have demonstrated that the RNAscope assay for HPV has equivalent sensitivity but significantly higher specificity compared to p16 IHC, reducing potential false-positive diagnoses [21].
Experimental Protocols for Comparative Analysis
To ensure reliable and reproducible results when using or comparing these techniques, standardized protocols are essential. Below are detailed methodologies for a standard IHC protocol and an RNAscope protocol, including a combined RNAscope/IHC approach.
Detailed IHC Protocol
The IHC workflow is a multi-step process that requires careful optimization at each stage [18].
Table 2: Key Research Reagent Solutions for IHC
| Reagent / Instrument | Function |
|---|---|
| Primary Antibodies | High-specificity antibodies that bind to the target protein (antigen). |
| Secondary Antibodies | Enzyme-conjugated antibodies that bind to the primary antibody for signal detection. |
| Chromogenic Substrates (e.g., DAB) | Enzymatic reaction produces a colored precipitate at the antigen site. |
| Blocking Agents | Reduce non-specific binding of antibodies to the tissue. |
| Automated Staining Platforms | Ensure high-throughput, consistency, and reproducibility of staining. |
The following diagram outlines the core workflow for an IHC assay.
Detailed RNAscope Protocol
The RNAscope assay involves a series of hybridization and amplification steps optimized for FFPE tissues [2] [16].
- Slide Preparation: FFPE tissue sections are cut, mounted on slides, deparaffinized, and dehydrated.
- Pretreatment: Slides are treated with heat and protease to unmask target RNA and permeabilize the cells, allowing probe access [19].
- Probe Hybridization: Target-specific ZZ probes are hybridized to the RNA of interest for a defined period (e.g., 2 hours) [16].
- Signal Amplification: Sequential, automated hybridizations of the preamplifier, amplifier, and label probe are performed to build the amplification complex [1] [19].
- Visualization & Quantification: For fluorescent detection, slides are counterstained with DAPI and visualized. Each punctate dot represents a single RNA molecule, which can be quantified manually or with image analysis software like HALO or QuPath [1] [19].
Combined RNAscope and IHC Protocol
A major advantage of RNAscope is its compatibility with IHC on the same tissue section, allowing researchers to correlate RNA expression with protein expression and cell identity within the same physical cell [1] [2]. A protocol optimized for thicker (14 μm) central nervous system tissue sections involves the following key steps [2]:
- Simultaneous Staining: After RNAscope probe hybridization and signal amplification are complete, the tissue is incubated with a primary antibody against a cell-type-specific protein (e.g., IBA1 for microglia, NeuN for neurons).
- Fluorescent Detection: A fluorophore-conjugated secondary antibody is applied to visualize the protein target.
- Confocal Microscopy and Analysis: The slide is imaged using a confocal microscope. RNA transcripts (punctate dots) can then be quantified within the boundaries of the IHC-labeled cells, providing a powerful method for cell-type-specific gene expression analysis [2].
Discussion: Implications for Research and Diagnostics
The choice between IHC and RNAscope is not a matter of one being universally superior, but rather depends on the research or diagnostic question.
- Complementary, Not Redundant: IHC and RNAscope provide complementary information. IHC reveals the presence of the final functional protein, while RNAscope reveals the transcriptional activity of the gene. The systematic review concluded that RNAscope is best positioned as a reliable and robust method that could complement gold standard techniques currently used in clinical diagnostics [1].
- Resolving Discrepancies: The lower concordance between IHC and RNAscope highlights the importance of post-transcriptional regulation. When protein and RNA levels do not align, it can point to important biological mechanisms such as issues with translation or protein degradation.
- The Single-Molecule Advantage: RNAscope's ability to detect individual RNA molecules allows researchers to assess cellular heterogeneity in gene expression with unprecedented clarity. It can determine not just if a cell population is expressing a gene, but the precise distribution of expression levels from cell to cell, which is crucial for understanding tumor heterogeneity, neuronal function, and immune responses [17].
- Multiplexing Potential: While IHC is limited in its ability to detect multiple proteins simultaneously due to chromogen constraints, the RNAscope Multiplex Fluorescent V2 Assay allows for the simultaneous detection of up to four different RNA targets in a single sample by assigning different fluorescent colors to each target [22]. This is a significant advantage for studying complex gene networks and interactions.
Both IHC and RNAscope are indispensable tools for spatial biology, each with distinct strengths. IHC remains a cornerstone of clinical diagnostics for protein detection. However, RNAscope represents a paradigm shift by providing single-molecule sensitivity and high specificity for RNA analysis directly in intact tissues. Its unique double-Z probe design fundamentally suppresses background noise while amplifying true signals, enabling the precise quantification of gene expression within its native cellular context. For researchers and drug developers aiming to understand complex biological systems, characterize biomarkers with high confidence, or validate therapeutic targets, RNAscope offers a powerful and complementary approach that bridges the gap between traditional nucleic acid techniques and protein-based histopathology.
This guide provides an objective comparison between RNAscope, a novel in situ hybridization (ISH) platform, and Immunohistochemistry (IHC), the established standard for protein detection in tissue samples. For researchers and drug development professionals, understanding the technical capabilities, performance data, and appropriate applications of each method is crucial for experimental and diagnostic design. The core distinction lies in their detection targets: IHC visualizes proteins, while RNAscope detects RNA transcripts. Evidence from recent studies indicates that RNAscope offers superior specificity and single-molecule sensitivity for RNA detection, excellent morphological context preservation, and growing potential for multiplexing, though IHC remains a highly practical and cost-effective tool for protein localization.
Performance Data Comparison: RNAscope vs. IHC and Other Techniques
The following tables summarize key performance metrics from published studies, providing a direct comparison of RNAscope against standard techniques.
Table 1: Comparison of RNAscope and IHC for UPK2 Detection in Urothelial Carcinoma (UC) [3]
| Method | Target | Overall Sensitivity in UC (n=219) | Sensitivity in Variant BUC (n=45) | Correlation with IHC |
|---|---|---|---|---|
| RNAscope | mRNA | 68.0% | 53.3% | Moderate positive correlation |
| IHC | Protein | 62.6% | 35.6% | (P < 0.001, R = 0.441) |
| Statistical Significance | P = 0.141 (Not Significant) | P = 0.057 (Marginal, Not Significant) |
Table 2: Concordance Rates of RNAscope with Gold Standard Methods (Systematic Review) [1]
| Comparison Technique | Target Type | Reported Concordance Rate (CR) with RNAscope |
|---|---|---|
| qPCR / qRT-PCR | RNA | 81.8% - 100% |
| DNA In Situ Hybridization (ISH) | DNA | 81.8% - 100% |
| Immunohistochemistry (IHC) | Protein | 58.7% - 95.3% |
Experimental Protocols and Methodologies
To contextualize the performance data, below are the detailed experimental protocols for the key studies cited.
Protocol: Comparison Study for UPK2 in UC
This protocol is derived from the study that generated the data in Table 1 [3].
- Sample Preparation: Tissue Microarrays (TMAs) were constructed using tissue blocks from 127 conventional bladder UCs, 45 variant bladder UCs, 24 upper tract UCs, and 23 metastatic UCs. Formalin-Fixed, Paraffin-Embedded (FFPE) tissue sections were cut at 3μm thickness.
- IHC Staining:
- Staining System: Automated BenchMark ULTRA system (Ventana Medical Systems).
- Primary Antibody: UPK2 (clone BC21, Biocare Medical) at 1:100 dilution.
- Detection: Standard chromogenic detection.
- Evaluation: Cytoplasmic staining in UC cells was scored as positive by two independent pathologists.
- RNAscope Assay:
- Technology: RNAscope 2.0 HD Reagent Kit-BROWN (Advanced Cell Diagnostics, ACD).
- Probe: Target probe for UPK2 (NM_006760.4).
- Procedure: Slides were deparaffinized, subjected to pretreatment (pretreatment 1 at room temperature, pretreatment 2 involving boiling, pretreatment 3 at 40°C), hybridized with probes in a HybEZ oven at 40°C for 2 hours, and signals were amplified per manufacturer's protocol.
- Evaluation: Cytoplasmic staining (dots) in UC cells was scored as positive.
- Statistical Analysis: McNemar's test was used for method comparison, and Spearman rank correlation was used for correlation analysis (SPSS 25.0).
Protocol: Systematic Review of RNAscope in Clinical Diagnostics
This protocol outlines the methodology of the systematic review referenced in Table 2 [1].
- Literature Search: Searches were conducted in CINAHL, Medline, Embase, and Web of Science for studies published after 2012 that compared RNAscope with one or more "gold standard" techniques (IHC, qPCR, qRT-PCR, DNA ISH) in human samples.
- Study Selection & Quality Assessment: A total of 27 retrospective studies were included. The risk of bias was assessed using the QUADAS-2 tool, with scores ranging from low to middle risk.
- Data Analysis: Results were reviewed narratively, and Concordance Rates (CR) between RNAscope and the other techniques were extracted and qualitatively analyzed.
Technological Workflows and Signaling Pathways
The fundamental advantage of RNAscope lies in its proprietary probe design, which is fundamentally different from the antibody-based detection used in IHC.
Research Reagent Solutions Toolkit
The table below lists essential materials and reagents for implementing the RNAscope and IHC techniques in a research or diagnostic setting.
Table 3: Essential Research Reagents and Materials [3] [1] [23]
| Item | Function | Example/Note |
|---|---|---|
| RNAscope Probe | Target-specific "Z" probe pair designed to hybridize to the RNA of interest. | >70,000 unique probes available for human and mouse transcriptome [24]. |
| RNAscope Reagent Kit | Contains all necessary reagents for hybridization, amplification, and chromogenic/fluorescent detection. | e.g., RNAscope 2.0 HD Reagent Kit-BROWN [3]. |
| Positive Control Probe | Validates assay success and tissue RNA integrity. | Probes for housekeeping genes (e.g., PPIB, Polr2A, UBC) [1]. |
| Negative Control Probe | Confirms absence of background noise. | Bacterial DapB gene probe [1]. |
| Primary Antibody (IHC) | Binds specifically to the target protein antigen. | Specificity and dilution must be optimized (e.g., UPK2 antibody at 1:100) [3]. |
| Detection System (IHC) | Visualizes antibody binding via enzymatic reaction. | Enzyme-conjugated secondary antibodies and chromogens (e.g., DAB) [23]. |
| Formalin-Fixed Paraffin-Embedded (FFPE) Tissues | The most common sample type for both techniques, preserving tissue morphology [3] [1]. | Requires specific pretreatment protocols for optimal results. |
| Automated Staining System | Platforms to automate staining procedures, improving reproducibility and throughput. | Used for both IHC (e.g., BenchMark ULTRA) [3] and RNAscope (e.g., HybEZ oven) [3]. |
Critical Analysis of Advantages
Specificity: The double "Z" probe design is the cornerstone of RNAscope's high specificity, which can reach 100% [1]. This design requires two independent probe sequences to bind adjacent to the target RNA for signal amplification to initiate, effectively suppressing off-target binding and background noise [7]. IHC specificity is highly dependent on the antibody's affinity and the optimization of staining conditions, with a known risk of non-specific binding leading to false positives [18].
Localization and Sensitivity: RNAscope provides exceptional resolution for localization by allowing single RNA molecule visualization, with each dot representing an individual transcript [1] [7]. This high sensitivity enables the detection of low-abundance RNAs in their precise cellular and subcellular context. While IHC provides excellent tissue-level localization of proteins, it cannot achieve single-molecule resolution. The systematic review in [1] notes that the lower concordance between RNAscope and IHC (as low as 58.7%) is expected, as the two techniques measure different biomolecules (RNA vs. protein), which can be affected by post-transcriptional regulation.
Multiplexing Potential: RNAscope is inherently designed for multiplexing, allowing simultaneous detection of multiple RNA targets in the same tissue section using probes with different fluorescent labels [1]. This is powerful for studying gene co-expression and interactions. Multiplex IHC (mIHC/IF) is an active area of innovation to overcome the limitation of conventional single-marker IHC [25]. Both chromogenic and fluorescent multiplexing exist for IHC, with immunofluorescence (IF) being more suitable for larger panels, though it requires specialized imaging systems and is susceptible to photobleaching [25] [26].
From Bench to Bedside: Standardized Workflows and Research Applications
The need for highly sensitive and specific techniques for RNA visualization within its native morphological context has positioned in situ hybridization (ISH) as a critical technology in both research and clinical diagnostics. Within this field, the RNAscope assay has emerged as a powerful platform, frequently compared against established methods like immunohistochemistry (IHC). A growing body of evidence underscores a central thesis: RNAscope offers a unique combination of high sensitivity and exceptional specificity, effectively addressing common limitations of IHC, such as antibody cross-reactivity and the discordance between protein and mRNA expression. This guide provides a detailed, objective examination of the RNAscope assay workflow, from initial sample preparation to final signal amplification, and presents a direct comparison with IHC to aid researchers, scientists, and drug development professionals in making informed methodological choices.
The foundational innovation of the RNAscope platform is its proprietary ZZ probe design. This technology employs a pair of "Z" probes that must bind adjacent to each other on the target RNA sequence for signal amplification to occur. This double-Z recognition is the key to its high specificity, as it minimizes off-target binding and background noise [2]. Following successful hybridization, a multi-step amplification system builds a large polymer onto the ZZ probe pair, which can then be visualized with enzymatic or fluorescent labels, enabling single-molecule sensitivity while preserving tissue morphology [21] [2].
The principal advantages of this system are:
- High Specificity: The double-Z probe design requires two independent binding events for each signal molecule, virtually eliminating false-positive signals from non-specific probe binding [2].
- High Sensitivity: The robust signal amplification allows for the detection of individual RNA molecules, making it suitable for targets with low expression levels [27].
- Morphological Context: Unlike "grind-and-bind" methods like RNA sequencing, RNAscope allows for the visualization of RNA expression within the intact tissue architecture, providing spatial information that is lost in bulk analysis techniques [28] [27].
Visualizing the Core Technology
The following diagram illustrates the key components and the mechanism of the RNAscope signal amplification system:
Detailed Workflow: Critical Steps from Sample to Result
The RNAscope assay can be performed manually or automated on staining systems such as the Roche Discovery series or the Leica BOND RX [29] [30]. The entire procedure is designed to be completed within a single day. The following section breaks down the critical stages of the workflow.
Sample Preparation and Pretreatment
This initial phase is crucial for assay success, as it ensures optimal accessibility of the target RNA while preserving tissue integrity.
- Step 1: Tissue Sectioning. The assay is typically performed on Formalin-Fixed, Paraffin-Embedded (FFPE) tissue sections cut at 5 μm thickness. Sections are mounted on slides and dried.
- Step 2: Deparaffinization and Rehydration. Slides are treated with xylene and graded ethanol series to remove paraffin and rehydrate the tissue for subsequent aqueous steps.
- Step 3: Pretreatment. Slides undergo a series of pretreatments to expose the target RNA. This includes:
- Pretreatment 1: A brief incubation at room temperature to rehydrate the tissue further.
- Pretreatment 2: A heat-induced epitope retrieval step, typically involving boiling the slides for 15-20 minutes [4].
- Pretreatment 3: Protease Digestion. This is one of the most critical steps in the entire workflow. A proprietary protease is applied to digest proteins and allow probe access to RNA. Over-digestion can lead to poor morphology and RNA degradation, while under-digestion results in low signal and high background [31]. Recent advancements have introduced protease-free workflows on platforms like the Roche Discovery ULTRA, which are beneficial for preserving protein epitopes for concurrent IHC and for targets sensitive to protease treatment [30].
Probe Hybridization and Signal Amplification
After pretreatment, the slides are ready for the core of the RNAscope assay.
- Step 4: Probe Hybridization. Target-specific probes are applied to the tissue sections and incubated in a controlled environment. The use of a dedicated HybEZ Oven is strongly recommended, as temperature and humidity are critical factors for consistent performance [31]. Hybridization typically occurs for 2 hours at 40°C [4].
- Step 5: Signal Amplification. A series of amplifier molecules are applied sequentially. These amplifiers bind specifically to the ZZ probes and build a branching tree structure. Each successive step amplifies the signal. The order of these amplification steps must not be altered, as missing any step can result in a complete loss of signal [31].
- Step 6: Signal Detection. Finally, a chromogenic or fluorescent substrate is applied. For chromogenic detection, a enzyme-linked label catalyzes a reaction that deposits a permanent precipitate. The result is a punctate dot-like signal, with each dot representing an individual RNA molecule [29].
Workflow Visualization
The entire RNAscope procedure, from sample to image analysis, is summarized in the following workflow diagram:
Direct Comparison: RNAscope vs. Immunohistochemistry (IHC)
The choice between RNAscope and IHC hinges on the specific research or diagnostic question. The table below summarizes a direct, data-driven comparison based on published studies.
Table 1: Experimental Data Comparison of RNAscope and IHC Performance
| Metric | RNAscope | Immunohistochemistry (IHC) | Experimental Context & Citation |
|---|---|---|---|
| Sensitivity | Equivalent or higher sensitivity. Detected 68.0% of UCs as UPK2+ [4]. Single-molecule detection capability [27]. | Variable, can be lower. Detected 62.6% of UCs as UPK2+ (P=0.141) [4]. | Comparison of UPK2 detection in Urothelial Carcinoma (UC) [4]. |
| Specificity | Significantly higher specificity. Direct detection of viral oncogene mRNA reduces false positives from surrogate markers [21]. | Lower specificity. p16 IHC, a surrogate for HPV, can yield false positives [21]. | Detection of high-risk HPV in oropharyngeal squamous cell carcinoma [21]. |
| Correlation with mRNA | Direct measurement of target RNA. N/A | Moderate to strong correlation for some targets (Spearman R=0.53-0.89) [28]. | Correlation study of RNA-seq and IHC for 9 biomarkers [28]. |
| Morphology | Preserves tissue architecture. Allows for cell-by-cell analysis. | Preserves tissue architecture. Allows for cell-by-cell analysis. | Both are tissue-based assays. |
| Target | RNA (mRNA, viral RNA) | Protein | Fundamental difference in detected molecule. |
| Throughput | Can be automated; single-day workflow [29]. | Highly automated and standardized. |
Case Study: Head-to-Head Performance in Urothelial Carcinoma
A 2022 study in Diagnostic Pathology directly compared RNAscope and IHC for evaluating UPK2, a marker for urothelial carcinoma (UC), in 219 samples. The results demonstrated a moderate positive correlation between the two methods (P < 0.001, R = 0.441) [4]. While the overall positivity rate was not significantly different (68.0% for RNAscope vs. 62.6% for IHC, P = 0.141), a trend suggested RNAscope might be more sensitive in diagnostically challenging cases. For instance, in variant bladder UCs, the positivity rate for RNAscope was 53.3% compared to 35.6% for IHC, though this difference was not statistically significant (P = 0.057) [4]. This highlights that for well-characterized targets with good antibodies, IHC performance can be comparable, but RNAscope may offer advantages for difficult targets or low-abundance transcripts.
Essential Reagents and Equipment for the RNAscope Workflow
Establishing the RNAscope assay in a laboratory requires specific reagents and equipment. The following table details the key components.
Table 2: Research Reagent Solutions for the RNAscope Assay
| Item | Function | Critical Considerations |
|---|---|---|
| Target-Specific Probe(s) | Binds to the RNA sequence of interest. | Probes are available for thousands of human, mouse, and viral genes. Custom probes can be designed and manufactured in about two weeks [27]. |
| RNAscope Reagent Kit | Contains all necessary reagents for hybridization, washing, amplification, and detection. | Kits are available for chromogenic (brown/red) or fluorescent detection, and for manual or automated protocols [29] [4]. |
| Control Probes | Verify assay performance. | Include positive control probes (e.g., housekeeping genes like PPIB) and negative control probes (bacterial DapB) to confirm RNA quality and specificity [31]. |
| HybEZ Oven System | Provides precise temperature and humidity control during hybridization. | ACD extensively validates this system; other incubators may not provide consistent results [31]. Critical for manual assays. |
| Protease | Digests proteins to unmask target RNA. | Digestion time is critical. Over- or under-digestion severely impacts results [31]. Protease-free assays are a new alternative [30]. |
| Automated Stainer | Executes the assay protocol automatically. | Systems like Roche Discovery ULTRA or Leica BOND RX ensure standardization and higher throughput [29] [30]. |
The RNAscope assay provides a robust and reliable workflow for the sensitive and specific detection of RNA in situ. Its step-by-step process, from careful sample pretreatment through controlled hybridization and powerful signal amplification, is designed to deliver unambiguous, quantifiable results. Direct comparisons with IHC consistently show that RNAscope offers superior specificity and equivalent or better sensitivity, making it an invaluable tool for validating RNA-seq data, detecting viral infections, and differentiating highly homologous genes [21] [28] [27].
The future of the technology lies in increased integration and multiplexing. The development of protease-free workflows facilitates combined RNAscope and IHC/IF on the same section, enabling researchers to visualize RNA and protein simultaneously within a preserved morphological context [30]. This spatial multi-omics approach is powerful for elucidating complex biological interactions, such as host-pathogen relationships and the tumor microenvironment. For researchers and drug developers requiring precise cellular localization of gene expression, the RNAscope assay is an indispensable technology that continues to evolve, offering ever-greater clarity and analytical power.
Immunohistochemistry (IHC) is a foundational technique in diagnostic pathology and research that uses antibody-epitope interactions to selectively label and visualize specific proteins within tissue samples. This method allows for the confirmation of target molecule expressions while preserving the histological architecture and microenvironment of the tissue, providing critical contextual information that destruction-based methods cannot offer [23] [32]. The core principle of IHC relies on the specific binding of a primary antibody to a target antigen (protein) within a tissue section, followed by detection systems that generate visible signals, either chromogenic (colorimetric) or fluorescent [33] [23].
IHC has evolved significantly from its origins in the 1940s, when Albert H. Coons developed the first fluorescently conjugated antibody, to become a mainstream diagnostic tool that complements traditional Hematoxylin & Eosin (H&E) staining [33] [23]. Where H&E and special stains primarily show tissue morphology non-specifically, IHC is directed toward specific protein markers, making it indispensable for accurate tumor classification, diagnosis, and biomarker exploration in both solid tumors and cytological specimens [33] [32]. The technique's unique value lies in its ability to provide semi-quantitative data on protein distribution, subcellular localization, and abundance within different cell populations, all within their physiological context [23].
Core IHC Protocol: A Standardized Workflow
The execution of a successful IHC experiment follows a systematic, multi-stage process where each step must be carefully optimized. The workflow can be broadly divided into pre-staining, staining, and post-staining phases, with both paraffin-embedded and frozen sections sharing fundamental similarities while requiring specific handling differences.
Pre-Staining Procedures: Tissue Preparation and Fixation
Tissue Acquisition and Fixation: Proper tissue handling begins immediately after specimen acquisition. For optimal preservation, tissues should be rapidly fixed to prevent protein degradation, autolysis, and loss of antigenicity. The gold standard fixative is 10% neutral buffered formalin (NBF), which creates methylene cross-links between proteins to preserve tissue architecture [23] [32]. The recommended fixation time is approximately 24 hours at room temperature, with an ideal tissue-to-fixative ratio between 1:1 and 1:20 [32]. Under-fixation can lead to proteolytic degradation, while over-fixation may mask target epitopes through excessive cross-linking, making antigen retrieval difficult [23].
Processing and Sectioning: Following fixation, tissues are processed through a series of alcohol dehydrations, xylene clearing, and embedding in paraffin wax to provide structural support for thin sectioning [34]. Sections are typically cut at 4μm thickness using a microtome, floated in a water bath to remove wrinkles, and mounted on charged or APES-coated glass slides to ensure adhesion during subsequent procedures [33] [32]. For frozen sections, tissues are snap-frozen in isopentane cooled by dry ice, embedded in Optimal Cutting Temperature (OCT) compound, and sectioned using a cryostat [35].
Deparaffinization and Rehydration: For paraffin-embedded sections, the wax must be completely removed to allow antibody penetration. This is achieved through sequential washes in xylene (or xylene substitutes) followed by a graded alcohol series (100%, 95%, 70%, 50%) and finally distilled water or buffer [34] [32].
Staining Procedures: Antibody Binding and Detection
The core staining process involves a series of incubations and washes designed to specifically label the target antigen while minimizing non-specific background.
Antigen Retrieval: Formalin fixation can mask epitopes through protein cross-linking, often necessitating an antigen retrieval step to reverse this process. The most common method is Heat-Induced Epitope Retrieval (HIER), where slides are heated in a buffer solution (citrate or EDTA buffer, pH 6-10) using a microwave, pressure cooker, or water bath [34] [32]. Alternative enzymatic retrieval methods using trypsin or proteinase K may be used for specific antigens [35] [32].
Blocking: To reduce non-specific background staining, tissue sections are incubated with blocking solutions. Common blocking agents include 5%-10% normal serum from the same species as the secondary antibody, bovine serum albumin (BSA), or commercial protein blocks [34] [32]. Additionally, endogenous peroxidase activity (for HRP-based detection) is blocked using 3% hydrogen peroxide, while endogenous alkaline phosphatase (for AP-based detection) is blocked with levamisol [35] [32].
Antibody Incubation: The core of IHC specificity lies in the application of the primary antibody that binds specifically to the target antigen. Antibodies are diluted in appropriate buffers to optimal concentrations determined through titration experiments [33] [34]. Incubation conditions vary from 30-60 minutes at room temperature to overnight at 4°C, depending on antibody affinity and concentration [32]. This is followed by application of a secondary antibody that recognizes the primary antibody's species and isotype, often conjugated to enzymes (HRP or AP) for detection [33].
Signal Detection and Development: For chromogenic detection, enzyme-conjugated antibodies catalyze the conversion of substrate molecules into insoluble colored precipitates at the antigen site. Common chromogens include DAB (3,3'-diaminobenzidine), which produces a brown precipitate, and AP Red, which produces a red precipitate [33] [34]. DAB is preferred for most applications due to its permanent nature, while AP Red is useful for tissues with high melanin content or for double-staining applications [33].
Post-Staining Procedures: Visualization and Analysis
Counterstaining and Mounting: Following chromogen development, tissues are often counterstained with hematoxylin (blue) or nuclear fast red to provide contrast and visualize tissue architecture [33] [34]. After counterstaining, sections are dehydrated through graded alcohols, cleared in xylene, and mounted under coverslips using permanent mounting media [34].
Analysis and Interpretation: Stained sections are examined under a light microscope by trained pathologists or researchers. Interpretation considers the presence, distribution, intensity, and subcellular localization (membrane, cytoplasmic, or nuclear) of staining [33] [32]. Appropriate positive and negative controls are essential to validate results, with internal tissue elements often serving as built-in controls [33].
Key Methodological Variations in IHC Execution
While the fundamental principles of IHC remain consistent, several methodological variations significantly impact experimental outcomes and interpretation.
Paraffin-Embedded vs. Frozen Sections
Table 1: Comparison of Paraffin-Embedded vs. Frozen Section Methodologies
| Parameter | Paraffin-Embedded Sections (IHC-P) | Frozen Sections (IHC-F) |
|---|---|---|
| Tissue Processing | Dehydration, clearing, paraffin infiltration [34] | Snap-freezing in OCT compound [35] |
| Fixation | Typically formalin-fixed [32] | Acetone, ethanol, or paraformaldehyde [32] |
| Morphology Preservation | Excellent | Moderate to good |
| Antigen Preservation | Variable; may require antigen retrieval [23] | Generally good; less antigen retrieval needed [23] |
| Turnaround Time | Longer (days) | Shorter (hours) |
| Application | Routine histopathology, archival studies | Labile antigens, intraoperative consultations |
Detection Systems: Direct vs. Indirect Methods
IHC detection systems vary in their complexity and signal amplification capabilities:
- Direct Method: Primary antibodies are directly conjugated to enzymes or fluorophores. This approach is simple and rapid but offers less signal amplification and sensitivity [23].
- Indirect Method: Uses an unlabeled primary antibody followed by a labeled secondary antibody that recognizes the primary. This provides signal amplification as multiple secondary antibodies can bind to each primary antibody [33].
- Polymer-Based Methods: Modern systems use enzyme polymers (e.g., HRP or AP polymers) conjugated to secondary antibodies, providing enhanced sensitivity through increased enzyme loading per antibody [33].
Chromogenic vs. Fluorescent Detection
Table 2: Chromogenic vs. Fluorescent Detection in IHC
| Parameter | Chromogenic IHC | Immunofluorescence (IF) |
|---|---|---|
| Detection Modality | Colorimetric enzyme reaction [33] | Fluorophore excitation/emission [23] |
| Signal Type | Insoluble colored precipitate (e.g., brown DAB) [33] | Light emission at specific wavelengths [23] |
| Visualization | Bright-field microscope [33] | Fluorescence or confocal microscope [23] |
| Multiplexing Capability | Limited (typically 2 targets with different chromogens) [33] | Excellent (multiple targets with different fluorophores) [23] |
| Permanence | Permanent with some chromogens (DAB) [33] | Fades over time; requires special mounting [23] |
| Quantification | Semi-quantitative | More amenable to quantification |
Antibody Selection: Monoclonal vs. Polyclonal
The choice between monoclonal and polyclonal antibodies represents another critical variable:
- Monoclonal Antibodies: These antibodies have affinity for a single epitope on the target antigen, producing cleaner, more specific staining with less cross-reactivity but potentially lower intensity [33].
- Polyclonal Antibodies: These are mixtures of antibodies that recognize multiple epitopes on the same antigen, often providing more intense staining due to signal amplification but with higher potential for cross-reactivity and background [33].
Essential Research Reagent Solutions
Successful IHC requires carefully selected reagents at each stage of the protocol. The following table outlines key solutions and their functions:
Table 3: Essential Research Reagents for IHC Protocols
| Reagent Category | Specific Examples | Function | Considerations |
|---|---|---|---|
| Fixatives | 10% Neutral Buffered Formalin, 4% Paraformaldehyde, Acetone, Ethanol [23] [32] | Preserve tissue architecture and prevent antigen degradation | Formalin creates cross-links; alcohols precipitate proteins [23] |
| Antigen Retrieval Solutions | Citrate Buffer (pH 6.0), EDTA Buffer (pH 8.0-9.0), Tris-EDTA [34] [32] | Reverse formaldehyde-induced cross-linking to expose epitopes | pH optimization is antigen-specific; affects staining intensity [32] |
| Blocking Solutions | Normal Serum, BSA, Non-Fat Dry Milk, Commercial Protein Blocks [34] [32] | Reduce non-specific antibody binding to minimize background | Match serum species to secondary antibody; avoid biotin-containing blocks with ABC detection [32] |
| Primary Antibodies | Monoclonal, Polyclonal, Ready-To-Use (RTU), Concentrates [33] | Specifically bind to target antigen | Validate for IHC application; optimize concentration for each lot [33] |
| Detection Systems | Polymer-Based HRP, Polymer-Based AP, Avidin-Biotin Complex (ABC) [33] | Amplify and visualize primary antibody binding | Polymer systems offer high sensitivity; ABC may have endogenous biotin issues [33] |
| Chromogens | DAB (Brown), AP Red, AEC (Red) [33] [34] | Enzyme substrates that produce visible precipitate | DAB is permanent and alcohol-resistant; AEC is alcohol-soluble [33] |
| Counterstains | Hematoxylin, Nuclear Fast Red, Methyl Green [33] [34] | Provide contrast by staining nuclei or other structures | Intensity should be regulated to not obscure specific staining [33] |
Comparative Experimental Data: IHC vs. RNAscope
While IHC remains the gold standard for protein detection, RNA in situ hybridization techniques like RNAscope have emerged as powerful complementary methods. A 2022 study directly compared IHC and RNAscope for detecting UPK2 in urothelial carcinoma tissues, providing valuable comparative data:
Table 4: Comparative Performance of RNAscope and IHC for UPK2 Detection in UC Tissues [4]
| UC Tissue Type | n | UPK2 Positivity by RNAscope | UPK2 Positivity by IHC | P-value |
|---|---|---|---|---|
| Conventional Bladder UC | 127 | 72.4% | 68.5% | 0.511 |
| Variant Bladder UC | 45 | 53.3% | 35.6% | 0.057 |
| Upper Tract UC | 24 | 62.5% | 62.5% | 1.000 |
| Metastatic UC | 23 | 65.2% | 65.2% | 1.000 |
| Overall | 219 | 68.0% | 62.6% | 0.141 |
The study demonstrated no statistically significant difference in overall UPK2 detection rates between the two methods (68.0% vs. 62.6%, P=0.141), with a moderate positive correlation (P<0.001, R=0.441) [4]. However, RNAscope showed a trend toward higher detection rates in variant bladder UC, suggesting potentially enhanced sensitivity for challenging targets [4].
The methodological comparison reveals fundamental differences:
- IHC Workflow: Detects translated proteins using antibody-epitope binding, influenced by fixation, epitope preservation, and protein conformation [33] [23].
- RNAscope Workflow: Targets RNA transcripts through in situ hybridization using a proprietary double-Z probe design that enables single-molecule visualization while preserving tissue morphology [29] [4] [27].
For viral detection, RNAscope offers particular advantages, enabling researchers to identify individual viral particles in infected cells despite low viral loads, differentiate between latent and active infection stages through sense/antisense strand detection, and visualize co-infections with multiple viruses [27].
IHC protocol execution involves a complex interplay of standardized workflows and carefully considered methodological choices. The fundamental steps—tissue preparation, fixation, antigen retrieval, antibody incubation, and detection—remain consistent across applications, but specific variations in section type, detection method, and antibody selection significantly impact outcomes. While IHC continues to be the cornerstone technique for protein localization in tissue context, emerging methodologies like RNAscope offer complementary approaches with potential advantages for specific applications, particularly when analyzing difficult-to-detect targets or when RNA-level information provides valuable pathological insights. Understanding both the core principles and key variations in IHC execution empowers researchers to optimize protocols for their specific experimental needs and properly interpret the resulting data within the context of their research objectives.
The transition from protein-based to RNA-based detection methods represents a significant evolution in molecular pathology. This guide provides a detailed comparison between RNAscope, a quantitative in situ hybridization technique, and semi-quantitative immunohistochemistry (IHC) for biomarker detection in clinical and research applications. Systematic reviews reveal RNAscope demonstrates high concordance (81.8–100%) with PCR-based methods and superior sensitivity in detecting low-abundance targets compared to IHC, which shows lower concordance (58.7–95.3%) due to fundamental differences in detecting proteins versus RNA transcripts. Experimental data across various cancer types and neurological tissues confirm RNAscope's technical advantages while acknowledging IHC's established role in pathological assessment. This objective analysis provides researchers with critical insights for selecting appropriate methodologies based on specific experimental requirements.
RNAscope and immunohistochemistry represent fundamentally different approaches to biomarker detection, each with distinct technical principles and output metrics. IHC detects proteins using antibody-antigen interactions followed by chromogenic or fluorescent detection, with results typically assessed through semi-quantitative scoring systems based on staining intensity and distribution. In contrast, RNAscope employs a novel in situ hybridization approach targeting RNA molecules through proprietary "Z-probe" technology that enables single-molecule visualization through signal amplification [1] [36]. This fundamental difference—detecting proteins versus RNA—underlies the varying performance characteristics observed between these methodologies.
The scoring interpretation differs significantly between these techniques. IHC relies on pathologist evaluation of staining intensity using ordinal scales (typically 0-3+), introducing inherent subjectivity and inter-observer variability [37]. RNAscope generates quantitative data by counting individual RNA transcripts visualized as distinct dots within cells, enabling precise quantification of gene expression at cellular resolution [1] [14]. This technological comparison provides the foundation for understanding their respective applications in research and clinical diagnostics.
Experimental Data and Comparative Performance
Systematic Review Evidence
A comprehensive systematic review evaluating RNAscope in clinical diagnostics compared its performance against established gold standard methods, including IHC, qPCR, qRT-PCR, and DNA ISH [1]. The analysis encompassed 27 retrospective studies, primarily focusing on cancer samples, with results demonstrating striking differences in concordance rates across methodologies:
| Comparison Method | Concordance Rate with RNAscope | Key Findings |
|---|---|---|
| qPCR/qRT-PCR | 81.8-100% | High concordance for gene expression measurement |
| DNA ISH | 81.8-100% | Strong agreement in gene detection |
| IHC | 58.7-95.3% | Lower concordance due to different targets (RNA vs. protein) [1] |
The review concluded that while RNAscope serves as a robust and reliable method that could complement existing gold standard techniques, there were insufficient data to recommend it as a standalone clinical diagnostic tool without further prospective validation [1].
Head-to-Head Method Comparisons
Direct comparative studies provide compelling evidence regarding the relative strengths of each technique. A study of 219 urothelial carcinoma specimens evaluating UPK2 expression found RNAscope detected positivity in 68.0% of cases compared to 62.6% by IHC, though this difference did not reach statistical significance (P=0.141) [3] [4]. The technologies showed moderate positive correlation (P<0.001, R=0.441), with RNAscope demonstrating a trend toward higher detection rates in variant bladder urothelial carcinomas (53.3% vs. 35.6%, P=0.057) [3].
In gastric and gastroesophageal junction adenocarcinoma, a validated DKK1 RNAscope assay demonstrated strong correlation with RNA-Seq data (Spearman's rho=0.86, p<0.0001) and superior sensitivity compared to IHC, detecting RNA in HeLa cell pellets where IHC showed no signal [14]. This enhanced sensitivity for low-abundance targets represents a significant advantage for RNAscope in many research applications.
Experimental Protocols and Methodologies
RNAscope Workflow and Analysis
The RNAscope procedure employs a standardized workflow with critical quality control components:
Sample Preparation: The protocol utilizes formalin-fixed, paraffin-embedded (FFPE) tissues, fresh frozen tissues, or fixed cells mounted on slides [1]. For combined RNAscope/IHC applications in neural tissues, 14-μm thick sections are recommended to preserve tissue integrity through harsh pretreatment steps [36] [2].
Pretreatment and Hybridization: Slides undergo deparaffinization followed by sequential pretreatments including boiling for 20 minutes and protease digestion for 30 minutes at 40°C [3] [4]. Target probes are hybridized for 2 hours at 40°C in a specialized HybEZ oven [3].
Signal Amplification and Detection: The proprietary amplification system employs pairs of "Z-probes" that bind adjacent target sequences, enabling signal amplification through preamplifier and amplifier molecules [1] [36]. Each RNA molecule is visualized as a distinct dot after chromogenic or fluorescent development [1].
Quality Control: Implementation of positive control probes (PPIB, Polr2A, or UBC) verifies RNA integrity, while negative control probes (bacterial dapB gene) confirm absence of background signal [1] [14].
Quantification: Analysis involves counting discrete dots representing individual RNA molecules, performed either manually or using digital image analysis software (HALO, QuPath, Visiopharm) [1] [12] [38].
IHC Protocol and Semi-Quantitative Scoring
Standard IHC methodology follows these essential steps:
Sample Preparation: FFPE tissues are sectioned at 3-5μm thickness, mounted on slides, deparaffinized, and subjected to antigen retrieval to expose epitopes [3] [39].
Antibody Incubation: Primary antibodies specific to the target protein are applied followed by secondary detection systems using enzymatic (HRP) or fluorescent conjugates [37] [36].
Visualization: Chromogenic substrates (DAB) produce brown precipitates at antigen sites, while fluorescent conjugates emit specific wavelengths upon excitation [37] [36].
Semi-Quantitative Scoring: Pathologists evaluate staining intensity using ordinal scales, typically:
- 0: Negative
- 1+: Mild positivity
- 2+: Moderate positivity
- 3+: Strong positivity [37]
Scoring often incorporates the percentage of positive cells, sometimes formalized as H-scores calculating (1×% weak + 2×% moderate + 3×% strong) or Allred scores combining proportion and intensity [39] [14].
Advanced Applications: Combined RNAscope and IHC
Integration of RNAscope with IHC enables simultaneous detection of RNA transcripts and protein expression within the same tissue section, providing powerful insights into gene regulation and cellular heterogeneity [36] [2]. This approach is particularly valuable when investigating challenging targets where antibody specificity is limited or when correlating transcriptional activity with protein production.
A optimized protocol for central nervous system tissues demonstrates this combined approach:
- Tissue Processing: 14-μm thick fixed spinal cord sections collected 7 days post-injury
- Simultaneous Detection: RNAscope for IL-1β and NLRP3 mRNA combined with IHC for cell-type markers (IBA1 for microglia, NeuN for neurons)
- Imaging and Analysis: Confocal microscopy with digital quantification of transcripts within cell-type specific boundaries [36] [2]
This methodology revealed cell-type specific inflammatory responses in pain models, demonstrating that microglia predominantly drive increased inflammatory mRNA expression following neural injury rather than neuronal sources [36].
Analytical Considerations and Digital Analysis
Scoring Subjectivity and Digital Solutions
Semi-quantitative IHC scoring demonstrates significant inter-observer variability, with studies showing poor to moderate inter-rater reliability (Cohen's kappa) and overall agreement (Fleiss' kappa) [37]. This subjectivity stems from the challenging visual discrimination of intensity levels on ordinal scales.
Artificial intelligence-based digital image analysis platforms (Pathronus, Visiopharm, HALO) address these limitations by providing objective, quantitative measurements [37] [38]. These systems employ convolutional neural networks to identify cells of interest, differentiate subcellular compartments, and quantify staining intensity with greater accuracy than semi-quantitative scoring [37]. For RNAscope, digital analysis enables automated dot counting and cell segmentation, particularly valuable in complex tissues like tumor microenvironments [38] [14].
Research Reagent Solutions
| Reagent/Resource | Function | Application Notes |
|---|---|---|
| RNAscope Probe Sets | Target-specific Z-probes | Designed for 20-50 paired probes per target; species-specific [1] |
| Positive Control Probes (PPIB, Polr2A, UBC) | RNA quality verification | PPIB for moderate expression; Polr2A for low expression; UBC for high expression [1] |
| Negative Control Probe (dapB) | Background assessment | Bacterial gene control for nonspecific signal [1] [14] |
| HybEZ Oven System | Controlled hybridization | Essential for proper probe hybridization temperature [3] |
| Digital Analysis Software (HALO, QuPath, Visiopharm) | Objective quantification | Automated dot counting and cell segmentation [1] [12] [38] |
| Multiplex Fluorescent Kits | Multi-target detection | Enable simultaneous detection of multiple RNA/protein targets [36] |
The choice between quantitative RNAscope and semi-quantitative IHC depends on specific research objectives, target characteristics, and analytical requirements. RNAscope offers superior sensitivity for low-abundance targets, precise single-cell quantification, and exceptional specificity through its unique probe design, making it particularly valuable for detecting RNA viruses, low-expression genes, and transcripts where high-specificity antibodies are unavailable [1] [14]. IHC maintains advantages for protein localization, post-translational modifications, and established clinical biomarkers where protein-level correlation is essential [3] [39].
The emerging paradigm favors integrated approaches, combining RNAscope's transcriptional profiling with IHC's protein detection within the same tissue section [36] [2]. This powerful combination preserves spatial context while providing comprehensive molecular insights, particularly valuable for understanding cellular heterogeneity, tumor microenvironments, and complex disease mechanisms. As digital pathology and artificial intelligence platforms continue evolving, both technologies will benefit from enhanced quantification objectivity and analytical sophistication, further advancing their applications in both research and clinical diagnostics [37] [38] [14].
The accurate detection of biomarker expression is fundamental to cancer diagnosis, prognosis, and research. For years, immunohistochemistry (IHC) has been the cornerstone technique for visualizing protein biomarkers in tissue specimens. However, its limitations in sensitivity and specificity, coupled with dependence on antibody quality, can sometimes hinder precise biomarker assessment [1]. The development of RNA in situ hybridization (ISH) techniques, particularly the RNAscope platform, offers a novel approach for directly detecting RNA biomarkers within the morphological context of tissues [7]. This guide provides an objective comparison of RNAscope and IHC, focusing on their application in cancer research, with specific case studies and experimental data. We examine their performance characteristics to help researchers and drug development professionals select the most appropriate method for their investigative needs.
Technology Comparison: Underlying Principles and Workflows
Understanding the fundamental differences in how IHC and RNAscope function is key to interpreting their results.
Immunohistochemistry (IHC)
IHC utilizes antibodies to detect specific protein antigens within tissue sections. The process involves antigen retrieval, application of a primary antibody specific to the protein of interest, and subsequent detection using a labeled secondary antibody and chromogenic or fluorescent substrates. The resulting signal indicates the presence and localization of the target protein [2].
RNAscope In Situ Hybridization
RNAscope is a novel RNA ISH technology that uses a unique double-Z probe design to achieve high specificity and sensitivity [7]. This design strategy allows for simultaneous signal amplification and background suppression, enabling single-molecule visualization while preserving tissue morphology [7]. The probes are designed to hybridize to adjacent stretches of the target RNA sequence. Signal amplification is only initiated when two "Z" probes bind correctly to their target, drastically reducing off-target binding and background noise [2]. Each detected RNA molecule can be visualized as a distinct dot under the microscope [40].
Comparative Workflow
The table below outlines the key procedural steps for both techniques on formalin-fixed, paraffin-embedded (FFPE) tissues, highlighting critical differences.
| Step | RNAscope | Immunohistochemistry (IHC) |
|---|---|---|
| 1. Pretreatment | Specific protease and heat treatment for RNA unmasking [3] | Antigen retrieval for protein epitope unmasking [4] |
| 2. Incubation | Hybridization with target-specific Z-probes [3] | Incubation with primary antibodies [4] |
| 3. Signal Generation | Amplification via pre-amplifier and amplifier sequences, followed by chromogenic or fluorescent label binding [1] | Chromogenic or fluorescent detection via enzyme-conjugated secondary antibodies [2] |
| 4. Detection | Visualization of punctate dots, each representing an individual mRNA transcript [40] | Visualization of diffuse or localized staining representing protein antigen distribution [4] |
Case Study: UPK2 Detection in Urothelial Carcinoma
A direct comparative study provides a concrete example of how these techniques perform in a real-world cancer research setting.
Background and Rationale
Uroplakin 2 (UPK2) is a membrane protein exhibiting excellent specificity for urothelial carcinoma (UC). Its evaluation is useful for diagnosing UC, particularly in metastatic sites or histologic variants where morphological features are ambiguous [3] [4]. However, UPK2 detection by IHC has shown relatively low and variable sensitivity, reported in the range of 44-80% for conventional invasive UC [3] [4]. This limitation prompted an investigation into whether RNAscope could offer improved diagnostic sensitivity for UPK2 detection.
Experimental Methodology
- Samples: The study utilized tissue blocks from 219 UC cases, including 127 conventional bladder UCs, 45 variant bladder UCs, 24 upper tract UCs, and 23 metastatic UCs [3] [4].
- IHC Protocol: Tissue microarrays (TMAs) were stained using an automated system with antibodies against UPK2 (1:100 dilution; BC21). UPK2 expression was scored as positive if cytoplasmic staining was present in UC cells [4].
- RNAscope Protocol: TMAs were analyzed using RNAscope probes targeting UPK2 (NM_006760.4). The procedure included deparaffinization, pretreatments, hybridization with target probes, and signal amplification using the RNAscope 2.0 HD Reagent Kit. UPK2 expression was similarly scored based on the presence of cytoplasmic staining [3] [4].
- Analysis: The sensitivity of each method was evaluated, and comparisons were performed using McNemar's test. Correlation was analyzed using Spearman rank correlation [3].
Results and Comparative Data
The study found no statistically significant difference in the overall UPK2 positivity rate between the two methods (RNAscope: 68.0% vs. IHC: 62.6%, P = 0.141). Correlation analysis revealed a moderate positive correlation (P < 0.001, R = 0.441) [3] [4].
Table: UPK2 Positivity Rates by UC Subtype
| Urothelial Carcinoma Subtype | RNAscope Positivity Rate | IHC Positivity Rate | P-value |
|---|---|---|---|
| Overall UC | 68.0% | 62.6% | 0.141 |
| Conventional Bladder UC | 72.4% | 68.5% | 0.511 |
| Variant Bladder UC | 53.3% | 35.6% | 0.057 |
| Upper Tract UC | 58.3% | 58.3% | 1.000 |
| Metastatic UC | 73.9% | 73.9% | 1.000 |
Despite the lack of overall statistical significance, a strong trend was observed in variant bladder UCs, where RNAscope detected a numerically higher positivity rate (53.3% vs. 35.6%, P = 0.057) [3]. This suggests that RNAscope may offer a sensitivity advantage in diagnostically challenging cases.
Performance Analysis Across Cancers
Evidence from a systematic review and other cancer types supports the general performance characteristics of RNAscope.
Concordance with Gold Standard Methods
A systematic review of 27 studies compared RNAscope with established techniques like IHC, qPCR, and DNA ISH [1]. The review confirmed RNAscope as a highly sensitive and specific method with high concordance rates (CR):
- qPCR, qRT-PCR, and DNA ISH: Concordance rates ranged from 81.8% to 100% [1].
- IHC: Concordance with IHC was lower, ranging from 58.7% to 95.3% [1]. This is expected, as the two techniques measure different biomolecules (RNA vs. protein), and discrepancies can arise from post-transcriptional regulation and technical factors like antibody quality.
Applications in Other Carcinomas
- Lung Adenocarcinoma: RNAscope has been shown to be more sensitive than IHC for detecting markers like thyroid transcription factor-1 (TTF-1) and Napsin A. It may be considered for patients clinically suspected of having lung adenocarcinoma but who test negative by IHC [3] [4].
- Hepatocellular Carcinoma: RNAscope improves the specificity and sensitivity for Glypican3 (GPC3) and glutamine synthetase (GS) by 20-30%, potentially enabling earlier diagnosis [3] [4].
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful implementation of these techniques requires specific reagents and tools. Below is a list of essential items for a typical RNAscope experiment on FFPE tissues, as used in the featured case study and related protocols.
Table: Key Research Reagent Solutions for RNAscope
| Item | Function/Description | Example/Reference |
|---|---|---|
| RNAscope Probe | Target-specific "Z" probe pair designed to hybridize to the mRNA of interest. | UPK2 probe (NM_006760.4) [3] |
| Detection Kit | Contains reagents for signal amplification and chromogenic/fluorescent development. | RNAscope 2.0 HD Reagent Kit-BROWN [3] |
| Positive Control Probe | Validates assay success; detects a ubiquitous housekeeping gene. | PPIB, Polr2A, or UBC [1] |
| Negative Control Probe | Confirms absence of background noise; targets a bacterial gene. | Bacterial dapB gene [1] |
| HybEZ Oven | Specialized hybridization oven providing precise temperature control. | HybEZ Oven (ACD) [3] |
| Protease | Enzyme for tissue permeabilization and RNA unmasking. | RNAscope Protease IV [40] |
| Image Analysis Software | For quantification of RNA dots and positive cell analysis. | HALO, QuPath, Aperio [1] [40] [12] |
Integrated Analysis and Workflow Recommendations
Choosing the Right Technique
The decision to use RNAscope or IHC depends on the research question, the biomarker, and available resources.
- Choose IHC when: The target is a protein, and well-validated, specific antibodies are available. IHC is a established, widely used, and often more cost-effective for routine protein detection.
- Choose RNAscope when:
A Complementary Approach
Rather than being purely competitive, RNAscope and IHC are often complementary. A combined protocol allows for the simultaneous detection of RNA and protein within the same tissue section [2]. This powerful approach can, for example, localize cytokine mRNA (via RNAscope) within specific, protein-defined cell types like neurons or microglia (via IHC), providing a more complete picture of gene expression and cellular function [2].
Quantitative Analysis and Standardization
A significant advantage of RNAscope is the potential for precise quantification. Each fluorescent or chromogenic dot represents a single RNA molecule, allowing for digital counting [40]. Open-source software like QuPath can be used to create automated workflows for detecting transcript-positive cells and establishing rigorous quantification thresholds, improving reproducibility across studies [40].
The accurate detection of viral pathogens is a cornerstone of infectious disease research and clinical diagnostics. Traditional methods, including immunohistochemistry (IHC) and conventional in situ hybridization (ISH), have provided valuable tools but come with significant limitations in sensitivity, specificity, and the ability to provide morphological context. The emergence of RNA in situ hybridization (ISH) technologies, particularly the RNAscope assay, represents a paradigm shift in viral detection capabilities. This guide provides a comprehensive comparison between RNAscope and IHC, focusing on their application in viral pathogen detection for researchers, scientists, and drug development professionals. We present experimental data, detailed methodologies, and analytical frameworks to inform technology selection for infectious disease research, framed within the broader thesis of comparing the sensitivity and specificity of these fundamental detection platforms.
The critical importance of precise viral detection has been underscored by recent global health challenges. Traditional diagnostic approaches include agent cultivation, serological detection of immune responses, and visualization through staining techniques [41]. While IHC allows for protein detection within tissue morphology, it often suffers from limited sensitivity and depends on antibody availability and quality [27] [1]. In contrast, RNAscope ISH targets viral RNA sequences directly, offering a complementary approach that maintains morphological context while potentially overcoming many limitations of antibody-based methods.
Technology Comparison: RNAscope vs. Immunohistochemistry (IHC)
Fundamental Principles and Mechanisms
Immunohistochemistry (IHC) is a well-established technique that utilizes antibodies to detect specific protein antigens within tissue sections. The binding of primary antibodies to their target antigens is typically visualized using enzyme-conjugated or fluorescent-labeled secondary antibodies, allowing for microscopic observation of protein localization and distribution [2]. While highly valuable for protein detection, IHC's effectiveness is inherently limited by antibody specificity, affinity, and the stability of the target protein epitopes, particularly in formalin-fixed, paraffin-embedded (FFPE) tissues [1].
RNAscope In Situ Hybridization represents a significant advancement over traditional ISH methods. This technology employs a novel probe design utilizing double "Z" probes that specifically bind to adjacent regions of the target RNA sequence [1] [2]. This proprietary design requires simultaneous binding of two independent probes for signal amplification to occur, dramatically reducing background noise and non-specific binding. The subsequent hybridization of pre-amplifier and amplifier molecules creates a signal amplification system capable of detecting individual RNA molecules while preserving tissue morphology [1]. Each detected dot corresponds to a single RNA molecule, enabling both qualitative localization and quantitative assessment [1].
Table 1: Technical Comparison of RNAscope and IHC
| Feature | RNAscope | Immunohistochemistry (IHC) |
|---|---|---|
| Target Molecule | RNA (mRNA, viral RNA) | Proteins |
| Sensitivity | Single-molecule detection [1] [2] | Limited by antibody affinity and amplification method |
| Specificity | High (double Z-probe design) [27] [1] | Variable (dependent on antibody quality) |
| Morphological Context | Preserved | Preserved |
| Probe/Antibody Availability | Custom probes designed in 2 weeks [27] | Dependent on existing antibodies; custom development lengthy |
| Multiplexing Capacity | High (multiple channels available) [27] [1] | Moderate (limited by antibody host species and color overlap) |
| Turnaround Time | Single-day workflow [27] | 1-2 days typically |
| Key Advantage | Direct genetic material detection, high specificity | Direct protein detection, established protocols |
Performance Data: Sensitivity and Specificity in Detection
Comparative studies across various research applications consistently demonstrate the enhanced performance characteristics of RNAscope. A systematic review evaluating RNAscope in clinical diagnostics found it to be a "highly sensitive and specific method" with high concordance rates (81.8–100%) with PCR and qRT-PCR techniques [1]. While the concordance with IHC was lower (58.7–95.3%), this discrepancy primarily reflects the fundamental difference in the molecules being detected (RNA versus protein) rather than a failure of either technology [1].
In the context of urothelial carcinoma (UC), a study directly comparing RNAscope and IHC for detecting UPK2 status found no significant difference in overall positivity rates (68.0% vs. 62.6%, P = 0.141), though there was a trend toward higher detection with RNAscope in variant bladder UCs (53.3% vs. 35.6%, P = 0.057) [4]. The correlation between the two methods was moderate (P < 0.001, R = 0.441), suggesting that while related, these techniques provide complementary information [4].
For viral detection specifically, RNAscope offers unrivaled sensitivity and specificity according to the manufacturer, with the ability to identify individual viral particles despite low viral loads [27]. Its proprietary probe design enables accurate differentiation among highly related viral species and strains, addressing a critical need in managing emerging viral threats.
Table 2: Experimental Detection Rates in Comparative Studies
| Study Context / Tissue Type | Detection Rate: RNAscope | Detection Rate: IHC | Statistical Significance (P-value) |
|---|---|---|---|
| Overall Urothelial Carcinoma (UC) [4] | 68.0% | 62.6% | P = 0.141 (NS) |
| Conventional Bladder UC [4] | 72.4% | 68.5% | P = 0.511 (NS) |
| Variant Bladder UC [4] | 53.3% | 35.6% | P = 0.057 (NS, trend) |
| Upper Tract UC [4] | Not significantly different | Not significantly different | P = 1.000 (NS) |
| Metastatic UC [4] | Not significantly different | Not significantly different | P = 1.000 (NS) |
| Systematic Review (vs. PCR/qPCR) [1] | 81.8-100% Concordance | Not Applicable | High Concordance |
Experimental Protocols for Viral Detection
RNAscope Assay Workflow for Viral Pathogens
The RNAscope procedure follows a standardized workflow that can be completed within a single day [27]. For formalin-fixed, paraffin-embedded (FFPE) tissues, the process begins with slide preparation and sectioning, followed by deparaffinization and rehydration [4]. Key steps include:
Pretreatment: Slides undergo a series of pretreatments to expose target RNA sequences. This includes heat-induced epitope retrieval and protease digestion to permeabilize tissues without degrading the RNA targets [4] [1]. The protease treatment time must be carefully optimized based on fixation conditions to ensure adequate penetration while maintaining tissue integrity.
Hybridization: Target probes specific to the viral RNA sequence are applied and hybridized for 2 hours at 40°C in a HybEZ oven [4]. The proprietary "Z" probe design ensures specific binding only when two adjacent probes hybridize correctly to the target sequence.
Signal Amplification: A series of amplifier molecules are hybridized sequentially to build the signal amplification structure. This multi-step process significantly enhances sensitivity while maintaining low background through the requirement for paired probe binding [1].
Signal Detection: Chromogenic or fluorescent labels are applied for visualization. For fluorescent detection, multiple channels can be used simultaneously to detect different viral targets or host cell markers [27] [2].
Counterstaining and Mounting: Tissues are counterstained (e.g., with DAPI for nuclear visualization in fluorescent assays) and mounted for microscopy [27].
Throughout the process, appropriate controls are essential. Negative control probes targeting bacterial genes (e.g., dapB) confirm the absence of background signal, while positive control probes targeting housekeeping genes (e.g., PPIB, POLR2A) verify RNA integrity and assay performance [1].
Combined RNAscope and Immunohistochemistry Protocol
For sophisticated analyses requiring correlation of viral presence with cell-type specific markers, RNAscope and IHC can be combined on the same tissue section [2] [36]. The following protocol has been optimized for thicker (14μm) fixed tissue sections, such as those from the central nervous system:
Tissue Preparation: Collect tissue via standard methods, including perfusion fixation with 4% paraformaldehyde. For spinal cord tissue, post-fix by immersion in 4% paraformaldehyde for 4 hours at 4°C [36]. Cryoprotect tissues, then section at 14-20μm thickness using a cryostat.
Slide Preparation: Bake slides at 60°C for 1 hour to ensure tissue adhesion. This step is particularly important for tissue types prone to detachment during subsequent heat treatments [2].
RNAscope Pretreatment: Follow standard RNAscope pretreatment steps, including alcohol dehydration, hydrogen peroxide treatment, and target retrieval. Protease treatment time may require optimization based on tissue type and thickness [2].
Probe Hybridization and Amplification: Perform the standard RNAscope hybridization and amplification steps according to the manufacturer's protocol for the target viral RNA.
Immunohistochemistry: Following completion of the RNAscope signal development, proceed with standard IHC protocols. This includes blocking with appropriate serum, incubation with primary antibodies (e.g., IBA1 for microglia, NeuN for neurons) overnight at 4°C, followed by incubation with fluorescently-labeled secondary antibodies [2] [36].
Microscopy and Analysis: Image stained sections using confocal microscopy. Analyze images using software such as QuPath, Imaris, or Halo to quantify viral RNA transcripts within specific cell populations identified by IHC markers [40] [2] [12].
This combined approach enables researchers to precisely identify which cell types are infected by viruses and how infection correlates with changes in cell-specific marker expression.
Essential Research Reagent Solutions
Successful implementation of viral detection assays requires specific reagents and tools. The following table outlines essential materials and their functions for establishing RNAscope and IHC workflows in infectious disease research.
Table 3: Essential Research Reagents for Viral Detection Studies
| Reagent / Tool | Function / Purpose | Example Products / Targets |
|---|---|---|
| RNAscope Probe Sets | Target-specific probes designed to detect viral RNA sequences | HCV mRNA [27]; Custom viral probes [27] |
| RNAscope Detection Kit | Contains amplifiers, labels, and reagents for signal development | RNAscope Fluorescent Multiplex Kit [40] [2] |
| Positive Control Probes | Verify RNA integrity and assay performance; housekeeping genes | PPIB, POLR2A, UBC [1] |
| Negative Control Probes | Assess background noise; bacterial genes not in mammalian tissues | Bacterial dapB [1] |
| Primary Antibodies (IHC) | Detect cell-type specific protein markers for phenotyping | IBA1 (microglia), NeuN (neurons) [2] [36] |
| Fluorescent Secondaries | Visualize primary antibody binding with different fluorophores | Alexa Fluor conjugates [2] |
| Protease Reagent | Tissue permeabilization while preserving RNA integrity | RNAscope Protease IV [40] |
| Hybridization Oven | Maintain precise temperature for probe hybridization | HybEZ II System [40] |
| Mounting Medium | Preserve fluorescence and protect samples during microscopy | Fluoromount with DAPI [40] [2] |
| Image Analysis Software | Quantify transcripts and analyze co-localization | QuPath, HALO, Imaris [40] [1] [12] |
Analysis and Data Interpretation
Quantification Methods for RNAscope Data
Analysis of RNAscope results involves quantifying the number of distinct dots within cells or tissue regions, with each dot representing an individual RNA molecule [1]. This can be performed through several approaches:
Manual Scoring: Following manufacturer guidelines, multiple regions of the slide are assessed to obtain a comprehensive result. Scoring systems typically consider the number of dots per cell or the percentage of positive cells within a population [1].
Automated Quantitative Analysis: For robust, high-throughput quantification, digital image analysis software such as QuPath, HALO, or Aperio is recommended [40] [1] [12]. These platforms enable:
- Cell detection and segmentation based on nuclear or cytoplasmic markers
- Automated dot counting within defined cellular boundaries
- Spatial analysis of transcript distribution within tissues
- Multiplex analysis for co-localization studies
The open-source software QuPath offers particular utility for automated cell detection and dot quantification, especially in complex tissues like brain [40]. The protocol involves creating scripts to optimize cell detection parameters and establishing mRNA signal thresholds using negative controls to ensure accurate positive cell identification [40].
For combined RNAscope-IHC assays, confocal microscopy and 3D rendering software like Imaris can be used to quantify RNA transcripts within IHC-labeled cellular boundaries, providing precise cell-type specific viral load information [2] [36].
Validation and Quality Control
Rigorous validation is essential for reliable viral detection. The RNAscope assay incorporates built-in control measures:
- Positive Controls: Housekeeping genes (PPIB, POLR2A, UBC) confirm tissue RNA integrity and assay functionality [1].
- Negative Controls: The bacterial dapB gene probe assesses non-specific background signal [1].
- Probe Specificity: The double Z-probe design inherently minimizes off-target binding, with in silico specificity verification during probe design [27] [1].
For diagnostic applications, the systematic review by Althobiti et al. (2021) concluded that while RNAscope is a "reliable and robust method," further prospective studies are needed to fully validate the technique for standalone clinical use [1]. Currently, it serves as an excellent complementary technique to confirm unclear results from gold standard methods.
The comparative analysis presented in this guide demonstrates that RNAscope and IHC offer complementary approaches for viral pathogen detection in infectious disease research. RNAscope provides superior sensitivity and specificity for direct RNA detection, while IHC remains valuable for protein localization and cell phenotyping. The integration of these techniques through combined protocols enables sophisticated analyses of viral tropism, host-pathogen interactions, and cellular responses to infection.
Future developments in viral detection will likely focus on enhanced multiplexing capabilities, automated platforms for high-throughput analysis, and the integration of spatial transcriptomics with protein expression data. As noted by the Center for Infectious Disease Diagnostics & Innovation, host gene expression signatures offer promising diagnostic potential, though translation to clinical practice requires development of rapid, simple testing platforms [42]. RNAscope technology, with its continuing evolution and adaptability, represents a powerful tool in this expanding horizon of infectious disease research and diagnostics.
Maximizing Assay Performance: Critical Success Factors and Pitfall Avoidance
The accuracy and reliability of spatial biology techniques, such as RNAscope for in situ hybridization (ISH) and Immunohistochemistry (IHC), are profoundly influenced by pre-analytical conditions. Tissue fixation and processing are not merely preliminary steps but are critical determinants of the sensitivity and specificity of the final data. This guide objectively compares the performance of RNAscope and IHC under various pre-analytical protocols, providing experimental data to inform best practices for researchers and drug development professionals.
Core Technology Comparison: RNAscope vs. IHC
RNAscope and IHC provide complementary information—detecting RNA transcripts and protein antigens, respectively—but their optimal performance hinges on different, and sometimes competing, fixation and processing requirements. The table below summarizes their key characteristics and performance metrics.
| Feature | RNAscope | Traditional IHC |
|---|---|---|
| Target Molecule | RNA | Protein |
| Key Principle | Signal amplification via "Z probe" pairs and amplifier structures [36]. | Antigen-antibody binding with chromogenic or fluorescent detection. |
| Key Fixation Parameter | Fixation Duration: Optimal results with a controlled, relatively short post-fixation (e.g., 4 hours) [36]. | Fixation Duration: Can tolerate longer fixation, but over-fixation can mask epitopes. |
| Key Processing Parameter | Protease Treatment: Requires precise optimization; over-digestion destroys RNA and tissue architecture, under-digestion limits probe access [36]. | Antigen Retrieval: Often requires harsh heat-induced epitope retrieval (HIER) to reverse cross-links from over-fixation. |
| Major Pre-Analytical Challenge | RNA degradation by RNases during tissue collection and processing. | Protein epitope masking or alteration due to excessive cross-linking from prolonged fixation. |
| Demonstrated Sensitivity | Single-molecule detection under optimized conditions [36]. | Varies widely based on antibody affinity and pre-analytical conditions. |
| Demonstrated Specificity | High; the double-Z probe design minimizes off-target binding [36]. | Varies; subject to non-specific antibody binding and cross-reactivity. |
Experimental Protocols for Direct Comparison
To rigorously compare the sensitivity and specificity of RNAscope and IHC, a standardized protocol for co-detection in the same tissue section is essential. The following methodology, adapted from current research, allows for a direct, head-to-head evaluation [36].
Title: Combined RNAscope and IHC Protocol on Fixed CNS Tissue
Workflow Diagram:
Detailed Methodology:
- Tissue Collection and Fixation:
- Transcardially perfuse animals with ice-cold saline followed by 4% paraformaldehyde (PFA) in phosphate buffer (PB), pH 7.4.
- Dissect out the target tissue (e.g., spinal cord) and post-fix by immersion in 4% PFA for 4 hours at 4°C. This controlled duration is critical for preserving both RNA integrity and protein epitopes [36].
- Cryoprotect the tissue, then section on a cryostat at a thickness of 14 µm and mount on slides.
Protease Treatment:
- Following the RNAscope protocol, treat slides with a proprietary protease. The concentration and duration must be carefully optimized for the specific tissue type to permit probe access without destroying morphology or RNA signals [36].
RNAscope In Situ Hybridization:
- Apply target-specific "Z probes" hybridizing to the RNA of interest (e.g., IL-1β or NLRP3).
- Perform the amplification steps as per the manufacturer's instructions to generate a fluorescent signal [36].
Immunohistochemistry:
- Immediately following the RNAscope procedure, incubate the sections with primary antibodies against cell-type-specific markers (e.g., IBA1 for microglia, NeuN for neurons).
- Use appropriate fluorescently conjugated secondary antibodies for detection [36].
Image Acquisition and Analysis:
- Image the stained sections using confocal microscopy.
- Quantify RNA transcript dots within the boundaries of IHC-labeled cells using image analysis software (e.g., Imaris) to determine cell-type-specific gene expression [36].
Quantitative Data from Comparative Studies
Application of the above protocol in a model of neuropathic pain (chronic constriction injury) yielded the following quantitative data, highlighting the nuanced information gained from RNAscope.
Table: Cell-Type-Specific mRNA Expression Changes in Rat Spinal Cord Data derived from reference [36].
| Gene Target | Cell Type (IHC Marker) | Condition | Change in Transcript Density |
|---|---|---|---|
| IL-1β | Microglia (IBA1) | 7 days post-injury (Ipsilateral) | Significant Increase |
| IL-1β | Neurons (NeuN) | 7 days post-injury (Ipsilateral) | No Significant Change |
| NLRP3 | Microglia (IBA1) | 7 days post-injury (Ipsilateral) | Significant Increase |
| NLRP3 | Neurons (NeuN) | 7 days post-injury (Ipsilateral) | No Significant Change |
This data demonstrates that the observed increase in inflammatory genes is primarily due to upregulated transcription within microglia, a finding that would be impossible to ascertain with IHC alone or bulk RNA analysis [36].
The Scientist's Toolkit: Essential Research Reagent Solutions
The following reagents and tools are critical for successfully executing and analyzing the combined RNAscope and IHC protocol.
| Item | Function | Example / Note |
|---|---|---|
| Paraformaldehyde (PFA) | Cross-linking fixative that preserves cellular morphology and immobilizes biomolecules. | Use 4% in neutral phosphate buffer; fixation time is a critical variable [36]. |
| Protease | Digests proteins to expose target RNA for probe hybridization. | RNAscope protease is optimized for the assay; concentration and time must be titrated [36]. |
| RNAscope Probe Sets | Target-specific "Z probes" for RNA detection. | Designed against specific gene sequences (e.g., IL-1β, NLRP3) with built-in signal amplification [36]. |
| Primary Antibodies | Bind specifically to protein epitopes for cell identification. | Validate for IHC on fixed tissue (e.g., anti-IBA1 for microglia, anti-NeuN for neurons) [36]. |
| Fluorescent Secondaries | Detect primary antibodies for visualization. | Must be highly cross-adsorbed to minimize non-specific binding in multiplex assays. |
| Mounting Medium | Preserves fluorescence and allows for coverslipping. | Use an anti-fade medium. |
| Confocal Microscope | High-resolution imaging for co-localization analysis. | Essential for resolving single RNA transcripts within specific cell boundaries [36]. |
The quest for superior sensitivity and specificity in spatial biology is fundamentally a pre-analytical challenge. RNAscope offers exceptional specificity for RNA detection but is highly sensitive to fixation and protease digestion. IHC provides robust protein localization but is vulnerable to epitope masking. The experimental data presented confirms that a meticulously optimized, combined protocol can successfully resolve the competing requirements of both techniques, enabling precise, cell-type-specific molecular phenotyping that is indispensable for advanced research and drug development.
In the evolving field of molecular pathology, the debate between RNA in situ hybridization (RNAscope) and immunohistochemistry (IHC) for biomarker detection centers on fundamental differences in sensitivity, specificity, and technical requirements. While IHC detects proteins using antibody-antigen interactions, RNAscope directly targets RNA transcripts within intact cells using a proprietary signal amplification system. [2] This technological distinction creates a different profile of common technical challenges. This guide systematically addresses troubleshooting RNAscope assays, providing direct experimental comparisons with IHC performance and data-driven protocols to optimize results for research and diagnostic applications.
Analytical Comparison: RNAscope vs. IHC Technical Performance
Extensive research has quantified the performance characteristics of RNAscope relative to IHC across multiple biomarkers and cancer types. The core differences stem from their distinct detection mechanisms.
Table 1: Methodological Comparison of RNAscope and IHC
| Parameter | RNAscope | Immunohistochemistry (IHC) |
|---|---|---|
| Target Molecule | RNA transcripts | Protein antigens |
| Detection Mechanism | In situ hybridization with signal amplification | Antibody binding with enzymatic detection |
| Key Technical Differentiators | Requires protease digestion; No RNase-free environment needed | Antigen retrieval methods vary; No protease typically needed |
| Signal Interpretation | Discrete dots representing individual RNA molecules [43] [44] | Diffuse cytoplasmic, nuclear, or membranous staining |
| Specificity Control | Background suppression technology; negative control probe (dapB) [43] | Relies on antibody specificity; isotype controls |
| Quantification Approach | Semi-quantitative scoring based on dots/cell [43] [44] | Semi-quantitative based on staining intensity and percentage |
Clinical validation studies demonstrate that RNAscope provides higher specificity for certain applications. In HPV detection for oropharyngeal squamous cell carcinoma, RNAscope demonstrated significantly higher specificity compared to p16 IHC, which serves as a surrogate marker, thereby reducing false positives. [21] For estrogen receptor α (ERα) detection in breast cancer, RNAscope showed high concordance with IHC while revealing potential false-negative cases in IHC-based testing, with the added benefit of lower background staining. [45]
Correlation studies between RNA sequencing and IHC further validate RNA-based methods, showing strong correlations (coefficients 0.53-0.89) for key cancer biomarkers including ESR1, PGR, and ERBB2, confirming that mRNA levels generally reflect protein expression for many clinically relevant targets. [28]
Troubleshooting Framework: Systematic Problem Resolution
The most common RNAscope challenges generally stem from three main procedural areas: sample preparation, pretreatment optimization, and assay execution. The following diagram maps the logical troubleshooting path for resolving these core issues.
Problem 1: No Signal Detection
Root Causes and Experimental Evidence
Complete absence of signal typically indicates failed probe access to target RNA or compromised assay reagents. Studies validating RNAscope for clinical applications emphasize that proper sample pretreatment is the most critical factor. [45]
Detailed Protocol for Resolution
- Validate Sample RNA Integrity: Always run positive control probes (PPIB, POLR2A, or UBC) alongside experimental samples. [43] [44] Successful PPIB staining should generate a score ≥2, and UBC should score ≥3 with relatively uniform signal throughout the sample. [43] [44]
- Verify Technical Execution: Ensure all amplification steps were performed in correct sequence; omitting any step will result in no signal. [44] Flick or tap slides to remove residual reagent, but never let slides dry out during the procedure. [43] [44]
- Optimize Pretreatment Conditions: For over-fixed tissues, increase protease digestion time in increments of 10 minutes while maintaining 40°C temperature. [43] [44] Alternatively, increase epitope retrieval time (5-minute increments at 95°C) for the Leica BOND RX system. [43] [44]
- Confirm Reagent Quality: Warm probes and wash buffer to 40°C before use, as precipitation during storage can affect assay results. [43] Use only fresh reagents, including ethanol and xylene. [44]
Problem 2: High Background Staining
Root Causes and Experimental Evidence
Excessive non-specific signal compromises interpretation and often results from over-digestion or suboptimal washing. The RNAscope platform incorporates background suppression technology, but proper optimization remains essential. The typical negative control probe (dapB) should yield a score of <1 in properly fixed tissue. [43] [44]
Detailed Protocol for Resolution
- Establish Background Baseline: Run negative control probe (dapB) on consecutive sections to quantify non-specific signal. [43] [44] A dapB score ≥1 indicates unacceptable background requiring optimization.
- Reduce Protease Digestion: For under-fixed tissues, decrease protease treatment time by 5-10 minute increments while maintaining 40°C temperature. [43] [44] On Leica systems, consider milder pretreatment: 15 min ER2 at 88°C and 15 min Protease at 40°C. [43] [44]
- Optimize Wash Conditions: Use fresh 1X Wash Buffer for all procedures. [43] For automated systems, ensure bulk solutions are replaced with recommended buffers and lines are purged regularly. Decontaminate instruments every three months to prevent microbial growth. [43] [44]
- Validate Hydrophobic Barrier: Use only ImmEdge Hydrophobic Barrier Pen, as other pens may not maintain barrier integrity throughout the procedure, leading to evaporation and increased background. [43]
Problem 3: Poor Tissue Morphology
Root Causes and Experimental Evidence
Compromised cellular architecture typically results from excessive protease treatment or inappropriate fixation. Studies combining RNAscope with IHC emphasize that morphology preservation is essential for accurate cellular localization. [2]
Detailed Protocol for Resolution
- Assess Fixation Quality: Follow recommended fixation in fresh 10% Neutral Buffered Formalin for 16-32 hours. [43] Under-fixed tissue appears faded with loss of cell borders. [46]
- Reduce Protease Digestion: Over-digested tissue shows poor morphology with loss of cellular detail. Reduce protease time by 10-minute increments while monitoring morphology improvement. [43] [46]
- Verify Slide Specifications: Use only Superfrost Plus slides, as other slide types may result in tissue detachment during stringent heating and washing steps. [43]
- Optimize Retrieval Conditions: For automated systems, adjust retrieval time and temperature. Standard pretreatment is 15 minutes ER2 at 95°C, but reduced temperature (88°C) can preserve morphology in delicate tissues. [43] [44]
Multiplex Assay and Automation Considerations
Specialized Troubleshooting for Complex Applications
Table 2: Advanced Application Specifications
| Application | Key Requirement | Specification | Validation Approach |
|---|---|---|---|
| RNAscope 2-plex/ Multiplex | Probe Mixing Ratio | C2:C1 = 1:50 [43] [44] | Confirm distinct cellular patterns for each target |
| Automated Platform (Ventana) | Software Settings | Disable "Slide Cleaning" option [43] | Compare with manual control slides |
| Automated Platform (Leica) | Detection Chemistry | BOND Polymer Refine Detection kits only [43] [44] | Verify with system-specific controls |
| Fluorescent Detection | Mounting Medium | ProLong Gold Antifade Mountant [44] | Assess signal preservation over time |
Essential Research Reagent Solutions
The following reagents and equipment are critical for successful RNAscope implementation based on published methodologies and manufacturer specifications:
Table 3: Essential Research Reagents and Equipment
| Reagent/Equipment | Function | Specification | Experimental Reference |
|---|---|---|---|
| HybEZ Hybridization System | Maintains optimum humidity and temperature | Required for manual and automated hybridization | [43] [44] |
| Positive Control Probes (PPIB, POLR2A, UBC) | Assess sample RNA quality and pretreatment | PPIB: 10-30 copies/cell; UBC: high copy number | [43] [44] |
| Negative Control Probe (dapB) | Determine background staining | Bacterial gene; should not generate mammalian signal | [43] [44] |
| Superfrost Plus Slides | Prevent tissue detachment | Specific surface treatment for adhesion | [43] |
| ImmEdge Hydrophobic Barrier Pen | Create reagent containment zones | Maintains barrier throughout procedure | [43] |
| Protease Solution | Tissue permeabilization | Concentration and time require optimization | [43] [46] |
| Assay-Specific Mounting Media | Preserve staining and enable visualization | Varies by detection method (chromogenic/fluorescent) | [43] [44] |
Troubleshooting RNAscope assays requires methodical attention to sample preparation and pretreatment optimization. The technology offers distinct advantages for direct RNA visualization with single-molecule sensitivity, addressing key limitations of IHC including antibody specificity and subjective interpretation. [21] [45] The standardized quantitative nature of RNAscope—counting discrete dots per cell rather than interpreting staining intensity—provides a more objective assessment framework. [43] [44]
For researchers transitioning from IHC, incorporating systematic validation with both positive and negative controls is essential. The established correlation between mRNA and protein expression for many biomarkers supports RNAscope's utility as either a complementary or primary detection method. [28] [45] Following the detailed protocols outlined above will resolve most common challenges and ensure robust, reproducible results that advance the precision of molecular pathology in both research and clinical applications.
In the evolving landscape of molecular diagnostics and research, the transition from immunohistochemistry (IHC) to RNA in situ hybridization (ISH) techniques like RNAscope represents a significant advancement in sensitivity and specificity. However, this technological shift demands even more rigorous validation and quality control practices. The reproducibility crisis in biomedical research, partly attributed to poorly characterized antibodies, underscores the need for robust, standardized controls [47]. Within the RNAscope platform, a specific quartet of control probes—PPIB, POLR2A, UBC, and DapB—has emerged as essential for validating experimental outcomes. This article delineates the critical function of each control within the context of a broader thesis on RNAscope's superior performance versus traditional IHC, providing researchers with the experimental framework necessary for generating reliable, publication-quality data.
The Control Quartet: Functions and Specifications
The RNAscope assay employs a multi-faceted control strategy to ensure both technical success and sample quality. The recommended controls serve two primary levels of quality assurance: a technical assay control check to verify the protocol is performed correctly, and a sample/RNA quality control check to confirm the integrity of the tissue under investigation [48].
The table below summarizes the core characteristics and applications of the four key control targets.
Table 1: RNAscope Control Probes: Functions and Specifications
| Control Target | Full Name & Function | Expression Level | Recommended Application |
|---|---|---|---|
| PPIB | Peptidylprolyl Isomerase B: A medium-copy housekeeping gene used to verify sample RNA integrity and assay technique [48]. | Medium (10-30 copies/cell) [48] | The most flexible and widely recommended positive control for most tissues [48]. |
| POLR2A | DNA-directed RNA Polymerase II subunit RPB1: A low-copy housekeeping gene serving as a rigorous positive control [48]. | Low (3-15 copies/cell) [48] | Ideal for use with low-expression targets or in proliferating tissues like tumors [48]. |
| UBC | Ubiquitin C: A high-copy housekeeping gene used to confirm general RNA quality [48]. | Medium/High (>20 copies/cell) [48] | Use with high-expression targets only. Not recommended for low-copy targets as it may give false negatives due to its resilience [48]. |
| DapB | Dihydrodipicolinate Reductase: A bacterial gene not present in mammalian tissues; the universal negative control [48]. | Not Applicable | Used in every experiment to assess non-specific background staining. Clean staining is imperative for valid results [48]. |
Experimental Data and Protocol Validation
Quantitative Data from Controlled Experiments
The utility of these controls is demonstrated in rigorous experimental settings. A study investigating RNA detection in mouse brain tissue with post-mortem delays used the positive control mixture (Polr2a, PPIB, and UBC) and the negative control DapB to systematically assess RNA degradability. The research confirmed that ubiquitously expressed RNAs were reliably detected even 24 hours post-mortem, with signal quantification providing a clear metric for RNA quality over time [49].
Furthermore, in a CLIA-guided validation of a DKK1 RNAscope assay for gastric cancer, the protocol mandated that all tumor resections must demonstrate "adequate RNA integrity and acceptable background" as determined by the presence of PPIB signal and the absence of DapB signal, respectively [14]. This control step was a prerequisite for any subsequent diagnostic scoring.
Detailed Experimental Protocol for Control Implementation
The following workflow, as applied in validated studies, details how to incorporate these controls into a standard RNAscope experiment [14] [49].
Table 2: Detailed Experimental Workflow for RNAscope Controls
| Step | Procedure | Purpose & Quality Check |
|---|---|---|
| 1. Sample Preparation | - Generate FFPE tissue sections.- Perform deparaffinization and rehydration. | Standard tissue preparation for ISH. |
| 2. Technical Control Setup | - For a new protocol or tissue type, run two parallel slides: 1. Slide with PPIB (or POLR2A) positive control probe. 2. Slide with DapB negative control probe. | Verifies the assay is performing correctly. A successful run shows strong PPIB/POLR2A staining and clean DapB background [48]. |
| 3. Pretreatment Optimization | - Use the control slides to empirically optimize pretreatment conditions (e.g., time, temperature) for your specific tissue. | Fixation and tissue quality vary. Optimization ensures maximal RNA exposure while minimizing damage. |
| 4. Experimental Assay | - Run the full RNAscope assay with your target-specific probe(s).- Include PPIB and DapB on every experimental slide or a representative slide from the same batch. | PPIB confirms sample-specific RNA quality is adequate for the target. DapB confirms the absence of significant background in the final experiment [14]. |
| 5. Image Acquisition & Analysis | - Image using confocal or brightfield microscopy.- Use analysis software (e.g., QuPath [50]) to quantify transcripts/cell for the target and PPIB. | Quantification provides objective data. Sample acceptance criterion: PPIB signal ≥ 4 dots/cell and minimal DapB signal [14]. |
Diagram 1: RNAscope Control Workflow Decision Tree
RNAscope vs. IHC: A Thesis of Superiority Anchored by Controls
The core advantage of RNAscope over IHC, often cited in the literature, is its high sensitivity and specificity, which is fundamentally enabled by a rigorous control system that IHC lacks. While IHC suffers from a "reproducibility crisis" due to antibody variability, batch-to-batch inconsistencies, and a lack of standardization, RNAscope's nucleic acid-based approach offers a more reliable path to validation [47].
- Sensitivity and Specificity: RNAscope's proprietary probe design allows for single-molecule detection in a cellular context, a level of sensitivity difficult to achieve with IHC [14]. This is validated by the low-copy positive controls like POLR2A. Studies comparing the two methods directly, such as one evaluating UPK2 in urothelial carcinoma, found that while RNAscope showed a trend towards a higher detection rate, its correlation with IHC was only moderate, suggesting it may detect true signals missed by IHC [4].
- Antibody Validation: RNAscope ISH is increasingly used as a gold standard to validate IHC antibodies. As noted by researchers, the struggle with unreliable antibodies can be circumvented by using RNAscope as an alternative or validation method [47]. The clear binary outcome of the control probes (PPIB must be positive, DapB must be negative) provides a level of confidence that is often unattainable with IHC.
- Morphological Context with Molecular Precision: Unlike "grind and bind" methods like PCR or NGS that lose spatial information, RNAscope, like IHC, preserves tissue morphology. However, it adds a layer of molecular precision through its controlled, sequence-based detection, allowing researchers to localize gene expression to specific cell types without ambiguity [27] [50].
Diagram 2: IHC Challenges vs RNAscope Advantages
The Scientist's Toolkit: Essential Research Reagent Solutions
Successful implementation of the RNAscope assay and its controls requires a suite of specific reagents and tools. The following table details the essential components for a controlled experiment.
Table 3: Essential Research Reagent Solutions for RNAscope with Controls
| Category | Product/Kit Examples | Critical Function |
|---|---|---|
| Core Detection Kit | RNAscope Multiplex Fluorescent Detection Kit v2 [49] | Provides all necessary reagents for signal amplification and detection in a standardized, optimized format. |
| Positive Control Probes | RNAscope 3-plex Positive Control Probe Mix (e.g., Mm-Polr2a-Ppib-Ubc for mouse) [49] | Validates the entire assay workflow and assesses sample RNA integrity simultaneously. |
| Negative Control Probe | RNAscope Negative Control Probe dapB [48] | Determines the level of non-specific background staining; essential for interpreting specificity. |
| Target-Specific Probes | Made-to-Order Probes (e.g., against DKK1, Gpr34) [27] [14] | Enable detection of the gene of interest; can be designed for any gene with 300+ base pairs in 2-3 weeks. |
| Cell Line Controls | FFPE Cell Pellet Arrays (CPA) [14] | Provide standardized positive and negative biological controls for initial assay validation and troubleshooting. |
| Image Analysis Software | QuPath [50] [14], FISHtoFigure [50] | Open-source or specialized software for automated cell segmentation and transcript quantification, enabling high-throughput, objective analysis. |
The integration of PPIB, POLR2A, UBC, and DapB is not merely a recommended step but a foundational requirement for robust RNAscope experimentation. These controls provide the empirical evidence needed to trust the generated data, distinguishing true signal from artifact and confirming that sample quality supports the experimental conclusions. As the field moves further toward precise molecular localization and away from the limitations of antibody-based methods, the disciplined application of this control quartet will remain paramount. They are the critical safeguards that ensure the superior sensitivity and specificity of RNAscope translate into reliable, reproducible scientific and diagnostic outcomes.
The evolution of precision oncology is intrinsically linked to advancements in diagnostic technologies, particularly immunohistochemistry (IHC) and RNA in situ hybridization. Automated IHC platforms, such as those from Ventana (Roche) and Leica Biosystems, have become cornerstones of clinical pathology, enabling robust biomarker detection for patient stratification. Concurrently, RNAscope has emerged as a powerful molecular technique offering single-molecule sensitivity and single-cell resolution within morphologically preserved tissue contexts. This guide provides a objective comparison of Ventana and Leica Biosystems automated platforms, framing their performance within a broader thesis comparing the sensitivity and specificity of RNAscope versus traditional IHC. For researchers and drug development professionals, understanding the capabilities, optimal applications, and limitations of these technologies is paramount for assay development and validation in both basic research and clinical diagnostics.
Ventana BenchMark Series platforms are renowned for their extensive menu of pre-optimized, Ready-To-Use (RTU) assays, which reduce validation burdens and ensure consistency. The system is a leader in companion diagnostics, with key assays like the VENTANA ALK (D5F3) CDx and PD-L1 (SP263) assays being FDA-approved for specific therapeutic indications [51]. The integrated ecosystem allows for seamless IHC and in situ hybridization (ISH) staining on a single platform.
Leica Biosystems BOND Series offer flexibility for both standardized RTU assays and laboratory-developed tests (LDTs) using concentrated antibodies. Recent developments include the introduction of novel antibodies like the PD-L1 clone 73-10, which has shown high sensitivity in exploratory evaluations [52]. The platform's open architecture is advantageous for research environments requiring custom protocol development.
Performance Data Comparison in Key Biomarkers
Direct performance comparisons can be drawn from recent peer-reviewed literature and proficiency testing data. The following tables summarize key quantitative findings for critical biomarkers.
Table 1: Comparative Performance in ALK (Lung) and PD-L1 Testing
| Biomarker | Platform/Assay | Performance Metric | Result | Notes |
|---|---|---|---|---|
| ALK (Lung) | Ventana (D5F3 RTU) | Pass Rate (NordiQC 2025) [53] | 55% | Significant decline from previous assessments. |
| Dako/Agilent (OTI1A4 RTU) | Pass Rate (NordiQC 2025) [53] | 100% | Most successful assay in the assessment. | |
| LYNX480 PLUS (BP6165) [54] | Sensitivity/Specificity | 98.3%/100% | Compared to FISH in lung adenocarcinoma. | |
| PD-L1 | Dako/Agilent (22C3) | Pass Rate (NordiQC 2025) [53] | 100% | For KEYTRUDA; most successful. |
| Leica (73-10 clone) | Concordance with SP263 (Kappa) [52] | 0.59 - 0.95 (TPS≥1%) | Varies by tumor type; exploratory status. | |
| Leica (73-10 clone) | Sensitivity (TPS≥1%) [52] | 78.3% - 100% | High specificity (97.9%-100%) across tumor types. |
Table 2: Performance in Other Key Diagnostic Biomarkers
| Biomarker | Platform/Assay | Performance Metric | Result | Notes |
|---|---|---|---|---|
| p40 | Multiple RTU Systems (Ventana, Dako, Leica) | Pass Rate (NordiQC 2025) [53] | 94% | High performance; robust across platforms. |
| ER | Ventana/Roche (RTU) | Pass Rate (NordiQC 2025) [53] | 77% (Overall) | Low pass rate; clone EP1 most robust. |
| MLANA | Clone A103 (Multiple Platforms) | Pass Rate (NordiQC 2025) [53] | 26% | Widespread insufficient results. |
| Clones EP43 & BS52 | Pass Rate (NordiQC 2025) [53] | 100% | Superior alternatives for melanoma. |
RNAscope vs. IHC: A Sensitivity and Specificity Framework
The choice between IHC and RNAscope is central to assay optimization. While IHC detects protein expression, RNAscope provides a highly sensitive and specific method for visualizing RNA transcripts within individual cells in FFPE tissue [27] [2]. Its proprietary "Z probe" design minimizes off-target binding and enables single-molecule detection [2].
Comparative Performance in Clinical Assays
- ERα in Breast Cancer: RNAscope shows high concordance with IHC for ERα (ESR1) status. It offers lower background staining and can help resolve false-negative cases, serving as a valuable complementary diagnostic tool [45].
- UPK2 in Urothelial Carcinoma: RNAscope and IHC perform similarly for detecting UPK2, with a trend towards a higher positive rate with RNAscope (68.0% vs. 62.6%, P=0.141), though not statistically significant in one study [4]. The techniques show a moderate positive correlation (R=0.441).
- Inflammatory Genes in CNS: For challenging targets like IL-1β and NLRP3, where specific antibodies are lacking, RNAscope combined with IHC allows precise cell-type-specific quantification of mRNA in neurons and microglia, a task intractable with IHC alone [2].
Integrated Workflow for Combined Analysis
Combining these techniques provides a more comprehensive biological picture. The following diagram illustrates a protocol for simultaneous RNAscope and IHC analysis on thicker CNS tissue sections, as used in neuroinflammatory research [2]:
Experimental Protocols for Key Studies
- Tissue Specimens: 3-μm FFPE sections from 87 lung adenocarcinoma specimens.
- Primary Antibody: BP6165 concentrated antibody (Biolynx), dilution 1:200.
- Automated Staining: Performed on LYNX480 PLUS System.
- Antigen Retrieval: EDTA, pH 9.0, 60 min at 100°C.
- Detection: BXV visualization system with DAB chromogen (no amplification).
- Counterstaining: Hematoxylin.
- Quality Control: Automated application of ALK Controls in Liquid Form (CLFs) via integrated QC module.
- Interpretation: Five blinded pathologists assessed granular cytoplasmic staining.
- Tissue Preparation: 14-μm fixed spinal cord sections; baked onto slides.
- RNAscope:
- Probes: Target-specific Z probes for IL-1β and NLRP3.
- Hybridization: 2 hours at 40°C in HybEZ oven.
- Signal Amplification: RNAscope 2.0 HD Reagent Kit.
- Immunohistochemistry:
- Primary Antibodies: IBA1 (microglia), NeuN (neurons).
- Visualization: Species-appropriate secondary antibodies with Alexa Fluor conjugates.
- Imaging & Analysis: Confocal microscopy; quantification of RNA transcripts within IHC-defined cell boundaries.
- Tissue Specimens: 208 unique TMA cores each for breast, colorectal, and hepatocellular carcinomas.
- Staining Platforms & Clones:
- Leica (73-10): BOND-III platform.
- Ventana (SP263): BenchMark ULTRA platform.
- Dako (22C3, 28-8): Autostainer Link 48 platform.
- Scoring: Three board-certified pathologists evaluated TPS (≥1%, ≥50%) and IPS (≥1%).
- Analysis: Inter-assay concordance (Cohen's Kappa), sensitivity, and specificity were calculated.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagent Solutions for Automated IHC and RNAscope
| Item | Function/Description | Example Products/Clones |
|---|---|---|
| Ready-To-Use (RTU) IHC Assays | Pre-optimized antibodies; reduce protocol variability and validation time. | VENTANA ALK (D5F3) CDx [51], Dako/Agilent PD-L1 22C3 [53], Leica p40 (BC28) [53] |
| Concentrated Antibodies for LDTs | Enable development of custom assays; require in-house validation. | BP6165 anti-ALK [54], Various PD-L1 clones [52] |
| RNAscope Probe Sets | Target-specific Z probes for RNA in situ hybridization. | Probes for UPK2 [4], IL-1β, NLRP3 [2], ESR1 (ERα) [45] |
| Automated Quality Controls | Cell-based controls to monitor staining variation; essential for QC. | ALK Controls in Liquid Form (CLFs) [54] |
| Detection & Visualization Kits | Chromogenic or fluorescent systems for signal generation. | BXV visualization system [54], DAB chromogen, RNAscope amplification kits [2] |
Optimizing automated IHC platforms requires a nuanced understanding of their respective strengths. Ventana systems excel with their extensive menu of validated, FDA-approved RTU assays, providing reliability in clinical diagnostics. Leica systems offer valuable flexibility for research and development, supporting both RTU and LDTs with robust performance, as evidenced by novel antibodies like the PD-L1 73-10 clone.
The integration of RNAscope as a complementary tool enhances the diagnostic and research arsenal. Its superior sensitivity and specificity for nucleic acid detection can resolve equivocal IHC results, detect low-abundance targets, and provide insights into gene expression at the single-cell level. The choice between IHC and RNAscope—or their combined application—should be guided by the specific research question, the biomarkers of interest, and the required balance between protein and RNA expression data. As the field advances, the continued objective evaluation of these platforms and techniques will be crucial for driving innovations in precision medicine.
The transition from traditional immunohistochemistry (IHC) to advanced RNA in situ hybridization techniques like RNAscope represents a significant paradigm shift in pathology and biomarker research. This evolution is further accelerated by the integration of artificial intelligence and quantitative image analysis platforms that enable precise, reproducible quantification at cellular and subcellular levels. For researchers and drug development professionals, selecting the appropriate analytical method is crucial for generating reliable data in studies of gene expression, protein localization, and tissue microenvironment characterization. The growing "reproducibility crisis" linked to antibody variability has intensified the search for more reliable quantification methods, with RNAscope emerging as a powerful alternative or complementary technique to traditional IHC [47]. This guide objectively compares the performance of HALO image analysis software and other methodologies within the context of RNAscope versus IHC sensitivity and specificity research, providing experimental data and protocols to inform platform selection for different research scenarios.
Technology Comparison: RNAscope vs. IHC
Fundamental Principles and Technical Basis
IHC (Immunohistochemistry) relies on antibody-antigen interactions to detect protein expression in tissue sections. Despite its widespread use as a "gold standard," IHC faces challenges including antibody specificity issues, batch-to-batch variability, and extensive validation requirements. The reproducibility crisis in biomedical research has been partially attributed to these antibody-related limitations, with studies indicating that a significant percentage of commercial antibodies lack sufficient specificity for their intended applications [47].
RNAscope (In Situ Hybridization) utilizes a novel double "Z" probe design that enables highly sensitive and specific detection of RNA molecules within intact tissue sections while preserving morphological context. This technology employs a proprietary signal amplification system that attaches to pairs of adjacent probes hybridized to the target RNA, resulting in dramatic signal amplification (up to 8,000 times) while minimizing background noise [1] [2]. Each detected RNA molecule appears as a distinct dot, allowing for precise quantification at the single-cell and single-transcript level.
Quantitative Performance Comparison
Table 1: Comparative Performance of RNAscope and IHC Across Multiple Studies
| Target/Biological Context | RNAscope Sensitivity | IHC Sensitivity | Concordance Rate | Key Findings | Citation |
|---|---|---|---|---|---|
| UPK2 in Urothelial Carcinoma | 68.0% | 62.6% | Moderate (R=0.441) | No significant difference in positivity rates (P=0.141); trend toward higher detection in variants with RNAscope | [4] |
| PD-L1 in Anaplastic Meningioma | N/A | N/A | Lower than expected | RNAscope demonstrated superior signal-to-noise ratio compared to IHC | [47] |
| COL11A1 in Ovarian Cancer | N/A | N/A | Consistent pattern | RNAscope provided higher resolution signal at cellular level | [47] |
| Various Targets (Systematic Review) | Highly sensitive | Variable | 58.7-95.3% | RNAscope shows high specificity and sensitivity; concordance lower due to measuring different molecules (RNA vs. protein) | [1] |
Image Analysis Platforms: HALO and Alternative Approaches
HALO Platform Capabilities and Specifications
HALO image analysis platform (Indica Labs) provides comprehensive quantitative tissue analysis solutions for both IHC and RNAscope applications. Key features include:
- AI-Powered Analysis: Integration of pre-trained deep learning networks for optimized nuclear and membrane segmentation in both brightfield and fluorescence applications [55]
- High-Throughput Processing: Multi-core processing and batch analysis capabilities for efficient workflow management
- Spatial Analysis: Tools for neighborhood analysis, proximity measurements, and tumor infiltration assessment
- Compatibility: Broad support for digital slide formats from major scanner manufacturers (Aperio, Hamamatsu, Leica, Zeiss, etc.) [55]
- Quantification Precision: Cell-by-cell analysis across entire tissue sections while maintaining interactive links between cell data and original images
For RNAscope-specific analysis, HALO offers specialized modules for quantifying transcript dots within cellular compartments, enabling precise gene expression measurement in the context of tissue architecture.
Comparative Platform Performance
Table 2: Image Analysis Platform Comparison for IHC and RNAscope Applications
| Platform/Technique | Primary Applications | Strengths | Limitations | Validation Status |
|---|---|---|---|---|
| HALO | Quantitative IHC, RNAscope, multiplex analysis | High-throughput, AI integration, extensive validation | Commercial license required | 2,000+ peer-reviewed publications [55] |
| Deep Learning CNN | Tumor detection, grading on H&E | Morphological pattern recognition, objective classification | Requires extensive training data | Validated for prostate cancer detection (Sensitivity: 0.971-1.000) [56] |
| QuPath | Digital pathology, IHC quantification | Open-source, customizable algorithms | Steeper learning curve | Research use, growing validation |
| Conventional Microscopy | Manual scoring and quantification | Accessibility, no special equipment needed | Subjective, labor-intensive, variable reproducibility | Established but limited by inter-observer variability |
Research indicates that deep learning approaches can achieve diagnostic performance comparable to human experts. In prostate cancer detection, CNN-based algorithms demonstrated sensitivity ranging from 0.971-1.000 and specificity of 0.875-0.976 across multiple validation cohorts, with Gleason grading agreement (kappa = 0.72-0.77) indistinguishable from experienced genitourinary pathologists [56].
Experimental Protocols and Methodologies
Combined RNAscope and IHC Protocol for Co-detection
The integration of RNAscope with IHC enables simultaneous detection of gene expression and protein localization within the same tissue section. The following protocol has been optimized for nervous system tissue but can be adapted to other tissue types [2]:
Tissue Preparation and Pre-treatment:
- Collect and fix tissues with appropriate fixative (e.g., 4% PFA for CNS tissue)
- Cut sections at 14μm thickness using cryostat
- Bake slides at 60°C for 1 hour to ensure adhesion
- Perform RNAscope protease treatment (15-30 minutes) for probe accessibility
Hybridization and Signal Development:
- Hybridize with target-specific RNAscope Z probes for 2 hours at 40°C
- Develop signal using RNAscope amplification system and fluorescent dyes
- Block tissue with appropriate serum (30 minutes) to reduce non-specific antibody binding
- Incubate with primary antibodies for cell-type specific markers (e.g., IBA1 for microglia, NeuN for neurons) overnight at 4°C
- Apply fluorophore-conjugated secondary antibodies (2 hours at room temperature)
Image Acquisition and Analysis:
- Acquire images using confocal microscopy with appropriate laser settings and sequential scanning to minimize bleed-through
- Process images using HALO or comparable analysis software
- Quantify RNA transcripts within cell-type specific boundaries defined by IHC staining
This combined approach has successfully demonstrated cell-type specific expression of inflammatory genes (IL-1b and NLRP3) in spinal cord neurons and microglia under neuropathic pain conditions [2].
Quantitative Image Analysis Workflow
The following diagram illustrates the comprehensive workflow for quantitative bioimaging experiments, from sample preparation through data interpretation:
Essential Research Reagent Solutions
Table 3: Key Reagents and Materials for RNAscope and IHC Experiments
| Reagent/Material | Function | Application Notes | Quality Control |
|---|---|---|---|
| RNAscope Probes | Target-specific RNA detection | Design requires 300bp unique sequence; 3-week development time | Positive (PPIB, Polr2A, UBC) and negative (dapB) controls essential [1] |
| Primary Antibodies | Protein target detection | Extensive validation required; batch-to-batch variability concerns | Validate with RNAscope or other orthogonal methods [47] |
| Protease Pretreatment | Tissue permeability for probe access | Critical step requiring optimization for different tissue types | Over-digestion reduces morphology; under-digestion lowers signal |
| Signal Amplification Systems | Enhanced detection sensitivity | RNAscope amplifier enables single-molecule detection | Minimizes background while maximizing specific signal [2] |
| Fluorophore-Conjugated Secondaries | Multiplex detection | Species-specific; requires spectral optimization | Check cross-reactivity; use sequential application for multiple targets |
| Mounting Media with DAPI | Nuclear counterstain and preservation | Photo-stable formulation for long-term storage | Essential for cell segmentation in image analysis |
Best Practice Recommendations
Platform Selection Guidelines
Choose image analysis platforms based on specific research needs:
- For high-throughput, standardized analyses: HALO provides optimized workflows with AI integration for consistent, batch-processable results [55]
- For algorithm development and customization: Open-source platforms like QuPath offer flexibility but require programming expertise
- For specialized applications: Consider technique-specific optimized modules (e.g., HALO FISH-IF for RNAscope analysis)
Validation and Quality Control
Implement rigorous validation protocols regardless of platform selection:
- Cross-validation with orthogonal methods: Use RNAscope to validate IHC results and vice versa [47]
- Include appropriate controls: Both positive and negative controls in every experiment [1] [57]
- Algorithm validation: Assess sensitivity, specificity, and reproducibility against manual scoring by experts
- Inter-platform consistency: Verify that results are consistent across different analysis systems
Experimental Design Considerations
- Begin with the end in mind: Define analysis requirements before starting experiments [57]
- Pilot studies are essential: Test entire workflow on small sample sets before full implementation
- Standardize sample processing: Minimize technical variability through parallel processing and consistent reagents
- Plan for proper statistical power: Include sufficient replicates for robust quantitative analysis
The evolving landscape of image analysis and quantification presents researchers with multiple sophisticated options for biomarker detection and quantification. RNAscope technology offers advantages in specificity and single-molecule detection for RNA targets, while IHC remains valuable for protein localization despite challenges with antibody validation. HALO image analysis platform provides comprehensive, AI-enhanced solutions for both methodologies, with performance comparable to expert pathologists in validated applications. The optimal approach depends on specific research questions, with combined methodologies often providing the most comprehensive biological insights. By implementing rigorous experimental protocols, appropriate controls, and validated analysis platforms, researchers can generate quantitative, reproducible data advancing drug development and basic research.
Evidence-Based Comparison: Sensitivity, Specificity, and Diagnostic Concordance
The accurate assessment of biomarker expression is a cornerstone of modern molecular pathology, directly influencing diagnostic, prognostic, and therapeutic decisions in oncology and other fields. For decades, immunohistochemistry (IHC) has served as the primary gold standard for visualizing protein expression within the morphologic context of tissue specimens. However, IHC faces significant challenges, including interobserver variability, dependence on antibody quality and affinity, and an inability to distinguish between closely related protein isoforms [1] [14]. The emergence of RNA in situ hybridization (ISH) techniques, particularly the RNAscope platform, offers a novel approach for directly detecting RNA transcripts within intact tissues. This systematic review synthesizes current evidence on the concordance between RNAscope and established gold standard methodologies, evaluating its potential as a complementary or alternative diagnostic tool.
RNAscope is a novel RNA ISH technology that utilizes a unique probe design strategy to achieve simultaneous signal amplification and background suppression, enabling single-molecule visualization while preserving tissue morphology [7]. The core of this technology is the use of so-called "Z-probes" [1] [2]. These probes are designed in pairs that must bind to adjacent sequences on the same target RNA molecule. Only when both probes hybridize successfully can the subsequent signal amplification cascade proceed. This dual-Z-probe requirement is the fundamental mechanism that confers exceptional specificity by minimizing off-target binding [2].
The following diagram illustrates the key mechanism and workflow of the RNAscope technology:
RNAscope Workflow and Analysis
The typical RNAscope workflow begins with slide preparation from formalin-fixed paraffin-embedded (FFPE) tissues, followed by three key steps: permeabilization, hybridization, and signal amplification [1]. The process can be automated and concludes with visualization using bright-field or fluorescent microscopy.
Analysis of RNAscope results involves quantifying the number of distinct dots within the tissue, where each dot represents a single RNA molecule [1] [14]. Scoring can be performed manually or using digital image analysis software such as Halo, QuPath, or Aperio, which offer improved precision, accuracy, and removal of pathologist bias [14].
Concordance with Gold Standard Methods
Systematic Review Evidence
A 2021 systematic review provides comprehensive evidence regarding the concordance between RNAscope and established techniques [1]. This review, which included 27 retrospective studies, evaluated RNAscope against IHC, quantitative PCR (qPCR), quantitative reverse transcriptase PCR (qRT-PCR), and DNA ISH.
Table: Concordance Rates Between RNAscope and Gold Standard Methods from Systematic Review
| Comparison Method | Concordance Rate Range | Key Factors Influencing Concordance |
|---|---|---|
| IHC (Immunohistochemistry) | 58.7% - 95.3% | Different products measured (RNA vs. protein); antibody quality; target stability |
| qPCR/qRT-PCR | 81.8% - 100% | Both measure RNA; RNAscope preserves spatial context while qPCR is grind-and-bind |
| DNA ISH | 81.8% - 100% | High specificity of probe design for both techniques |
The review concluded that RNAscope is a highly sensitive and specific method with high concordance rates against molecular techniques like qPCR and DNA ISH. The lower concordance with IHC was expected, as these techniques measure different biomolecules (RNA versus protein) that may have differing expression levels due to post-transcriptional regulation [1].
RNAscope vs. IHC for Specific Biomarkers
HER2 in Breast Cancer
Multiple studies have directly compared RNAscope and IHC for assessing HER2 status in breast cancer, with significant implications for treatment with antibody-drug conjugates like trastuzumab deruxtecan.
Table: Concordance Data for HER2 Assessment in Breast Cancer
| Study | Sample Type | Key Finding | Quantitative Correlation |
|---|---|---|---|
| Modern Pathology (2024) [58] | 526 breast cancer TMA cores | HER2 RNA levels by RNAscope strongly correlated with HER2 protein levels | P < 0.0001 |
| 32 metastatic biopsies | RNA levels were significantly higher in responders to T-DXd | 6.4 ± 8.2 dots/cell (responders) vs. 2.6 ± 2.2 (non-responders) | |
| AI-assisted HER2 Study (2025) [59] | 53 breast cancer samples across 35 labs | Manual IHC interpretation showed poor concordance for HER2-low cases | Only 26.4% of cases showed complete concordance across all labs |
The 2024 study found that while RNAscope strongly correlated with HER2 protein levels overall, neither protein levels nor RNA levels significantly differed between cases scored 0, ultralow, and 1+ by IHC, highlighting the challenge of accurately classifying HER2-low breast cancers with current IHC assays [58]. This is particularly relevant given the 35.9% improvement in concordance achieved when using AI-assisted interpretation for previously discordant IHC cases [59].
DKK1 in Gastric/Gastroesophageal Junction Cancer
A 2021 validation study of a DKK1 RNAscope assay for gastric and gastroesophageal junction adenocarcinoma demonstrated strong correlation between RNAscope and other measurement techniques [14]. When compared with RNA-Seq data from the Cancer Cell Line Encyclopedia across 48 cell lines, the study found a significant correlation (Spearman's rho = 0.86, p < 0.0001) [14]. The study also noted that the RNAscope assay was more sensitive than IHC, detecting DKK1 RNA in HeLa cell pellets where no IHC signal was observed.
RNAscope as a Solution for IHC Limitations
RNAscope offers particular advantages over IHC in specific diagnostic scenarios:
- Detection of B-cell Clonality: RNAscope probes for kappa and lambda light chain mRNA provide assessment of light chain restriction in B-cell lymphomas, a task that can be challenging with IHC, especially when fresh tissue for flow cytometry is unavailable [60].
- HPV Status Determination: For detection of high-risk HPV in oropharyngeal squamous cell carcinoma, RNAscope targeting HPV E6/E7 viral oncogene mRNA demonstrates significantly higher specificity compared to p16 IHC, reducing potential false positives [21].
- Combination with IHC: Protocols successfully combine RNAscope and IHC on the same tissue section, allowing simultaneous detection of RNA and protein within specific cell types identified by immunohistochemical markers [2].
Experimental Protocols and Validation
RNAscope Validation Protocol
The validation of the DKK1 RNAscope assay followed Clinical Laboratory Improvement Amendments (CLIA) guidelines to assess specificity, sensitivity, accuracy, and precision [14]. The protocol included:
- Specificity Assessment: Evaluation of probe specificity using cell lines expressing related Dickkopf family members (DKK2, DKK3, DKK4, DKKL1) confirmed minimal cross-reactivity.
- Sensitivity Confirmation: Detection of tumor cells with a range of DKK1 expression, including cells with only a single dot (representing one RNA molecule).
- Digital Image Analysis: Development and validation of a QuPath-based digital image analysis algorithm to identify tumor cells and quantify DKK1 signal, reducing pathologist time and variability.
Protocol for Combined RNAscope and IHC
A 2023 study detailed a method for combining RNAscope and IHC in thicker (14-μm) fixed spinal cord sections [2]. Key modifications from the standard RNAscope protocol included:
- Tissue Preparation: Fixation at time of post-mortem collection, cryostat sectioning at 14-μm, and baking onto slides after heat treatment and protease steps to prevent tissue loss.
- Staining Sequence: Simultaneous visualization of FISH and IHC without RNase-removing reagents.
- Image Analysis: Confocal microscopy and image analysis to quantify cell-specific RNA expression within IHC-defined cell type boundaries.
Research Reagent Solutions
The following table details key reagents and materials essential for implementing RNAscope in a research or diagnostic setting:
Table: Essential Research Reagent Solutions for RNAscope Implementation
| Reagent/Material | Function | Examples/Specifications |
|---|---|---|
| Target-Specific Probes | Hybridize to RNA of interest | DKK1, HER2, IgK, IgL, IGLL5, HPV E6/E7; Catalog numbers provided by manufacturer [60] [14] |
| Control Probes | Validate assay performance | Positive: PPIB (moderate expression), Polr2A (low expression), UBC (high expression). Negative: dapB (bacterial gene) [1] [14] |
| Amplification Reagents | Signal generation | Chromogenic or fluorescent detection kits; compatible with bright-field or fluorescent microscopy [1] [7] |
| Digital Analysis Software | Quantification and analysis | Halo, QuPath, Aperio; enables objective quantification of dots/cell [1] [14] |
| Automated Staining Platforms | Standardization and throughput | Compatible with various autostainer platforms; enables standardized workflow [14] |
The body of evidence synthesized in this review demonstrates that RNAscope shows high concordance with molecular gold standard methods like qPCR and DNA ISH, with slightly more variable but generally strong concordance with IHC. This variability stems from fundamental differences in the biomarkers measured (RNA vs. protein) rather than technical deficiencies. RNAscope offers significant advantages in scenarios where IHC faces limitations: distinguishing HER2-low breast cancers, detecting B-cell clonality, determining HPV status, and when suitable antibodies are unavailable. The technique's high sensitivity and specificity, ability to be quantified digitally, and capacity for multiplexing make it a powerful tool for both research and clinical diagnostics. While the current evidence supports RNAscope as a robust complementary technique, further prospective studies validating diagnostic accuracy in accordance with regulatory standards will help define its role as a potential standalone clinical diagnostic.
Uroplakin 2 (UPK2) serves as a highly specific marker for urothelial lineage, making its accurate detection crucial for diagnosing urothelial carcinoma (UC), particularly in metastatic sites or histologic variants. This guide provides a direct clinical comparison between the established method of immunohistochemistry (IHC) and the emerging RNA in situ hybridization technique, RNAscope, for evaluating UPK2 status. Objective analysis of experimental data from recent clinical studies demonstrates that RNAscope performs with similar to marginally higher sensitivity compared to IHC, without compromising the exceptional specificity of UPK2. The findings suggest that RNAscope can serve as a reliable alternative or adjunct to IHC in diagnostic and research settings, potentially enhancing the detection of urothelial differentiation in challenging cases.
Uroplakin 2 (UPK2) is one of four membrane proteins (UPK1a, UPK1b, UPK2, and UPK3a) that are specific differentiation products of normal urothelial cells [3] [4]. Its expression is exceptionally restricted to the urothelium and is undetectable in non-urothelial tissues such as skin, prostate, ovary, and liver [3] [4]. This high tissue specificity is well-maintained in urothelial carcinomas (UCs), making UPK2 a valuable diagnostic marker for confirming urothelial origin, especially for metastatic carcinomas of unknown primary or for UC variants with divergent differentiation that can be morphologically distinct from conventional UC [3] [4] [61].
Despite its excellent specificity, a significant limitation of UPK2 has been the relatively low sensitivity of its detection via conventional immunohistochemistry (IHC), with reported positivity rates in conventional invasive UC ranging from 44% to 80% [3] [4]. This diagnostic gap has spurred the investigation of more reliable detection methods, such as the novel RNA in situ hybridization assay, RNAscope [3] [4]. This guide directly compares the performance of IHC and RNAscope for UPK2 detection, providing researchers and pathologists with objective experimental data to inform their methodological choices.
Comparative Performance Data: RNAscope vs. IHC
A comprehensive 2022 study by Lu et al. directly compared RNAscope and IHC for UPK2 detection in 219 UC samples, including conventional bladder UCs, variant bladder UCs, upper tract UCs, and metastatic UCs [3] [4] [62]. The following tables summarize the key quantitative findings from this comparative analysis.
Table 1: Overall Comparison of UPK2 Detection by RNAscope and IHC in 219 UC Samples [3] [4]
| Detection Method | Overall Positivity Rate | Correlation with IHC | Statistical Significance (P-value) |
|---|---|---|---|
| Immunohistochemistry (IHC) | 62.6% (137/219) | Baseline | Reference |
| RNAscope | 68.0% (149/219) | Moderate positive correlation (P < 0.001, R = 0.441) | P = 0.141 (Not Significant) |
Table 2: Comparison of UPK2 Positivity Rates Across Urothelial Carcinoma Subtypes [3] [4]
| UC Subtype | Sample Size | IHC Positivity Rate | RNAscope Positivity Rate | Statistical Significance (P-value) |
|---|---|---|---|---|
| Conventional Bladder UC | 127 | 68.5% | 72.4% | P = 0.511 |
| Variant Bladder UC | 45 | 35.6% | 53.3% | P = 0.057 |
| Upper Tract UC | 24 | 70.8% | 70.8% | P = 1.000 |
| Metastatic UC | 23 | 69.6% | 69.6% | P = 1.000 |
Key Findings from Comparative Data
- Overall Performance: The study found no statistically significant difference in the overall UPK2 positivity rate between the two methods (68.0% vs. 62.6%, P=0.141) [3] [4]. The results showed a moderate positive correlation, indicating general concordance [3] [4].
- Performance in Challenging Subtypes: A notable, albeit not statistically significant (P=0.057), trend was observed in variant bladder UCs, where RNAscope detected a higher positivity rate (53.3%) compared to IHC (35.6%) [3] [4]. This suggests a potential advantage for RNAscope in diagnostically challenging cases where aberrant differentiation may affect protein expression.
- Sensitivity and Specificity Profile: While the study primarily focused on comparative positivity rates, it reaffirmed the excellent specificity of UPK2 for urothelial lineage, a characteristic inherent to the marker itself rather than the detection method [3] [4]. RNAscope demonstrated a marginally higher, but not statistically significant, sensitivity.
Experimental Protocols and Methodologies
To ensure reproducibility and provide context for the data, this section outlines the core experimental protocols used in the cited comparative study.
Tissue Sample Preparation
- Source: The study utilized tissue blocks from 219 UC cases retrieved from the Department of Pathology, Sun Yat-sen University Cancer Center (May 2000 - September 2017) [3] [4].
- Design: Tissue microarrays (TMAs) were constructed using a tissue arrayer, with two or three representative 1-mm cores taken from each donor block [3] [4]. This allowed for standardized and simultaneous processing of all samples under identical conditions for both IHC and RNAscope.
Immunohistochemistry (IHC) Protocol
The IHC protocol followed standardized, clinically applicable procedures [3] [4]:
- Sectioning: Three-micrometer sections were obtained from the TMA blocks.
- Staining: Automated staining was performed using a BenchMark ULTRA system.
- Antibody: Primary antibody against UPK2 (clone BC21, Biocare Medical) was used at a 1:100 dilution [3] [4].
- Evaluation: Two pathologists independently evaluated the slides. Cytoplasmic staining in UC cells was scored as positive [3] [4].
RNAscope Protocol
The RNAscope assay was performed according to the manufacturer's protocol (Advanced Cell Diagnostics) [3] [4]:
- Probe: A specific probe targeting UPK2 mRNA (NM_006760.4) was used on TMA sections [3] [4].
- Pretreatment: Slides were deparaffinized and subjected to a series of pretreatments (Pretreatment 1 at room temperature for 10 min, Pretreatment 2 by boiling for 20 min, Pretreatment 3 at 40°C for 30 min) to enable probe access [3] [4].
- Hybridization & Amplification: Slides were hybridized with the target probe for 2 hours at 40°C in a HybEZ oven. Signals were then amplified and generated using the RNAscope 2.0 HD Reagent Kit-BROWN [3] [4].
- Evaluation: As with IHC, cytoplasmic staining in UC cells was scored as positive [3] [4].
RNAscope is a novel, proprietary version of RNA in situ hybridization that achieves high sensitivity and specificity through a unique probe design and signal amplification system [1] [2].
Diagram 1: RNAscope Signal Amplification Principle
The core innovation of RNAscope is the use of a pair of "Z" probes that are designed to bind adjacent sequences on the same target mRNA molecule [1] [2]. This paired-probe system is the foundation of its high specificity:
- Specificity Mechanism: The signal amplification structure can only bind if both "Z" probes are correctly hybridized to their adjacent target sites. This requirement minimizes off-target binding and background noise, as single probe binding does not yield a signal [1] [2].
- Amplification Mechanism: Each pair of "Z" probes can bind a pre-amplifier, which in turn binds multiple amplifiers. Each amplifier then binds numerous enzyme-labeled probes. This cascade results in up to an 8,000-fold signal amplification, enabling the visualization of single RNA molecules under a microscope [1].
The Scientist's Toolkit: Essential Research Reagents
The following table details key reagents and materials required to perform UPK2 detection using either IHC or RNAscope, based on the protocols from the cited studies.
Table 3: Key Research Reagent Solutions for UPK2 Detection
| Item | Function / Description | Exemplar Product / Clone |
|---|---|---|
| Primary Antibody (for IHC) | Binds specifically to UPK2 protein for visual detection. | UPK2 Mouse Monoclonal Antibody (Clone BC21) [3] [4] |
| RNAscope Probe (for ISH) | A pool of oligonucleotide "Z" probes designed to hybridize to UPK2 mRNA. | UPK2 RNAscope Probe (Targeting NM_006760.4) [3] [4] |
| Detection Kit (IHC) | Automated system for antibody detection and visualization. | BenchMark ULTRA Staining System (Ventana) [3] [4] |
| Detection Kit (RNAscope) | Contains reagents for hybridization, amplification, and chromogenic signal development. | RNAscope 2.0 HD Reagent Kit-BROWN [3] [4] |
| Control Probes (RNAscope) | Essential for validating assay performance. | Positive Control Probe (e.g., PPIB), Negative Control Probe (dapB) [1] |
Discussion and Integrated Analysis
The direct clinical comparison reveals that RNAscope is a robust and reliable method for UPK2 detection, performing with statistically equivalent overall sensitivity to IHC while offering several distinct technical advantages.
- Advantages of RNAscope: The technology provides single-molecule visualization while preserving tissue morphology [3]. Its unique probe design confers very high specificity and low background noise [1] [2]. Furthermore, it is less susceptible to issues that can affect IHC, such as variable antibody affinity and protein conformation changes due to suboptimal tissue fixation [45].
- Advantages of IHC: IHC remains a widely established, familiar, and often more accessible technology in most clinical pathology laboratories. The protocol is generally less complex and time-consuming than the RNAscope procedure.
- Concordance and Discrepancy: The moderate positive correlation (R=0.441) indicates good but not perfect agreement between the methods [3] [4]. Discrepant cases, where one method is positive and the other negative, can occur due to biological reasons (e.g., transcriptional activity without immediate translation, or post-translational protein loss) or technical limitations of either assay [1]. The trend towards higher RNAscope detection in variant UCs suggests its potential utility in cases where IHC is negative but clinical suspicion for urothelial origin remains high.
Clinical and Research Implications
Within the broader thesis of RNAscope vs. IHC research, the data on UPK2 aligns with findings for other biomarkers, such as TTF-1 and Glypican 3, where RNAscope often demonstrates equivalent or superior sensitivity [3]. For clinical practice and drug development:
- Adjunct Use: RNAscope can be a valuable complementary tool to confirm UPK2 status in IHC-negative or equivocal cases, especially for variant or metastatic UCs.
- Biomarker Validation: In research settings, particularly clinical trials for UC, employing both methods could provide a more comprehensive understanding of UPK2 as a predictive or prognostic biomarker.
- Pitfall Avoidance: Researchers should be aware that neither method is 100% sensitive. A negative result with one technique does not definitively rule out urothelial origin, and clinical-pathological correlation remains paramount.
This direct clinical comparison demonstrates that RNAscope is a valid and sensitive method for detecting UPK2 expression in urothelial carcinoma tissues. It performs with comparable overall efficacy to IHC, with a promising trend towards improved detection in the diagnostically challenging variant subtypes of bladder cancer. While IHC remains a cornerstone of diagnostic pathology, RNAscope emerges as a powerful alternative or adjunct technique. Its high specificity and ability to directly target mRNA make it a valuable addition to the scientist's and pathologist's toolkit for confirming urothelial lineage, ultimately contributing to more accurate diagnosis and research in the field of urothelial carcinoma.
Immunohistochemistry (IHC) and RNA in situ hybridization (RNAscope) are powerful complementary techniques for biomarker analysis in clinical and research pathology. However, researchers frequently observe discrepant results between these methods, creating challenges for data interpretation. This review systematically examines the technological and biological factors underlying these divergences, drawing on comparative studies across multiple cancer types. We analyze the correlation data between RNA and protein detection methods, provide detailed experimental protocols for parallel validation, and visualize the mechanistic basis for observed differences. Understanding these discordances is essential for optimizing biomarker discovery and validation strategies in oncology research and drug development.
The correlation between RNA expression and protein abundance represents a fundamental challenge in molecular biology and diagnostic pathology. While IHC detects protein antigens using antibody-based recognition, RNAscope identifies RNA transcripts through in situ hybridization with proprietary signal amplification [63] [64]. These techniques measure different molecular entities with distinct biological regulation, leading to frequent discrepancies in research and clinical settings.
The clinical implications of these technical differences are significant. With IHC remaining the gold standard in clinical diagnostics and RNAscope emerging as a highly sensitive research tool, understanding the basis for divergent results is crucial for proper interpretation of biomarker data [20]. This review examines the fundamental principles underlying both techniques, presents comparative performance data across multiple biomarkers, and provides methodological guidance for researchers navigating these complementary technologies.
Fundamental Differences Between IHC and RNAscope
Technological Principles
IHC (Immunohistochemistry) relies on antibody-antigen interactions to localize specific proteins within tissue sections. The technique involves epitope recognition by primary antibodies, followed by signal amplification and chromogenic detection. IHC results can be influenced by numerous factors including antibody specificity, epitope availability, fixation conditions, and detection sensitivity [63].
RNAscope utilizes a novel in situ hybridization approach with proprietary "Z probe" pairs that hybridize to adjacent regions of the target RNA sequence. This double-Z probe design provides exceptional specificity, as signal generation requires two independent probes to bind correctly to the same RNA molecule. The method then employs a powerful signal amplification system that allows for single-molecule visualization while preserving tissue morphology [36]. Each dot in RNAscope represents an individual RNA molecule, enabling semi-quantitative assessment of gene expression.
Biological Basis for Divergence
The fundamental biological distinction between RNA and protein expression underlies many observed discrepancies:
- Temporal discordance: mRNA transcription precedes protein translation, creating natural time lags between RNA detection and protein accumulation [64]
- Post-transcriptional regulation: Mechanisms including miRNA-mediated silencing, RNA degradation, and translational control create disparities between mRNA abundance and protein output
- Post-translational modifications: Protein stability, modification, and turnover rates affect IHC detection independently of RNA levels
- Secreted proteins: For secreted factors, IHC detects accumulated protein in extracellular matrix while RNAscope identifies producing cells [64]
Diagram: Biological pathway showing points where RNA and protein detection may diverge. RNAscope detects early transcription events while IHC detects later protein expression, with multiple regulatory points creating discordance.
Comparative Performance Data
Concordance Studies Across Biomarkers
Multiple studies have systematically compared RNAscope and IHC for various biomarkers, demonstrating variable correlation depending on the target molecule:
Table 1: Concordance Rates Between RNAscope and IHC Across Studies
| Biomarker | Disease Context | Concordance Rate | Correlation Coefficient | Reference |
|---|---|---|---|---|
| UPK2 | Urothelial Carcinoma | 68.0% vs 62.6% (positivity rates) | R = 0.441 | [3] [4] |
| DKK1 | Gastric/GEJ Adenocarcinoma | Higher sensitivity for RNAscope | Spearman's rho = 0.86 (vs RNA-Seq) | [14] |
| Multiple Biomarkers* | Systematic Review | 58.7%-95.3% | Range across studies | [20] |
| ESR1, PGR, ERBB2, etc. | Multiple Solid Tumors | Strong correlations | 0.53-0.89 (RNA-seq vs IHC) | [28] |
Multiple biomarkers including hormone receptors, proliferation markers, and immune checkpoints across 27 studies
The systematic review by PMC found that RNAscope has high concordance with PCR-based methods (81.8-100%) but variable concordance with IHC (58.7-95.3%), highlighting the fundamental differences between RNA and protein detection [20].
Sensitivity and Specificity Comparisons
Studies directly comparing the sensitivity of both techniques reveal important performance differences:
Table 2: Sensitivity Comparison in Detection of UPK2 in Urothelial Carcinoma [3] [4]
| UC Subtype | RNAscope Positivity | IHC Positivity | P-value |
|---|---|---|---|
| Conventional Bladder UC | 72.4% | 68.5% | 0.511 |
| Variant Bladder UC | 53.3% | 35.6% | 0.057 |
| Upper Tract UC | 70.8% | 70.8% | 1.000 |
| Metastatic UC | 65.2% | 65.2% | 1.000 |
| Overall | 68.0% | 62.6% | 0.141 |
RNAscope demonstrated a trend toward higher detection rates in variant bladder urothelial carcinomas, though this did not reach statistical significance in the study of 219 samples [3]. This pattern suggests that for certain biomarkers and tissue contexts, RNAscope may offer improved sensitivity over IHC.
The DKK1 validation study demonstrated another advantage of RNAscope: detecting expression in HeLa cell pellets where IHC failed, confirming its superior sensitivity for low-abundance targets [14].
Experimental Protocols for Method Comparison
Parallel Testing Methodology
To ensure valid comparisons between IHC and RNAscope, researchers should implement standardized parallel testing protocols:
Tissue Preparation Considerations:
- Use consecutive sections from the same FFPE block for both assays
- Ensure section thickness consistency (typically 3-5μm)
- Maintain freshly cut sections to prevent RNA degradation [63] [14]
- Employ Superfrost Plus slides for RNAscope to prevent tissue detachment [63]
IHC Protocol Highlights:
- Automated staining systems (e.g., Ventana BenchMark ULTRA) recommended for consistency
- Antibody validation with appropriate positive and negative controls
- Antigen retrieval optimization based on tissue fixation [3] [4]
RNAscope Protocol Key Steps:
- Deparaffinization and rehydration with fresh xylene and ethanol [63]
- Pretreatment sequence:
- Pretreatment 1 (10 min at room temperature)
- Pretreatment 2 (boiling for 20 min)
- Pretreatment 3 (30 min at 40°C) [3]
- Protease digestion (critical step requiring optimization) [31]
- Hybridization with target probes (2 hours at 40°C in HybEZ oven) [3]
- Signal amplification using RNAscope 2.0 HD Reagent Kit [4]
Essential Research Reagent Solutions
Table 3: Key Reagents and Equipment for Comparative Studies
| Item | Function | Technical Considerations |
|---|---|---|
| HybEZ Oven System | Maintains optimum humidity and temperature during hybridization | Critical for assay performance; other incubators may not provide consistent results [31] |
| Positive Control Probes (PPIB, POLR2A, UBC) | Assess sample RNA quality and optimal permeabilization | Different copy number controls available (low to high) [63] |
| Negative Control Probe (dapB) | Assess background and specificity | Bacterial gene should not generate signal in properly fixed tissue [63] |
| ImmEdge Hydrophobic Barrier Pen | Maintains reagent containment on slides | Specific pen required; others may fail during procedure [63] |
| Protease Reagents | Tissue permeabilization for RNA access | Concentration and time critical; affects signal and morphology [31] |
| Specific Target Probes | Detection of genes of interest | Channel-specific (C1-C4) for multiplexing; C1 must be included [63] |
Analysis and Interpretation of Discordant Results
Technical Artifacts vs. Biological Significance
When facing discrepant results between IHC and RNAscope, researchers must systematically evaluate potential causes:
Technical Factors Favoring IHC Detection:
- Antibody cross-reactivity leading to false positive protein detection
- Epitope preservation affected by fixation conditions
- High protein stability with rapid RNA turnover
Technical Factors Favoring RNAscope Detection:
- Superior sensitivity for low-abundance targets [14]
- Resistance to RNA degradation through multi-probe design [14]
- No requirement for protein conformation or post-translational status
Biologically Meaningful Discordance:
- Active transcription with translational repression (positive RNAscope, negative IHC)
- Protein accumulation without ongoing transcription (positive IHC, negative RNAscope)
- Secreted proteins where producing cells and protein localization differ [64]
Integration with Complementary Methods
To resolve ambiguous cases, researchers can incorporate additional techniques:
- RNA sequencing provides orthogonal validation of transcript presence [28]
- Western blotting confirms protein expression independently
- Digital PCR offers quantitative RNA assessment
- Multiplexed protein detection validates IHC findings
The integration of spatial transcriptomics with RNAscope and IHC creates a powerful multimodal approach that overcomes limitations of individual methods [65]. This strategy leverages the genome-wide discovery capacity of sequencing with the cellular resolution and sensitivity of targeted detection.
The correlation between RNAscope and IHC results is influenced by a complex interplay of technical and biological factors. While these methods show moderate to strong concordance for many biomarkers, understanding the basis for their divergence provides valuable insights into gene expression regulation. RNAscope offers exceptional sensitivity and specificity for RNA detection, while IHC provides direct evidence of protein expression. Rather than viewing discrepancies as methodological failures, researchers should interpret them as opportunities to uncover meaningful biology, including post-transcriptional regulation, protein secretion dynamics, and translational control mechanisms. The strategic combination of both techniques, with awareness of their respective strengths and limitations, creates a powerful approach for comprehensive biomarker validation in research and clinical development.
In molecular pathology, accurate biomarker detection is fundamental for diagnosis, prognosis, and therapy selection. However, challenging samples—including tumors with variant histologies and those with low target antigen expression—present significant obstacles for conventional detection methods. Immunohistochemistry (IHC), while widely established, often struggles with sensitivity limitations in these scenarios, particularly when antigen expression is low or heterogenous. RNA in situ hybridization (ISH) technologies, specifically the RNAscope platform, have emerged as promising alternatives that operate on a different detection principle—targeting RNA rather than protein. This guide objectively compares the performance of RNAscope versus IHC in these diagnostically challenging contexts, providing researchers and drug development professionals with experimental data to inform their methodological choices.
Technology Comparison: Fundamental Principles and Workflows
Core Technological Differences
The fundamental difference between these techniques lies in their detection targets: IHC detects proteins using antibody-antigen interactions, while RNAscope detects RNA transcripts using a proprietary in situ hybridization process.
IHC Workflow: IHC relies on antibody binding to specific protein epitopes, followed by chromogenic or fluorescent detection. Its performance is heavily influenced by antibody affinity, specificity, and the preservation of protein epitopes through fixation and processing. Post-transcriptional and post-translational modifications can affect the correlation between mRNA and protein levels, potentially leading to discrepancies between IHC and RNA-based methods [1].
RNAscope Workflow: RNAscope utilizes a novel signal amplification and background suppression system. The technology employs paired "Z" probes that bind adjacent to each other on the target RNA sequence. This double-Z probe design requires both probes to bind correctly for signal amplification to occur, dramatically reducing non-specific background. The subsequent amplification steps allow for single-molecule visualization while preserving tissue morphology [1] [2].
Table 1: Core Technology Comparison
| Feature | Immunohistochemistry (IHC) | RNAscope ISH |
|---|---|---|
| Detection Target | Proteins | RNA transcripts |
| Signal Amplification | Enzyme-based (e.g., HRP) | Branched DNA amplification |
| Probe Design | Antibodies (polyclonal/monoclonal) | Paired "Z" probes |
| Key Limitation | Dependent on antibody quality and protein integrity | Susceptible to RNA degradation |
| Multiplexing Capability | Limited by antibody host species and color overlap | Designed for multiplex detection (up to 12-plex in some systems) |
RNAscope Workflow Visualization
The following diagram illustrates the key steps and decision points in the RNAscope experimental workflow, particularly highlighting the quality control measures essential for challenging samples:
Performance Data in Challenging Samples
Direct Comparison in Urothelial Carcinoma Variants
A comprehensive 2022 study directly compared RNAscope and IHC for detecting UPK2 (a marker for urothelial carcinoma) in 219 samples, including conventional urothelial carcinomas, variant histologies, upper tract carcinomas, and metastatic cases [4]. The results demonstrate important performance differences:
Table 2: UPK2 Detection in Urothelial Carcinoma Variants (n=45)
| Method | Positivity Rate | Statistical Significance | Key Findings |
|---|---|---|---|
| RNAscope | 53.3% | P = 0.057 | Trend toward higher detection rate in variant histologies |
| IHC | 35.6% | (Not statistically significant) | Lower sensitivity in morphologically distinct variants |
In the overall cohort of 219 UC samples, RNAscope detected UPK2 in 68.0% of cases compared to 62.6% by IHC, though this difference did not reach statistical significance (P = 0.141). Correlation analysis revealed a moderate positive correlation between the two methods (P < 0.001, R = 0.441), suggesting they provide complementary rather than identical information [4].
Addressing Low Antigen Expression
The challenge of low antigen expression is particularly evident in breast cancer HER2 testing. Recent clinical trials have expanded antibody-drug conjugate therapies to patients with HER2-low and HER2-ultra-low expression, creating an urgent need for more sensitive detection methods [66]. While not directly employing RNAscope, studies have demonstrated that quantitative IHC approaches coupled with artificial intelligence interpretation can significantly improve HER2 expression quantification in these low ranges. This suggests limitations in conventional IHC for detecting low-abundance targets—a challenge that RNAscope's signal amplification system is theoretically well-positioned to address.
Experimental Protocols for Challenging Samples
RNAscope Protocol for Variant Histologies
The following detailed methodology is adapted from the UPK2 comparison study and RNAscope technical resources [4] [63]:
Sample Preparation:
- Use formalin-fixed, paraffin-embedded (FFPE) tissue sections (4-5 μm thickness) mounted on SuperFrost Plus slides
- Fix tissues in fresh 10% neutral buffered formalin for 16-32 hours
- Bake slides at 60°C for 1 hour before use
Pretreatment Conditions:
- Deparaffinize slides in xylene and ethanol series
- Perform antigen retrieval by boiling in RNAscope Target Retrieval Reagents (15-30 minutes at 98-102°C)
- Digest with RNAscope Protease Plus (15-30 minutes at 40°C)
Hybridization and Detection:
- Hybridize with target-specific RNAscope probes (2 hours at 40°C)
- Apply signal amplification reagents according to manufacturer's protocol
- Develop chromogenic signal using DAB or fluorescent detection
- Counterstain with Gill's Hematoxylin (diluted 1:2) and mount
Critical Optimization Steps for Challenging Samples:
- For variant histologies with potential lower target expression, extend protease treatment incrementally (e.g., +10 minutes)
- Include positive control probes (PPIB, POLR2A, or UBC) and negative control (dapB) on adjacent sections
- For low-expressing targets, use POLR2A as a positive control rather than high-copy number genes [63]
Combined RNAscope and IHC Protocol
For maximum information from precious samples, a combined RNAscope and IHC protocol enables simultaneous detection of RNA and protein in the same tissue section [2]:
Sequential Staining Approach:
- Complete RNAscope protocol through signal development
- Block tissue with appropriate serum or protein block (10-15 minutes)
- Incubate with primary antibody (diluted in buffer, 1-2 hours at room temperature)
- Apply secondary detection system with different chromogen/fluorophore
- Counterstain and mount
Key Considerations:
- Thicker sections (14 μm) are recommended for combined protocols to preserve tissue integrity
- Perform RNAscope first to prevent RNase contamination from IHC reagents
- Use different detection channels (e.g., fluorescent colors) to distinguish signals
- Include controls for both methods separately to validate combined results
The Scientist's Toolkit: Essential Research Reagents
Successful application of these technologies in challenging samples requires specific reagents and controls:
Table 3: Essential Research Reagents for RNAscope in Challenging Samples
| Reagent Category | Specific Examples | Function in Challenging Samples |
|---|---|---|
| Positive Control Probes | PPIB, POLR2A, UBC | Verify RNA integrity; POLR2A preferred for low-expression targets |
| Negative Control Probes | dapB | Assess background noise and specificity |
| Pretreatment Reagents | Target Retrieval Reagents, Protease Plus | Optimize target accessibility without over-digestion |
| Signal Amplification Kits | RNAscope 2.5 HD Reagent Kit | Enhance sensitivity for low-copy targets |
| Detection Systems | Chromogenic (DAB) or Multiplex Fluorescent | Flexible detection based on expression level |
| Tissue Preservation | Fresh 10% NBF, RNase-free conditions | Preserve RNA quality in archival samples |
Technical Considerations and Limitations
Impact of Sample Quality on RNAscope Performance
A critical factor in RNAscope success, particularly with challenging samples, is RNA integrity. A 2025 study systematically assessed RNA degradation over archival time in FFPE tissues and found that RNAscope signals decrease in an archival duration-dependent fashion [67]. The degradation was most pronounced in highly expressed housekeeping genes (UBC and PPIB) compared to low-to-moderate expressors (POLR2A and HPRT1). This highlights the necessity of using appropriate positive controls that match the expected expression level of target genes, especially in archival samples or those with variant histologies that may have lower target expression.
Systematic Review Evidence
A 2021 systematic review comparing RNAscope with gold standard methods found high concordance rates with PCR-based methods (81.8-100%) but lower concordance with IHC (58.7-95.3%) [1]. This discrepancy underscores the fundamental differences between detecting RNA versus protein, including the effects of post-transcriptional regulation, protein turnover rates, and technical limitations of both methods. The review concluded that RNAscope could complement but not necessarily replace IHC in clinical diagnostics, though for research applications in challenging samples, it provides valuable orthogonal data.
The comparative data presented in this guide demonstrate that RNAscope offers particular advantages for detecting biomarkers in samples with variant histologies and potentially those with low target expression. While IHC remains the established standard for protein detection, its sensitivity limitations in these challenging contexts create opportunities for RNAscope to provide complementary information.
For researchers and drug development professionals, the decision between these technologies should consider:
- The biological question (RNA versus protein detection)
- Sample quality and characteristics
- Required sensitivity level
- Need for multiplexing or combination with other techniques
As therapeutic options expand to include targets with lower expression levels (e.g., HER2-low breast cancers), more sensitive and quantitative detection methods like RNAscope may play increasingly important roles in both research and clinical translation.
In clinical diagnostics and research, the complete molecular picture of cellular activity often requires observing both the initial genetic instructions and their final executed forms. Gene expression involves the transcription of DNA into messenger RNA (mRNA), which is then translated into protein [1]. While proteins are the primary functional actors in most cellular processes, RNA analysis provides crucial insights into transcriptional regulation and can identify cells that are actively producing a protein, even before it is secreted. Immunohistochemistry (IHC), which detects proteins within tissues, has long been a cornerstone of clinical diagnostics. RNA in situ hybridization (ISH), particularly the highly sensitive RNAscope method, has emerged as a powerful complementary technology for visualizing RNA transcripts with single-molecule resolution while preserving tissue morphology [1] [36]. The integration of RNA and protein data offers a more comprehensive understanding of disease mechanisms, cellular identity, and regulatory processes, ultimately leading to more informed diagnostic and therapeutic decisions [64].
Technology Comparison: RNAscope vs. Immunohistochemistry (IHC)
Fundamental Principles and What They Measure
IHC relies on antibody-antigen interactions to visualize the spatial distribution and abundance of specific proteins within tissue sections. It reveals the end product of gene expression and provides information about post-translational modifications and protein localization. However, IHC can be limited by antibody availability, specificity, and its inability to distinguish the producing cell for secreted proteins [64].
RNAscope is a novel variant of RNA ISH that uses a proprietary double "Z" probe design to achieve high specificity and signal amplification. Each pair of Z probes binds to an adjacent region of the target RNA, and only when both are bound can a large amplification structure be assembled, resulting in a detectable punctate dot. Each dot represents a single RNA molecule, allowing for precise localization and quantification of gene expression at the single-cell level [1] [36] [2].
Table 1: Core Comparison of IHC and RNAscope Technologies
| Feature | Immunohistochemistry (IHC) | RNAscope |
|---|---|---|
| Target Molecule | Protein (post-translationally modified) | RNA (primarily mRNA) |
| Detection Principle | Antibody-antigen binding | Nucleic acid hybridization with "Z" probes |
| Key Output | Protein presence, localization, and abundance | RNA presence, localization, and transcript count |
| Single-Molecule Sensitivity | Typically not achievable | Yes, each dot represents one transcript |
| Ideal for Identifying | Functional protein endpoints, cellular architecture | Active gene transcription, producing cells of secreted factors |
Direct Performance Comparison: Sensitivity and Specificity
A direct head-to-head comparison of RNAscope and IHC for detecting UPK2, a marker for urothelial carcinoma (UC), illustrates their performance characteristics. A 2022 study on 219 UC samples found that while RNAscope showed a trend towards higher sensitivity, the overall difference was not statistically significant [3].
Table 2: Experimental Performance Data: UPK2 Detection in Urothelial Carcinoma [3]
| UC Tissue Type | Positive by IHC | Positive by RNAscope | P-value |
|---|---|---|---|
| All Urothelial Carcinomas (n=219) | 62.6% | 68.0% | P = 0.141 |
| Conventional Bladder UC (n=127) | 68.5% | 72.4% | P = 0.511 |
| Variant Bladder UC (n=45) | 35.6% | 53.3% | P = 0.057 |
The data reveals a moderate positive correlation between the two methods (P < 0.001, R = 0.441) [3]. The trend of higher sensitivity with RNAscope was particularly notable in variant bladder UCs, a context where diagnosis can be challenging. This suggests RNAscope could serve as a valuable alternative or adjunct to IHC in diagnostically difficult cases [3]. A 2021 systematic review further confirmed that RNAscope is a highly sensitive and specific method, though it noted that its concordance rate with IHC (58.7–95.3%) is lower than with PCR-based techniques, largely because IHC and RNAscope measure different biological molecules (protein vs. RNA) that are subject to different regulatory mechanisms [1].
Experimental Workflows and Protocols
Detailed Methodologies for a Combined Assay
Combining RNAscope and IHC on a single tissue section allows researchers to correlate transcriptional activity with protein expression and cell identity within a precise spatial context. The following protocol is adapted from a 2023 method optimized for central nervous system tissue, which successfully detected inflammatory genes in specific neuronal and microglial populations [36] [2].
Tissue Preparation and Pre-treatment:
- Fixation: Tissue is collected and fixed, typically with 4% paraformaldehyde (PFA). For spinal cord tissue, a 4-hour post-fixation at 4°C is often optimal for preserving both RNA integrity and protein epitopes [36].
- Sectioning: Tissues are cryosectioned at a thickness of 14 μm and mounted on slides. Thicker sections help preserve tissue architecture during the stringent assay steps.
- Baking and Protease Treatment: Slides are baked to adhere the tissue, followed by a protease treatment (e.g., RNAscope RTU Protease IV) to permeabilize the tissue and make the target RNA accessible. The protease concentration and time must be carefully optimized to avoid over-digestion, which can destroy protein epitopes for subsequent IHC [36].
RNAscope In Situ Hybridization:
- Hybridization: Slides are incubated with the target-specific RNAscope probes in a HybEZ hybridization oven at 40°C for 2 hours.
- Signal Amplification: A series of amplification steps are performed using the RNAscope Fluorescent Multiplex reagent kit. These steps build the signal amplification structure on the bound "Z" probes, ultimately attaching a fluorescent label to each target RNA molecule [40] [68].
Immunohistochemistry:
- Antibody Incubation: Following the RNAscope procedure, slides are directly incubated with primary antibodies against cell-type-specific protein markers (e.g., IBA1 for microglia, NeuN for neurons). This is followed by incubation with secondary antibodies conjugated to fluorophores distinct from those used in the RNAscope channels [36] [2].
- Mounting and Imaging: Slides are counterstained with DAPI, coverslipped, and imaged using a confocal or high-resolution fluorescence microscope capable of capturing all the fluorescence channels used.
The Scientist's Toolkit: Essential Research Reagent Solutions
The successful execution of a combined RNAscope/IHC experiment depends on key reagents and equipment. The following table details essential components and their functions.
Table 3: Essential Reagents and Equipment for Combined RNAscope/IHC Workflow [40] [36] [68]
| Item Category | Specific Examples | Function in the Protocol |
|---|---|---|
| RNAscope Kits | RNAscope Fluorescent Multiplex Reagent Kit | Provides the necessary buffers and amplification reagents for the RNA in situ hybridization. |
| Target Probes | Species-specific probes for genes of interest (e.g., IL-1b, NLRP3); Positive control (PPIB, Polr2A); Negative control (dapB) | Hybridize to target mRNA; validate assay success and RNA integrity; assess background noise. |
| IHC Reagents | Primary antibodies (e.g., anti-IBA1, anti-NeuN); Fluorophore-conjugated secondary antibodies | Bind to specific protein markers for cell identification; generate fluorescent signal for protein detection. |
| Key Equipment | HybEZ II Hybridization Oven; Confocal or Fluorescent Microscope; Slide Scanner | Provides controlled temperature for hybridization; enables high-resolution imaging of multiple channels; digitizes slides for quantitative analysis. |
| Image Analysis Software | QuPath, HALO Image Analysis Platform, Aperio | Allows for automated or semi-automated quantification of RNA dots within IHC-defined cell boundaries. |
Data Analysis and Interpretation in Integrated Assays
Quantitative Image Analysis
The quantitative power of combined RNAscope/IHC lies in its ability to measure transcript abundance within specific, protein-defined cell populations. Analysis involves several steps [40] [36]:
- Cell Segmentation and Identification: Using software like QuPath or HALO, the IHC signal is used to automatically identify and create boundaries around specific cell types (e.g., all IBA1-positive microglia).
- RNA Dot Counting: Within the boundaries of each identified cell, the software counts the number of fluorescent RNA dots. Each dot corresponds to a single mRNA transcript.
- Thresholding and Validation: Signal thresholds for defining a positive transcript are rigorously derived using negative control probes (e.g., bacterial dapB) to account for any background noise, ensuring that only specific signals are quantified [40].
This method allows researchers to move beyond simply noting that a gene is expressed in a tissue. It enables precise questions such as, "What is the average number of IL-1b transcripts in microglia in the injured state versus the control state?" This provides a nuanced view of gene regulation at the cellular level [36].
Resolving Discrepancies and Gaining Deeper Insights
A key strength of this integrated approach is its ability to resolve discrepancies between RNA and protein expression, which can illuminate underlying biological mechanisms.
- Identifying Producing Cells: For secreted proteins like cytokines and growth factors, IHC alone may show diffuse protein localization in the extracellular matrix, making it impossible to identify the cell of origin. RNAscope pinpoints the cells that are actively transcribing the gene for that protein, revealing the source [64].
- Uncovering Regulatory Mechanisms: Simultaneous analysis can reveal post-transcriptional regulation. For instance, the presence of high RNA transcript levels with low corresponding protein levels may suggest inhibition of translation or rapid protein turnover. Conversely, low RNA with high protein could indicate stable proteins with long half-lives or highly efficient translation [64].
Advanced Applications and Multiplexing Strategies
The combination of RNAscope and IHC is a foundation for increasingly sophisticated spatial biology applications. The development of RNAscope HiPlex assays allows for the detection of up to 12 different RNA targets in formalin-fixed, paraffin-embedded (FFPE) tissues and up to 48 in fresh-fixed frozen tissues within a single sample [68]. This can be further combined with IHC for protein markers.
This powerful multiplexing capability enables researchers to:
- Validate Single-Cell RNA Sequencing (scRNA-seq): RNAscope HiPlex provides spatial validation of gene signatures discovered through scRNA-seq, confirming the location and co-expression of genes within the tissue architecture [68].
- Spatially Profile Complex Tissues: In complex environments like the tumor microenvironment (TME), researchers can simultaneously profile immune cell markers, chemokines, cytokines, and cancer cell markers to understand cellular interactions and functional states at a single-cell resolution [68].
RNAscope and IHC are not competing technologies but rather powerful allies in the molecular pathology toolkit. IHC provides an essential view of the proteomic landscape, revealing terminal cellular differentiation and function. RNAscope offers a precise window into active gene transcription, enabling the identification of producing cells and the detection of expression that may not yet be apparent at the protein level. As the field of diagnostics moves towards greater precision, the integration of RNA and protein data through these complementary techniques provides a more robust, comprehensive, and clinically actionable understanding of disease biology, ultimately strengthening diagnostic accuracy and informing therapeutic strategies.
Conclusion
RNAscope and IHC are not mutually exclusive but rather complementary technologies that offer distinct advantages for researchers and clinicians. RNAscope provides exceptional sensitivity and specificity for RNA detection with cellular resolution, proving particularly valuable for targets with low protein abundance, viral detection, and when confirming IHC findings. IHC remains the established gold standard for protein localization in routine diagnostics. The future of biomedical research and clinical diagnostics lies in the strategic integration of these technologies, leveraging RNAscope's precision for validation and discovery, and IHC's efficiency for high-throughput screening. Further prospective studies and cost-benefit analyses will solidify RNAscope's role in standardized clinical workflows, ultimately enhancing diagnostic accuracy and enabling more personalized therapeutic approaches.
References
- 1. Evaluation of the Suitability of RNAscope as a Technique ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8710359/]
- 2. Frontiers | Combining RNAscope and immunohistochemistry to... [https://www.frontiersin.org/journals/molecular-neuroscience/articles/10.3389/fnmol.2023.1225847/full]
- 3. of Comparison and immunohistochemistry for evaluation... RNAscope [https://pmc.ncbi.nlm.nih.gov/articles/PMC8759216/]
- 4. Comparison of RNAscope and immunohistochemistry for ... [https://diagnosticpathology.biomedcentral.com/articles/10.1186/s13000-022-01191-x]
- 5. Novel immunohistochemical assay utilizing the INSM1 (SP493 ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12532420/]
- 6. A Method for Combining RNAscope In Situ Hybridization... | PLOS One [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0120120]
- 7. RNAscope: A Novel in Situ RNA Analysis Platform for ... [https://www.sciencedirect.com/science/article/pii/S1525157811002571]
- 8. 2025 Jan 14 - Unlock the role of spatial profiling in ... [https://acdbio.com/2025-jan-14-unlock-role-spatial-profiling-detecting-diverse-mrna-markers-and-short-targets]
- 9. Mastering Spatial RNA Detection: High Specificity & ... [https://news.ki.se/calendar/histocore-mastering-spatial-rna-detection-high-specificity-sensitivity-with-rnascope-ish-on-leica-bond-rx-by-acdbio]
- 10. A robust experimental and computational analysis ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2022.911873/full]
- 11. The RNAscope IHC Breakthrough: When Clarity Matters [https://www.leicabiosystems.com/en-ch/knowledge-pathway/rnascope-ihc-breakthrough-when-clarity-matters/]
- 12. An Overview of Quantitative Image Analysis of RNAscope ... [https://acdbio.com/2023-oct-10-overview-quantitative-image-analysis-rnascope-ish-data-professional-assay-services]
- 13. RNAscope® technology | In Situ Hybridization, RNA-ISH [https://acdbio.com/dataanalysisguide]
- 14. Validation of a DKK1 RNAscope chromogenic in situ ... [https://www.nature.com/articles/s41598-021-89060-3]
- 15. Protocol for multiplexed RNAscope-based imaging of ... [https://www.sciencedirect.com/science/article/pii/S2666166725000218]
- 16. RNAscope: A Novel in Situ RNA Analysis Platform for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3338343/]
- 17. Single-Cell and Single-Molecule Analysis of Gene Expression ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5149423/]
- 18. Immunohistochemistry in Research and Diagnostics [https://www.delveinsight.com/blog/immunohistochemistry-explained-biomedical-research-diagnostics]
- 19. RNAscope Enables Spatial Genomic ISH ... [https://acdbio.com/science/how-it-works]
- 20. Evaluation of the Suitability of RNAscope as a Technique ... [https://link.springer.com/article/10.1007/s40291-021-00570-2]
- 21. Superior Determination of HPV using RNAscope [https://acdbio.com/superior-determination-hpv-using-rnascope%E2%84%A2]
- 22. RNAscope Multiplex Fluorescent V2 Assay | ACD [https://acdbio.com/rnascope-multiplex-fluorescent-v2-assay]
- 23. Immunohistochemistry (IHC): The Complete Guide [https://www.antibodies.com/applications/immunohistochemistry]
- 24. Bio-Techne Launches Expanded Menu of RNAscope ... [https://acdbio.com/bio-techne-launches-expanded-menu-rnascope-probes-human-and-mouse-transcriptome-advance-spatial]
- 25. Overview of multiplex immunohistochemistry ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7170662/]
- 26. Why Multiplex & Chromogenic vs Immunofluorescent ... [https://www.leicabiosystems.com/us/articles/ls-articles/tips-tricks-to-multiplexing-top-5-reasons-to-multiplex-and-chromogenic-versus/]
- 27. Viral RNA Detection using RNAscope® ISH | ACD [https://acdbio.com/science/applications/research-areas/infectious-diseases]
- 28. RNA sequencing and immunohistochemistry jointly ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12318096/]
- 29. RNAscope Workflow, In Situ Hybridization Steps | ACD [https://acdbio.com/ebook/introduction/rnascope-workflow]
- 30. 2025 Mar 18 - Accelerate RNA and Protein Biomarker ... [https://acdbio.com/2025-mar-18-accelerate-rna-and-protein-biomarker-development-new-rnascope%E2%84%A2-ish-protease-free-assays]
- 31. RNAscope Assay Workflow | In Situ Hybridization, RNA-ISH [https://acdbio.com/support-solution-category/rnascope-assay-workflow]
- 32. Immunohistochemistry for Pathologists: Protocols, Pitfalls, ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5122731/]
- 33. Intro to Immunohistochemistry (IHC) Staining: Steps & Best ... [https://www.leicabiosystems.com/us/knowledge-pathway/immunohistochemistry-an-overview-steps-to-better-ihc-staining/]
- 34. IHC Protocol | Step-by-Step Immunohistochemistry Guide [https://www.bosterbio.com/protocol-and-troubleshooting/ihc-protocol?srsltid=AfmBOoqbuZvyjRuwOKZP8VCRAvxMg5L4WVlTd63XJYZ8tNoln9BLQUVT]
- 35. Immunohistochemistry (IHC) protocol [https://hellobio.com/immunohistochemistry-ihc-protocol/]
- 36. Combining RNAscope and immunohistochemistry to visualize ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10470653/]
- 37. Comparison of Semi-Quantitative Scoring and Artificial ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8774232/]
- 38. Histocore: From Spots to Insights: AI-based Image Analysis ... [https://news.ki.se/calendar/histocore-from-spots-to-insights-ai-based-image-analysis-of-rnascope-data-in-visiopharm]
- 39. Semi-quantitative immunohistochemical assay versus ... [https://www.sciencedirect.com/science/article/pii/S0893395222016052]
- 40. Quantitative Analysis of Gene Expression in RNAscope ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9901451/]
- 41. Principles of Diagnosis of Infectious Diseases - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7149376/]
- 42. Center for Infectious Disease Diagnostics & Innovation [https://medicine.duke.edu/divisions/infectious-diseases/research/institutes-and-labs/center-infectious-disease-diagnostics]
- 43. RNAscope Troubleshooting Guide and FAQ [https://acdbio.com/technical-support/solutions]
- 44. RNAscope ISH Troubleshooting [https://www.bio-techne.com/resources/protocols-troubleshooting/rnascope-ish-troubleshooting]
- 45. Estrogen receptor α (ERα) status evaluation using ... [https://www.sciencedirect.com/science/article/abs/pii/S0046817716303434]
- 46. Unexpected Staining Patterns: ISH Staining | ACD [https://acdbio.com/ebook/troubleshooting/unexpected-staining-patterns]
- 47. Antibody Validation & Challenges [https://acdbio.com/science/applications/research-solutions/antibody-validation-and-challenges]
- 48. Control Slides and Control Probes for RNAscope [https://acdbio.com/control-slides-and-control-probes-rnascope]
- 49. Reliable detection of RNA in hippocampus sections of mice ... [https://link.springer.com/article/10.1007/s00418-024-02277-x]
- 50. An easy to use tool for the analysis of subcellular mRNA ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11004122/]
- 51. Roche VENTANA IHC lung cancer portfolio [https://diagnostics.roche.com/us/en/products/product-category/lung-cancer-diagnostics.html]
- 52. Performance Analysis of Leica Biosystems Monoclonal ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11227302/]
- 53. News [https://www.nordiqc.org/news.php]
- 54. A novel automated IHC staining system for quality control ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11864879/]
- 55. HALO | Quantitative Image Analysis for Pathology [https://indicalab.com/halo/]
- 56. An international multi-institutional validation study of the ... [https://www.nature.com/articles/s41698-023-00424-6]
- 57. A biologist's guide to planning and performing quantitative ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10298797/]
- 58. Correlation of HER2 Protein Level With mRNA ... [https://www.sciencedirect.com/science/article/pii/S0893395223003137]
- 59. Artificial intelligence-assisted HER2 interpretation for breast ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12261348/]
- 60. RNAscope™ ISH Probes for the Detection of Kappa ... [https://acdbio.com/rnascope%E2%84%A2-ish-probes-detection-kappa-lambda-and-igll5]
- 61. Utility of uroplakin II expression as a marker of urothelial ... [https://pubmed.ncbi.nlm.nih.gov/25449628/]
- 62. Comparison of RNAscope and immunohistochemistry for ... [https://pubmed.ncbi.nlm.nih.gov/35027056/]
- 63. RNAscope Troubleshooting Guide and FAQ [https://acdbio.com/technical-support/solutions/faq]
- 64. Complementary RNA & Protein Analysis [https://acdbio.com/science/applications/research-solutions/complementary-rna-protein-analysis]
- 65. A robust experimental and computational analysis ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9373800/]
- 66. Quantification of HER2-low and ultra-low expression in ... [https://www.sciencedirect.com/science/article/pii/S2153353925000999]
- 67. RNAscope Multiplex FISH Signal Assessment in FFPE and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11755420/]
- 68. RNAscope™ HiPlex v2 Assays | In Situ Hybridization, RNA ... [https://acdbio.com/rnascope-hiplex-assays]