Signaling Dynamics in Gastrulation: Integrating Mechanics, Pathways, and Models for Developmental Biology and Drug Discovery
Gastrulation is a pivotal developmental stage where a uniform cell sheet transforms into the structured embryonic germ layers, a process long thought to be directed solely by biochemical signals.
Signaling Dynamics in Gastrulation: Integrating Mechanics, Pathways, and Models for Developmental Biology and Drug Discovery
Abstract
Gastrulation is a pivotal developmental stage where a uniform cell sheet transforms into the structured embryonic germ layers, a process long thought to be directed solely by biochemical signals. This article synthesizes recent breakthroughs revealing that mechanical forces and the precise temporal dynamics of signaling pathways are equally critical. We explore foundational principles, highlighting the interplay between pathways like Nodal, BMP, and PCP. We detail cutting-edge methodologies, from Brillouin microscopy mapping mechanical properties in real-time to optogenetic tools and deep learning models like EmbryoNet for high-throughput phenotyping. The content addresses challenges in modeling human development and validates findings across model organisms. This integrated view of signaling dynamics offers profound implications for understanding congenital disorders and advancing regenerative medicine strategies.
Core Principles: How Signaling Dynamics and Mechanical Forces Orchestrate Germ Layer Formation
Gastrulation represents a pivotal developmental event wherein a uniform sheet of embryonic cells undergoes large-scale morphogenetic reorganization to form the three primary germ layers—ectoderm, mesoderm, and endoderm—and establish the fundamental body plan. This process is governed by an intricate interplay between biochemical signaling pathways and physical mechanical forces. This whitepaper synthesizes current research to delineate the signaling dynamics, including the roles of BMP, WNT, and Nodal pathways, and the emerging understanding of how tissue geometry, tension, and mechanical competence integrate with molecular cues to direct gastrulation. We provide a detailed analysis of experimental models, quantitative data, and methodologies that are advancing our understanding of these fundamental developmental mechanisms, with implications for regenerative medicine and therapeutic development.
Gastrulation is the critical developmental process during which a single-layered embryo transforms into a multilayered structure called the gastrula, possessing the three germ layers that will give rise to all fetal tissues [1]. In humans, this process commences around week three of development and is marked by the appearance of the primitive streak, a groove at the caudal end of the epiblast layer that establishes the cranial-caudal axis [1]. The initiation and progression of gastrulation are directed by a conserved set of signaling pathways, including TGF-β (encompassing BMP and Nodal), WNT, and others, which create patterning gradients across the embryo [1].
Historically, research has focused on these biochemical signals. However, emerging evidence underscores that mechanical forces are equally critical; gastrulation proceeds only when cells are both chemically prepared and physically primed [2]. This whitepaper explores the dynamics of these signals and forces, providing a technical guide for researchers dissecting the fundamental mechanisms of human development.
Core Signaling Pathways and Their Dynamics
The coordination of gastrulation relies on the precise spatiotemporal activity of several key signaling pathways. The table below summarizes the primary functions and known inhibitors of these pathways.
Table 1: Core Signaling Pathways in Gastrulation
| Signaling Pathway | Primary Role in Gastrulation | Key Inhibitors/Regulators |
|---|---|---|
| BMP | Initiates symmetry breaking; specifies extraembryonic mesoderm (ExM) and other cell fates [2] [3]. | BMP inhibitors (e.g., from hypoblast) help confine activity [1]. |
| WNT | Induces primitive streak formation; regulates convergence and extension movements [1] [4]. | Dkk-1, Crescent [1]. |
| Nodal | Promotes mesendodermal fate; works with WNT to induce streak formation [1] [3]. | Antagonists released by the hypoblast [1]. |
| TGF-β (Vg1) | Induces primitive streak formation; acts on Nodal to continue chemical cascade [1]. | Not specified in search results. |
| Non-canonical WNT (PCP) | Regulates directed cell migration and polarity during convergent extension [4]. | Not specified in search results. |
The Interplay of BMP, WNT, and Nodal Signaling
The formation of the primitive streak is initiated by a system of signaling pathways that work to both positively and negatively regulate downstream expression. The combination of TGF-β, WNT, Nodal, and BMPs is instrumental in this process [1]. WNT and TGF-β signaling appear to be primary inducers, with factors like Vg1 (a TGF-β member) shown to induce streak formation, partly by acting on Nodal [1]. Furthermore, WNT activation is crucial, as evidenced by the fact that Wnt antagonists like Dkk-1 and Crescent can prevent streak formation [1]. BMP signaling also plays a regulatory role, with its concentration typically lower near the streak and higher in the surrounding embryo, creating a gradient that guides cell differentiation [1].
Recent research using human embryonic stem cells (hESCs) confirms that the modulation of BMP, WNT, and Nodal signaling can rapidly (within 4-5 days) and efficiently (≈90%) induce the differentiation of both naive and primed hESCs into extraembryonic mesoderm-like cells (ExMs) [3]. This specification predominantly proceeds through intermediates exhibiting a primitive streak-like gene expression pattern, delineating the regulatory roles of WNT and Nodal signaling [3].
Signaling in Model Organisms
Studies in model organisms like Drosophila and zebrafish have been instrumental in elucidating the conserved mechanisms of gastrulation signaling.
- Drosophila: Gastrulation is triggered by a gradient of the morphogen Spätzle, which peaks at the ventral midline and leads to the nuclear translocation of the transcription factor Dorsal [5]. Different thresholds of nuclear Dorsal concentration activate distinct target genes; high concentrations activate mesoderm-specific genes like twist and snail [5]. The connection between gene expression and morphogenesis is exemplified by mesoderm invagination, which is driven by apical constriction activated by secreted ligands and GPCR signaling [5].
- Zebrafish: Gastrulation involves large-scale morphogenetic processes like epiboly, internalization, and convergent extension [4]. The non-canonical Wnt/Planar Cell Polarity (PCP) pathway is a major regulator of these polarized cell movements, controlling processes like directed cell migration and the narrowing/extension of the body axis [4]. This pathway, which includes proteins like Frizzled, Dishevelled, and Prickle, regulates the cytoskeleton to orchestrate collective cell behavior [4].
The Role of Mechanical Forces in Gastrulation
A paradigm shift in gastrulation research is the recognition that biochemical signals alone are insufficient; physical forces are a necessary and integral component.
Mechanical Competence and Symmetry Breaking
Research using optogenetic tools to activate BMP4 in human embryonic stem cells revealed that chemical cues alone could not fully initiate gastrulation. In unconfined, low-tension environments, BMP4 activation gave rise to extra-embryonic cell types but failed to generate the mesoderm and endoderm layers that form the body's organs [2]. However, when the same BMP4 signal was activated in confined cell colonies or in tension-inducing hydrogels, the missing germ layers began to form [2]. This demonstrates that mechanical tension is a prerequisite for symmetry breaking and germ layer formation.
Further experiments revealed the molecular mechanism behind this phenomenon: the mechanosensory protein YAP1 acts as a molecular brake on gastrulation. Nuclear YAP1 prevents transformation until mechanical tension releases this brake, thereby fine-tuning the downstream biochemical signaling pathways mediated by WNT and Nodal [2]. This interdependence defines a state of mechanical competence that cells must achieve to progress through developmental milestones [2].
Cellular Mechanisms of Force Generation
At the cellular level, force generation is driven by the actomyosin cytoskeleton.
- Apical Constriction: In Drosophila, mesoderm invagination is driven by apical constriction, where cells contract their apical surfaces using actin and nonmuscle myosin 2 (myosin 2) [5]. This changes cell shape from columnar to wedge-shaped, promoting inward tissue curvature [5].
- Actomyosin Networks: The assembly of multicellular actomyosin networks allows force to propagate across hundreds of cells, enabling coordinated tissue sculpting. Such networks are conserved and also operate during gastrulation and neural tube closure in vertebrates [5].
Experimental Models and Methodologies
Advances in stem cell biology and bioengineering have provided powerful models to study human gastrulation, circumventing the ethical and technical challenges of working with human embryos.
hESC-Based Models of Extraembryonic Mesoderm Specification
An efficient culture system has been established to differentiate hESCs into expandable ExM-like cells (ExMs). The protocol below details the methodology.
Table 2: Experimental Protocol for ExM Specification from hESCs
| Step | Parameter | Details |
|---|---|---|
| 1. Cell Preparation | Cell Line | AIC-N human embryonic stem cells (hESCs) [3]. |
| Surface Coating | Matrigel-coated dishes [3]. | |
| 2. Culture Medium | Base Medium | FH-N2B27 (modified N2B27 medium containing FGF4 and heparin) [3]. |
| Key Additives | CHIR99021 (CHIR, a GSK3 inhibitor) and BMP4 [3]. | |
| 3. Process | Differentiation Duration | 4 days [3]. |
| Efficiency | ~90% conversion to ExM-like cells [3]. | |
| 4. Validation | Markers (Upregulated) | GATA6, SNAIL, VIM, KDR, FLT1, HAND1, PDGFRA [3]. |
| Markers (Downregulated) | POU5F1 (OCT4), NANOG, SOX2 [3]. | |
| Analysis Methods | Immunofluorescence, Flow Cytometry, Bulk RNA-seq, scRNA-seq [3]. |
Optogenetic Control and Synthetic Embryo Models
To dissect the interplay between biochemical signals and mechanical forces, an optogenetic tool was developed to activate developmental genes with spatiotemporal precision [2].
- Protocol: Human embryonic stem cells are engineered to express a light-sensitive switch that permanently turns on BMP4 upon exposure to a specific wavelength of light [2].
- Application: Researchers can shine light on specific regions of confined cell colonies to test how tissue geometry and mechanical stress influence developmental outcomes. This system revealed that gastrulation requires mechanical tension in concert with BMP4 signaling [2].
- Mathematical Modeling: A "digital twin" of a developing embryo, built using actual measurements of mechanical tension, can simulate how biochemical signals and physical forces interact to self-organize. These simulations closely match experimental observations [2].
Engineered Models of Peri-Gastrulation
The field has seen a rapid expansion of engineered models that replicate specific stages of development:
- Pre-gastrulation models (e.g., Blastoids): Replicate blastocyst formation and implantation [6].
- Gastrulation models (e.g., 2D micropatterned systems, 3D gastruloids): Provide insights into germ layer formation and tissue organization [6].
- Post-gastrulation models (e.g., Somitoids): Mimic early somitogenesis and axial elongation [6].
These systems are enhanced by engineering technologies like micropatterned substrates, microfluidic systems, and synthetic biology tools, which allow for precise control over the cellular microenvironment [6].
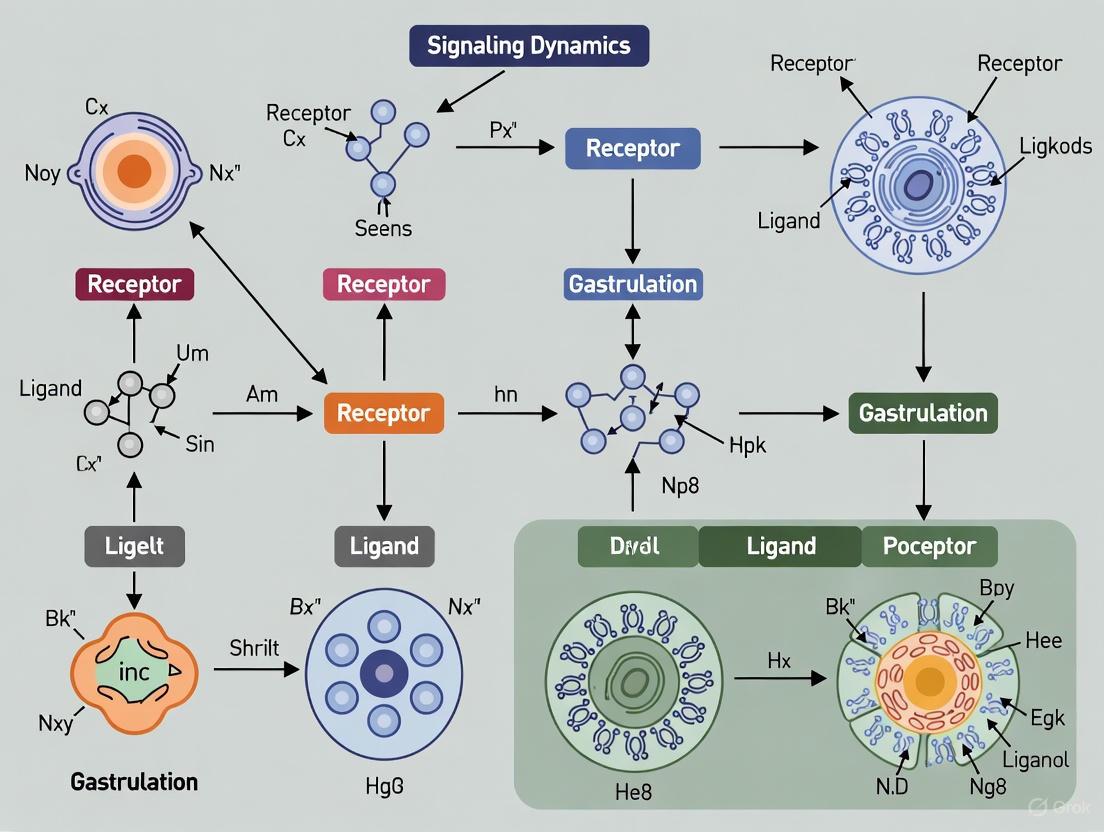
Visualization of Signaling Pathways and Workflows
BMP/WNT/Nodal Signaling in Gastrulation
Diagram Title: Signaling and Mechanical Control of Gastrulation
Experimental Workflow for ExM Specification
Diagram Title: Workflow for Directing ExM from hPSCs
The Scientist's Toolkit: Essential Research Reagents
The following table compiles key reagents and their applications for investigating signaling dynamics in gastrulation.
Table 3: Research Reagent Solutions for Gastrulation Studies
| Reagent / Tool | Function / Application | Example Use in Gastrulation Research |
|---|---|---|
| CHIR99021 | GSK-3 inhibitor; activates WNT signaling. | Used with BMP4 to efficiently differentiate hESCs into ExM cells [3]. |
| BMP4 | Morphogen; key signaling protein. | Induces symmetry breaking and specifies cell fates like ExM [3] [2]. |
| FGF4 & Heparin | Growth factor and co-factor; supports cell growth. | Component of the base FH-N2B27 medium for ExM differentiation [3]. |
| Optogenetic BMP4 Switch | Light-activated genetic switch for BMP4. | Precisely controls timing and location of BMP4 signaling to study force interplay [2]. |
| Micropatterned Substrates | Engineered surfaces to control cell geometry. | Controls colony shape and internal mechanical stresses in 2D gastruloid models [2] [6]. |
| Tension-Inducing Hydrogels | Synthetic extracellular matrices. | Provides mechanical confinement to study the role of tension in germ layer specification [2]. |
| AIC-N hESCs | Naive human embryonic stem cell line. | Cell line used for efficient ExM differentiation studies [3]. |
| 2,2,2',4'-Tetrachloroacetophenone | 2,2,2',4'-Tetrachloroacetophenone | High Purity | High-purity 2,2,2',4'-Tetrachloroacetophenone for research. A key synthon & photoinitiator. For Research Use Only. Not for human or veterinary use. |
| 2-hexan-3-yloxycarbonylbenzoic acid | 2-hexan-3-yloxycarbonylbenzoic Acid | High-Purity Reagent | 2-hexan-3-yloxycarbonylbenzoic acid is a key reagent for organic synthesis and pharmaceutical research. For Research Use Only. Not for human or veterinary use. |
The process of gastrulation is no longer viewed as a simple cascade of biochemical signals but as an integrated system where biochemical gradients and physical forces are inextricably linked. Core signaling pathways—BMP, WNT, and Nodal—provide the instructional code, but the physical context of the cells, their mechanical tension, and tissue geometry provide the necessary permissive conditions for execution. The emergence of sophisticated models, from hESC differentiation systems to optogenetically controlled synthetic embryos, is providing unprecedented insight into these dynamics. This deeper understanding not only illuminates the fundamental principles of human development but also paves the way for advances in regenerative medicine by providing controlled methods to specify cell fates and engineer tissues.
This whitepaper provides a comprehensive technical analysis of the Nodal, BMP/WNT, and Planar Cell Polarity (PCP) signaling pathways, focusing on their integrated roles during gastrulation. These evolutionarily conserved pathways form a critical signaling network that governs embryonic patterning, germ layer specification, and tissue morphogenesis. We examine the molecular mechanisms, pathway crosstalk, and dynamic signaling behaviors that coordinate these processes, with particular emphasis on quantitative signaling parameters and experimental approaches relevant to in vitro model systems. Additionally, we present a curated toolkit of research reagents and visualization resources to facilitate further investigation into the role of signaling dynamics in gastrulation research.
Gastrulation represents a pivotal phase in embryonic development where a homogeneous population of pluripotent epiblast cells self-organizes into the three primary germ layers—ectoderm, mesoderm, and endoderm—that form the entire embryo. Decades of research in model organisms and human stem cell models have revealed that a precisely orchestrated signaling cascade involving Bone Morphogenetic Protein (BMP), WNT, and NODAL pathways is indispensable for initiating gastrulation [7]. These pathways activate specific genetic programs that dictate cellular fate decisions through precise spatiotemporal dynamics rather than stable signaling gradients [7]. The planar cell polarity (PCP) pathway, while often associated with later morphogenetic events, also contributes to the coordinated cellular movements and polarity establishment during this critical developmental window. Understanding the integrated dynamics of these pathways provides crucial insights into the fundamental principles of embryonic patterning and offers valuable paradigms for directed differentiation of stem cells for regenerative medicine applications.
Pathway Mechanisms and Molecular Components
Nodal Signaling Pathway
The Nodal signaling pathway is a specialized branch of the transforming growth factor-β (TGF-β) superfamily that plays central roles in early embryonic patterning, including mesoderm specification, anterior-posterior axis patterning, and left-right axis determination [8]. Nodal signaling utilizes core TGF-β receptors and Smad-dependent signaling components but possesses several distinctive characteristics. The pathway requires EGF-CFC family co-receptors (Cripto and Cryptic) for effective signaling complex formation [8]. Following ligand binding to type I and type II receptor heterodimers, phosphorylated R-Smads form trimeric complexes with Smad4 that translocate to the nucleus. These complexes specifically cooperate with FoxH1 or Mixer transcription factors to regulate spatial and temporal expression of Nodal-dependent target genes [8]. The pathway is subject to unique negative regulation by Lefty proteins, which inhibit Nodal signaling through direct interaction with both the ligand and EGF-CFC co-receptors [8].
BMP and WNT Signaling Pathways
The BMP and WNT pathways function as interconnected regulators of cell fate decisions during gastrulation and organogenesis. BMP signaling, another TGF-β family pathway, exhibits concentration-dependent effects on cell fate specification, with lower levels promoting intermediate mesoderm formation and higher levels favoring lateral plate mesoderm development [9]. The canonical WNT/β-catenin pathway regulates target gene expression through a sophisticated protein degradation mechanism. In the absence of WNT ligands, β-catenin is phosphorylated by a multiprotein destruction complex comprising Axin, APC, GSK3β, and CK1α, marking it for proteasomal degradation [10]. WNT ligand binding to Frizzled receptors and LRP5/6 co-receptors disrupts this destruction complex, allowing β-catenin accumulation and nuclear translocation where it associates with TCF/LEF transcription factors to activate target genes [10]. Non-canonical WNT pathways, including the Wnt/PCP and Wnt/Ca²⺠pathways, function independently of β-catenin to modulate cell polarity, migration, and calcium signaling [10].
Planar Cell Polarity (PCP) Signaling
The Planar Cell Polarity pathway coordinates the polarization of cells within the epithelial plane, enabling uniform orientation of cellular structures and directional movements. Core PCP components include Frizzled (Fz), Flamingo (Fmi), Van Gogh (Vang), Prickle (Pk), and Dishevelled (Dsh) [11]. These proteins undergo asymmetric subcellular localization, forming distinct molecular complexes on opposite sides of cells [12]. The system employs intercellular feedback loops where Fmi homodimers bridge opposing complexes in adjacent cells, communicating polarity information across the tissue [12]. Recent research demonstrates that PCP signaling can polarize cells autonomously without intercellular communication, though tissue-wide coordination is lost under these conditions [12]. This pathway ensures properly oriented cell divisions, asymmetric cellular morphology, and directional cell migration—processes essential for neural tube closure, inner ear hair cell orientation, and other morphogenetic events [11].
Table 1: Core Components of Key Signaling Pathways in Gastrulation
| Pathway | Key Components | Regulators | Primary Functions |
|---|---|---|---|
| Nodal | Nodal, Type I/II TGF-β receptors, Smad2/3, Smad4, FoxH1 | Cripto, Cryptic, Lefty | Mesoderm specification, anterior-posterior patterning, left-right axis determination |
| BMP | BMP ligands, BMP receptors, Smad1/5/8, Smad4 | Noggin, Chordin | Germ layer patterning, intermediate mesoderm specification at low levels |
| Canonical WNT | Wnt ligands, Frizzled, LRP5/6, Dvl, β-catenin, GSK3β, APC, Axin, TCF/LEF | DKK1, IWP2 | Primitive streak formation, mesoderm induction, posterior patterning |
| Non-canonical WNT | Wnt ligands, Frizzled, ROR2, Dvl, RAC, RHOA, JNK, PLC | - | Cell polarity, migration, calcium signaling |
| PCP | Frizzled, Flamingo, Van Gogh, Prickle, Dishevelled, Diego | Ft, Ds, Fj | Tissue-scale cell alignment, directional cell movements, oriented cell divisions |
Quantitative Signaling Dynamics in Gastrulation
Signaling Dynamics and Thresholds
Quantitative analysis of signaling pathway dynamics reveals distinct temporal behaviors during gastrulation. BMP signaling initiates first, becoming restricted to colony edges within 12 hours in micropatterned human gastruloid systems [7]. This initial BMP activation triggers sequential waves of WNT and NODAL signaling activity that propagate toward the colony center at constant rates [7]. Mathematical modeling of these dynamics suggests they are inconsistent with reaction-diffusion-based Turing systems, indicating the absence of stable WNT/NODAL signaling gradients [7]. Instead, the final signaling state is homogeneous, with spatial differences arising primarily from boundary effects. The duration rather than the concentration of WNT and NODAL signaling appears critical for mesoderm differentiation, while BMP signaling duration controls extra-embryonic cell differentiation [7].
Table 2: Quantitative Parameters for Signaling Pathway Manipulation in Stem Cell Differentiation
| Signaling Pathway | Modulator | Concentration/Type | Effect on Differentiation | Experimental Context |
|---|---|---|---|---|
| WNT | CHIR99021 (GSK3β inhibitor) | 3-8 μM | Induces TBXT+/MIXL1+ mesoderm progenitors | Intermediate mesoderm differentiation [9] |
| BMP | BMP4 | 4-100 ng/mL | Low concentrations (4 ng/mL) with WNT promote OSR1+/GATA3+/PAX2+ intermediate mesoderm | Optimized IM differentiation protocol [9] |
| Nodal/Activin | Activin A | 100 ng/mL | Induces primitive streak and mesendoderm | Conventional differentiation protocol [9] |
| Nodal Inhibition | SB-431542 (ALK4/5/7 inhibitor) | Not specified | Blocks Activin/Nodal/TGFβ signaling | Primitive streak induction studies [13] |
| WNT Inhibition | DKK1 | Not specified | Inhibits canonical WNT signaling | Primitive streak induction studies [13] |
Pathway Crosstalk and Combinatorial Signaling
Extensive crosstalk occurs between the Nodal, BMP, and WNT pathways, forming an integrated signaling network that collectively orchestrates cell fate decisions. Studies in embryonic stem cells demonstrate that Activin/Nodal and WNT signaling are required for primitive streak induction, while BMP signaling exerts a posteriorizing effect on this population [13]. All three pathways regulate the induction of Flk1+ mesoderm, but their requirements shift during subsequent specification events [13]. The combinatorial interpretation of BMP and WNT signaling specifically controls the decision between primitive streak and extraembryonic fates [14]. This interplay extends to PCP signaling, which intersects with WNT pathways through shared components like Frizzled and Dishevelled, creating a complex regulatory network that coordinates tissue patterning with cellular polarization [10] [11].
Experimental Approaches and Methodologies
Gastruloid Models and Micropatterned Systems
Micropatterned in vitro models of human gastrulation provide a robust platform for quantifying signaling dynamics and their relationship to cell fate patterning. In these systems, human pluripotent stem cells are confined to circular micropatterns and treated with BMP4 to initiate self-organized differentiation [7]. This approach generates radial patterns of germ layer organization: an outer ring of CDX2+ extra-embryonic cells, followed by concentric rings of SOX17+ endoderm, BRACHYURY+ mesoderm, and a central region of NANOG+/SOX2+ pluripotent cells [7]. The reproducibility and geometric uniformity of these systems enable precise quantification of signaling dynamics using live-cell imaging of pathway reporters, mathematical modeling, and systematic perturbation studies. This experimental paradigm has demonstrated that self-organized patterning can occur in the absence of stable signaling gradients, highlighting the importance of dynamic signal interpretation in cell fate decisions [7].
Directed Differentiation Protocols
Optimized differentiation protocols for generating specific progenitor populations provide valuable insights into pathway requirements at distinct developmental stages. For intermediate mesoderm differentiation, a highly efficient two-step protocol has been established: treatment of human pluripotent stem cells with 3 μM CHIR99021 for 48 hours induces TBXT+/MIXL1+ mesoderm progenitors, followed by treatment with 3 μM CHIR99021 and 4 ng/mL BMP4 for an additional 48 hours to generate OSR1+/GATA3+/PAX2+ intermediate mesoderm cells [9]. This protocol emphasizes the importance of suppressing high Nodal signaling during the mesoderm step and employing low BMP4 concentrations for specific IM induction, contrasting with conventional approaches that use higher BMP4 concentrations (100 ng/mL) [9]. Stage-specific pathway requirements have also been delineated for hematopoietic specification, where WNT signaling is essential for commitment to the primitive erythroid lineage but not required for definitive hematopoietic lineages [13].
Research Reagent Solutions
Table 3: Essential Research Reagents for Investigating Gastrulation Signaling Pathways
| Reagent | Type | Function/Application | Example Use |
|---|---|---|---|
| CHIR99021 | Small molecule inhibitor | GSK3β inhibitor that activates WNT signaling | Induces mesoderm progenitors at 3-8 μM [9] |
| BMP4 | Recombinant protein | BMP pathway ligand; patterns mesoderm | Promotes IM fate at low concentrations (4 ng/mL) [9] |
| Activin A | Recombinant protein | Nodal pathway surrogate; induces mesendoderm | Primitive streak induction at 100 ng/mL [9] |
| IWP2 | Small molecule inhibitor | Inhibits WNT ligand secretion | Blocks WNT signaling in perturbation studies [7] |
| SB-431542 | Small molecule inhibitor | Inhibits Activin/Nodal/TGFβ signaling (ALK4/5/7 inhibitor) | Blocks Nodal signaling in primitive streak studies [13] |
| DKK1 | Recombinant protein | Canonical WNT signaling inhibitor | Blocks WNT signaling in primitive streak induction [13] |
| BMPR-Fc fusion proteins | Recombinant receptor | Sequesters BMP ligands to inhibit signaling | BMP pathway inhibition studies [13] |
| NODAL −/− cells | Genetically modified cells | CRISPR-Cas9 knockout of NODAL | Studying Nodal-independent differentiation [7] |
Signaling Pathway Visualizations
Nodal Signaling Pathway
Diagram Title: Nodal Signaling Mechanism
BMP/WNT/PCP Signaling Network
Diagram Title: BMP-WNT-PCP Signaling Cascade
Planar Cell Polarity Mechanism
Diagram Title: PCP Asymmetric Complex Formation
Gastruloid Differentiation Protocol
Diagram Title: Intermediate Mesoderm Differentiation Workflow
The Nodal, BMP/WNT, and PCP signaling pathways constitute an integrated regulatory network that coordinates cell fate specification and tissue patterning during gastrulation through dynamic signaling behaviors rather than static concentration gradients. The quantitative parameters, experimental models, and research reagents outlined in this technical guide provide a foundation for investigating the sophisticated signaling dynamics that govern embryonic development. Future research directions include elucidating the molecular mechanisms of pathway crosstalk at higher resolution, developing more sophisticated mathematical models that predict signaling outcomes across diverse cellular contexts, and applying these insights to improve the efficiency and fidelity of stem cell differentiation for regenerative medicine applications. The continued integration of experimental and computational approaches will be essential for unraveling the complex signaling dynamics that orchestrate gastrulation and embryonic patterning.
Gastrulation is a fundamental milestone in early embryogenesis, responsible for transforming a simple sheet of cells into a complex, multi-layered structure with defined body axes. While the genetic and biochemical signals governing this process have been extensively studied, the crucial role of mechanical forces has only recently been fully appreciated. The emerging paradigm in developmental biology recognizes that mechanical forces and tissue mechanics are not merely passive outcomes but active regulators that coordinate cell behaviors across thousands of cells to drive robust and reproducible morphogenesis [15]. This mechanobiological perspective reveals that gastrulation represents an intricate dance between biochemical signaling and physical forces, where mechanochemical feedback loops ensure precise spatial and temporal coordination of cell fate specification and tissue remodeling [15] [16].
The framework of gastrulation must account for how short-range and long-range signaling integrates cell behaviors at the tissue and organism scale. Mechanical stresses generated by cellular activities create local and global feedback, coordinating cell behaviors through mechanosensitive signaling pathways [15]. Within this context, the interplay between tissue mechanics and classic morphogen signaling pathways such as BMP4, WNT, and Nodal creates a sophisticated regulatory system that guides symmetry breaking and germ layer formation [2] [16]. Understanding these mechanochemical principles provides not only fundamental biological insights but also practical applications for regenerative medicine and therapeutic development.
Core Mechanical Processes Driving Gastrulation
Key Cell Behaviors and Their Mechanical Basis
Gastrulation is driven by active forces arising from energy-consuming molecular motor activity and filament polymerization. These molecular-scale activities manifest in specific cell behaviors that collectively drive large-scale tissue transformations [15].
Table 1: Mechanical Cell Behaviors in Gastrulation
| Cell Behavior | Mechanical Function | Molecular Mechanisms | Representative Systems |
|---|---|---|---|
| Intercalation | Tissue elongation through convergent extension; generates tissue flows | Junctional actin-myosin cables; super-cellular myosin structures | Chick epiblast; Drosophila gastrulation |
| Internalization/EMT | Cell ingression through primitive streak; tissue internalization | Myosin II-driven apical constriction; complete EMT | Amniote mesendoderm precursors; Drosophila ventral furrow |
| Cell Division | Stress relief; tissue fluidity; oriented divisions relieve anisotropic stress | Division orientation along convergence axis | Avian embryos; frog and fish models |
| Apical Constriction | Tissue folding and invagination | Medial-apical actomyosin accumulation | Drosophila ventral furrow formation |
Intercalation and Convergent Extension
Intercalation involves neighboring cells exchanging positions, driving tissue elongation through a process called convergent extension [15]. In chick embryos, epithelial epiblast cells intercalate by shortening and remodeling cell-cell junctions through junctional actin-myosin cables [15]. The mechanical signature of this process includes the formation of super-cellular myosin cables spanning 2-8 cell junctions that orient perpendicular to the elongating streak, creating aligned junctional contractions [15]. When myosin phosphorylation is inhibited, these intercalation-associated tissue flows and streak formation are blocked, demonstrating the essential mechanical nature of this process [15].
Internalization and Epithelial-Mesenchymal Transition
Internalization processes allow mesoderm and endoderm precursors to move from the surface to the interior of the developing embryo. In amniotes, this involves a complete epithelial-mesenchymal transition (EMT), where cells individually ingress through the primitive streak [15]. The mechanical driver of this process is myosin II-driven apical constriction, where apical cell surface areas shrink before ingression, eventually leading to full EMT and migration beneath the epiblast [15]. Similar processes occur in Drosophila gastrulation, where ventral cells accumulate medial-apical actomyosin, causing apical constriction, tissue folding, and invagination of the mesoderm [17].
Material Properties and Their Dynamics
The material properties of embryonic tissues play a crucial role in gastrulation, undergoing dynamic changes that facilitate specific morphogenetic events. Recent advances in Brillouin microscopy have enabled the characterization and spatial mapping of the dynamics of cell material properties during Drosophila gastrulation in three dimensions and with high temporal resolution [17].
Table 2: Quantitative Measurements of Material Properties During Gastrulation
| Measurement Parameter | Technical Approach | Key Findings | Biological Significance |
|---|---|---|---|
| Longitudinal Modulus | Line-scan Brillouin microscopy (LSBM) | Transient increase in Brillouin shift within sub-apical compartment of mesodermal cells during ventral furrow formation | Identifies rapid, spatially varying changes in material properties correlated with cell fate |
| Brillouin Shift Dynamics | GHz-frequency mechanical probing | Differential dynamics between mesodermal and ectodermal cells; peaks at initiation of invagination | Reveals fate-specific mechanical signatures; suggests microtubule involvement |
| Microtubule Contribution | Colcemid disruption experiments | Reduced Brillouin shift during ventral furrow formation after microtubule disruption | Identifies microtubules as potential mechano-effectors |
These studies reveal that blastoderm cells undergo rapid and spatially varying changes in their material properties that differ according to cell fate and behavior [17]. Specifically, mesodermal cells display a transient increase in Brillouin shift (a proxy for longitudinal modulus) within their sub-apical compartment during ventral furrow formation, coinciding with the reorganization of sub-apical microtubules [17]. This mechanical transition is functionally important, as physical models confirm that a dynamic sub-apical increase in longitudinal modulus is essential for proper fold formation [17].
Experimental Approaches and Methodologies
Optogenetic Control of Morphogen Signaling
The recently developed optogenetic tools for controlling morphogen signaling represent a breakthrough in dissecting the interplay between biochemical and mechanical signals. This approach enables precise spatial and temporal activation of developmental genes, allowing researchers to test how tissue geometry and mechanical stress influence developmental outcomes [2] [16].
Experimental Protocol: Light-Inducible BMP4 Signaling in hESCs
Cell Line Engineering: Insert the human BMP4 coding sequence downstream of a loxP-flanked stop cassette in a piggyBac vector, enabling controlled gene expression via light-induced loxP recombination [16].
Light Sensitivity Induction: Treat cells with DOX (doxycycline) to confer light sensitivity by inducing the expression of the Cre recombinase fused to the light-inducible Cry2/CIB1 system [16].
Spatiotemporal Activation: Expose cells to specific wavelengths of blue light (450-490 nm) to activate BMP4 signaling in precise patterns and locations [16].
Mechanical Context Manipulation: Apply the optogenetic stimulation in different mechanical environments:
- Low-tension environments: Unconfined cell cultures
- High-tension environments: Cells confined on micropatterns or embedded in tension-inducing hydrogels [2]
Outcome Assessment: Analyze the resulting patterns of cell differentiation, specifically the emergence of mesoderm and endoderm lineages, which require both BMP4 signaling and mechanical tension [2].
This methodology revealed that BMP4 signaling alone is insufficient to drive complete gastrulation - mechanical tension is equally essential. The combination of optogenetic control with mechanical manipulation demonstrated that YAP1 nuclear localization acts as a mechanical sensor, fine-tuning downstream biochemical signaling pathways mediated by WNT and Nodal [2].
Brillouin Microscopy for Material Property Mapping
Brillouin microscopy provides a non-invasive method for measuring material properties in living embryonic tissues with high spatial and temporal resolution. This technique exploits the inelastic interaction between light and biological matter, where the energy shift of scattered photons (Brillouin shift) correlates with the material's longitudinal modulus [17].
Experimental Protocol: Line-Scan Brillouin Microscopy (LSBM) for Gastrulation Studies
Sample Preparation: Mount live Drosophila embryos expressing appropriate fluorescent markers for cell fate identification in appropriate imaging chambers [17].
Instrument Setup: Configure the line-scan Brillouin microscope with:
- Laser source at 532 nm wavelength
- Scanning system for line illumination
- Tandem Fabry-Pérot interferometer for spectral analysis
- CCD camera for signal detection [17]
Data Acquisition:
- Perform sequential line scans across the region of interest
- Acquire data at multiple z-planes to generate 3D maps
- Maintain time intervals of 2-5 minutes between full volume acquisitions to capture dynamics [17]
Brillouin Shift Calculation:
- Process raw spectra to extract Brillouin shift values
- Convert shift values to longitudinal modulus using appropriate calibration (requires refractive index and mass density measurements) [17]
Spatiotemporal Analysis:
- Register Brillouin maps with fluorescence images to correlate mechanical properties with cell fate
- Track temporal evolution of mechanical properties in specific cell populations (mesodermal vs. ectodermal) [17]
Perturbation Experiments:
- Apply cytoskeletal drugs (e.g., Colcemid for microtubule disruption)
- Compare Brillouin shift dynamics in control vs. perturbed conditions [17]
This approach has revealed that mesodermal cells undergo a transient increase in longitudinal modulus during ventral furrow formation, while ectodermal cells display different mechanical dynamics, demonstrating fate-specific mechanical signatures [17].
Integrated Mechanochemical Signaling Pathways
The coordination of gastrulation emerges from the sophisticated interplay between mechanical forces and biochemical signaling pathways. Research across model systems has revealed conserved principles of mechanochemical integration that ensure robust patterning and morphogenesis.
The diagram above illustrates the core mechanochemical signaling network governing gastrulation. Mechanical forces, including tissue tension and geometric confinement, activate the YAP/TAZ mechanosensory system, which enables and fine-tunes the response to morphogen signaling including BMP4, WNT, and Nodal [2] [16]. These pathways collectively regulate cell fate specification, driving behaviors such as intercalation and EMT that generate further mechanical forces, creating a reinforcing feedback loop that ensures robust pattern formation [15] [2] [16].
This mechanochemical integration operates across multiple scales, from molecular mechanisms within individual cells to tissue-scale force generation and patterning. The presence of mechanical competence - where cells must be physically primed as well as chemically prepared - adds a crucial layer of regulation that ensures gastrulation proceeds with proper timing and spatial coordination [2].
Research Reagent Solutions Toolkit
Table 3: Essential Research Reagents for Mechanobiology Studies
| Reagent/Category | Specific Examples | Function/Application | Experimental Context |
|---|---|---|---|
| Optogenetic Systems | Light-inducible BMP4; Cry2/CIB1-Cre system | Spatiotemporal control of gene expression; tests biochemical-mechanical interplay | Human embryonic stem cells; synthetic embryo models [2] [16] |
| Mechanical Manipulation | Micropatterned substrates; tunable hydrogels; confinement devices | Controls tissue geometry and mechanical stress; tests force-response relationships | 2D gastruloids; embryonic explants [2] [16] |
| Live Imaging Reporters | Actin markers (Utrophin-GFP); Myosin reporters (Myosin-II-RLC-GFP); YAP localization biosensors | Visualizes cytoskeletal dynamics and mechanosensitive protein localization | Drosophila, chick, mouse embryos; live imaging [15] [17] |
| Cytoskeletal Drugs | Colcemid (microtubule disruption); Blebbistatin (myosin inhibition); Latrunculin (actin disruption) | Tests specific cytoskeletal contributions to mechanical properties | Drosophila embryos; tissue explants [17] |
| Mechanosensory Inhibitors | YAP/TAZ pathway inhibitors; ROCK inhibitors | Disrupts mechanotransduction pathways; tests mechanical signaling necessity | Human pluripotent stem cell models [2] [16] |
| Antibodies for Mechanobiology | Phospho-SMAD1/5; BRA/T; GATA6; SOX17; Phospho-myosin | Detects activation of biochemical and mechanical signaling pathways | Immunostaining across model systems [16] |
| Tetrabutylammonium diphenylphosphinate | Tetrabutylammonium diphenylphosphinate, CAS:208337-00-2, MF:C28H46NO2P, MW:459.6 g/mol | Chemical Reagent | Bench Chemicals |
| Diisopropyl chloromalonate | Diisopropyl Chloromalonate | High Purity | RUO Supplier | Diisopropyl chloromalonate: A versatile chloromalonate ester for organic synthesis & peptide mimicry. For Research Use Only. Not for human use. | Bench Chemicals |
This toolkit enables researchers to probe both the generation and sensing of mechanical forces, providing comprehensive approaches to dissect the mechanobiology of gastrulation. The combination of optogenetic control with mechanical manipulation and advanced imaging represents particularly powerful emerging methodology [2] [16] [17].
The mechanobiology perspective reveals gastrulation as a process orchestrated by the intimate interplay between physical forces and biochemical signals. Mechanical competence - where tissues must achieve specific physical states to respond to morphogenetic cues - emerges as a fundamental principle ensuring the robust spatial and temporal progression of development [2]. The integration of YAP/TAZ-mediated mechanosensation with classic morphogen pathways creates a sophisticated regulatory system that translates tissue-scale forces into precise cellular responses [2] [16].
Future research directions will likely focus on identifying the potential existence of a mechanical organizer - a force-based counterpart to classical signaling centers that shape the early embryo [2]. Additionally, the development of more sophisticated synthetic embryo models and advanced mechanical measurement techniques will continue to bridge scales from molecular mechanisms to embryo-scale morphogenesis. These advances will not only deepen our understanding of human development but also provide novel approaches to regenerative medicine and fertility treatments by revealing the fundamental rules governing how mechanical forces guide the emergence of form in early embryogenesis [2].
The transformation of a homogeneous cell population into diverse, specialized lineages is a cornerstone of development and disease. This whitepaper synthesizes current research to elucidate how cells interpret extracellular signals not just from their identity, but from their dynamic patterns—their intensity, rhythm, and duration. We explore the core principle that signaling dynamics are a critical mechanism for dictating cell fate decisions, with a specific focus on the paradigm of gastrulation. By integrating quantitative data, experimental protocols, and visual guides, this document provides researchers and drug developers with a technical framework for analyzing and manipulating these dynamic systems to control cellular outcomes.
Classical views of cell signaling often portrayed pathways as simple on-off switches in response to stimuli. However, the advent of single-cell live imaging and quantitative analysis has revealed a far more complex reality: signaling pathways exhibit a rich repertoire of dynamic behaviors, including oscillations, pulses, and sustained activation [18]. This temporal dimension is not mere biological noise; it is a fundamental feature of how cells process information and make reliable fate decisions amidst molecular stochasticity.
The concept of cell fate can be understood through the theoretical framework of Waddington's epigenetic landscape, modernized as a dynamical system of attractor states [18] [19]. In this model, distinct cell fates—such as survival, apoptosis, or differentiation—represent stable attractors. Signaling dynamics act as guiding forces, pushing cells from one basin of attraction to another, thereby determining the final developmental trajectory [18]. This review details how these principles operate during gastrulation, a pivotal developmental event, and provides the toolkit for their experimental dissection.
Molecular Mechanisms: Decoding Dynamics into Destiny
Several evolutionarily conserved signaling pathways have been characterized as prime examples of dynamic information encoders. Their temporal patterns are decoded by the cell's molecular machinery to activate specific transcriptional programs.
NF-κB Oscillations and Immune Cell Fate
The NF-κB pathway is a paradigm for understanding how oscillatory dynamics control gene expression. In the canonical pathway, stimuli such as TNFα lead to the degradation of the inhibitor IκB, allowing NF-κB to translocate to the nucleus. A key negative feedback loop, wherein NF-κB induces the transcription of IκBα, drives the oscillatory, nucleo-cytoplasmic shuttling of the transcription factor with a period of approximately 1.5 hours [18] [20].
- Functional Decoding: These oscillations are not just a byproduct of feedback; they are functional. Different genes are activated at different phases of the oscillation. Some responsive genes are rapidly induced by the first nuclear translocation peak, while others require repeated cycles or sustained activity for activation, enabling a single pathway to orchestrate a complex inflammatory response [18] [20]. The specific dynamic is shaped by the interplay of different IκB isoforms (IκBα, IκBβ, IκBε), with IκBα knockout cells showing persistent, non-oscillatory NF-κB activation [20].
p53 Pulsatile Dynamics in DNA Damage Response
In response to DNA damage, the tumor suppressor p53 exhibits pulsatile dynamics. The number, amplitude, and duration of these pulses are not uniform; they vary with the type and intensity of the stressor [18]. This allows p53 to function as a dynamic decoder: different pulse patterns activate distinct sets of target genes that dictate cell fate outcomes, such as transient cell cycle arrest versus irreversible senescence or apoptosis [18].
BMP/WNT/Nodal Dynamics in Gastrulation and ExM Specification
Gastrulation and the specification of extraembryonic mesoderm (ExM) are exquisitely controlled by the dynamic interplay of BMP, WNT, and Nodal signaling. Research using human embryonic stem cell (hESC) models has demonstrated that precise modulation of these pathways can efficiently direct cell fate.
- ExM Specification Protocol: A defined protocol involving the combined treatment of naive hESCs with CHIR99021 (a GSK3 inhibitor that activates WNT signaling) and BMP4 in a specific medium (FH-N2B27) can induce differentiation into expandable ExM-like cells with over 90% efficiency within 4-5 days [3].
- Developmental Trajectory: This specification process proceeds through a primitive streak-like intermediate (PSLI), delineating a clear developmental roadmap [3]. The initial pluripotent state of the hESCs (naive vs. primed) influences the cellular response to these signals, affecting the developmental progression and transcriptional characteristics of the resulting ExM [3].
Table 1: Key Signaling Dynamics and Their Functional Outcomes
| Signaling Pathway | Stimulus/Context | Dynamic Pattern | Decoding Mechanism | Cell Fate Decision |
|---|---|---|---|---|
| NF-κB | TNFα / Immune Response | Damped oscillations (∼1.5 hr period) | Kinetics of IκB negative feedback; gene-specific activation thresholds | Inflammatory gene expression vs. cell survival [18] [20] |
| p53 | DNA Damage / Genotoxic Stress | Discrete pulses | Pulse number & amplitude; promoter-specific response | Cell cycle arrest vs. senescence vs. apoptosis [18] |
| BMP/WNT/Nodal | Gastrulation / ExM Specification | Sustained, combinatorial activation | Pathway synergy; primitive streak intermediate | Specification of Extraembryonic Mesoderm [3] |
Experimental Approaches: Mapping the Dynamic Landscape
Understanding the link between signaling dynamics and cell fate requires methodologies that capture temporal information at the single-cell level and connect it to functional outcomes.
Live-Cell Imaging and Single-Cell Analysis
The direct observation of signaling dynamics relies on live-cell microscopy of fluorescently tagged proteins (e.g., RelA-NF-κB) [18]. This allows for the quantification of translocation kinetics and oscillatory behavior in real-time. To connect these dynamics to lineage decisions, this imaging is integrated with single-cell transcriptomics. Computational tools like Topographer can then be applied to this data to reconstruct a "quantitative Waddington's landscape," infer transition probabilities between cell states, and identify branching points where fate decisions occur [19].
A Protocol for Perturbing Dynamics to Establish Causality
To move from correlation to causation, researchers must experimentally perturb signaling dynamics. The following methodology, derived from studies on ExM specification, provides a template [3].
- Objective: To test the causal role of BMP and WNT signaling dynamics in ExM specification from naive hESCs.
- Workflow Diagram:
- Key Reagents & Steps:
- Cell Line: Naive hESCs (e.g., AIC-N line) [3].
- Baseline Medium: FH-N2B27 (N2B27 medium supplemented with FGF4 and Heparin) [3].
- Signaling Agonists: CHIR99021 (a GSK3 inhibitor, activating WNT signaling) and BMP4 [3].
- Culture Duration: 4-5 days to achieve ~90% efficiency of ExM differentiation.
- Validation: Immunofluorescence and flow cytometry for ExM markers (GATA6, SNAIL, VIM, KDR, FLT1); scRNA-seq to characterize heterogeneity and confirm trajectory through a primitive streak-like intermediate [3].
The Scientist's Toolkit: Essential Research Reagents
The following table compiles key reagents and tools essential for research in signaling dynamics and cell fate, as featured in the cited studies.
Table 2: Research Reagent Solutions for Signaling Dynamics Studies
| Reagent / Tool | Function / Target | Application in Context |
|---|---|---|
| CHIR99021 | GSK3 inhibitor; activates WNT signaling | Directly modulates WNT pathway dynamics to specify ExM from hESCs [3]. |
| BMP4 | Ligand for BMP receptors | Provides the BMP signal component in combination with WNT activation for ExM induction [3]. |
| Fluorescent Protein-Tagged TFs (e.g., RelA-p65) | Live-cell imaging of transcription factor localization | Enables real-time tracking of NF-κB or other TF dynamics in single cells via microscopy [18]. |
| Topographer Algorithm | Computational analysis of scRNA-seq data | Constructs quantitative developmental landscapes and infers fate/transition probabilities from static snapshot data [19]. |
| TNFα | Cytokine; activator of NF-κB pathway | Standard stimulus used to induce and study the oscillatory dynamics of the NF-κB system [20]. |
| 2-Chloro-3-hydroxy-1,4-naphthoquinone | 2-Chloro-3-hydroxy-1,4-naphthoquinone | High Purity | High-purity 2-Chloro-3-hydroxy-1,4-naphthoquinone for research applications. For Research Use Only. Not for human or veterinary use. |
| 5-Phenylpenta-2,4-dienal | 5-Phenylpenta-2,4-dienal, CAS:13466-40-5, MF:C11H10O, MW:158.2 g/mol | Chemical Reagent |
Signaling Pathway Diagrams
The following diagrams illustrate the core logic and dynamic feedback structures of the key pathways discussed.
The evidence is compelling: signaling dynamics are a fundamental and ubiquitous mechanism for controlling cell fate decisions from immune response to embryonic development. The quantitative study of these dynamics, powered by live-cell imaging, single-cell omics, and engineered stem cell models, has moved the field from observation to mechanistic understanding and predictive modeling.
Future research must focus on bridging the remaining gaps. This includes achieving a more comprehensive mechanistic understanding of how dynamic signals are precisely decoded at the level of chromatin and gene regulatory networks [18]. Furthermore, the innovative use of targeted therapies in oncology [21] underscores the translational potential of this field. As our ability to measure and model these complex temporal patterns improves, so too will our capacity to predict cellular behavior and design novel therapeutic interventions that manipulate cell fate in disease contexts such as cancer, degenerative disorders, and regenerative medicine. The journey from signal to fate, once a black box, is now a frontier of quantitative, dynamic, and therapeutic science.
Cell fate commitment during embryonic development is orchestrated by dynamic epigenetic mechanisms that regulate gene expression without altering DNA sequences. This whitepaper examines how coordinated histone modification dynamics serve as central regulators of transcriptional programs governing cell lineage specification. Within the context of gastrulation research, we explore the sophisticated signaling dynamics that direct epigenetic reprogramming, with particular emphasis on zygotic genome activation (ZGA) and the establishment of germ layer identities. Integrating recent multi-omics findings, we present quantitative analyses of histone modification patterns, detailed experimental methodologies for profiling epigenetic dynamics, and essential reagent solutions for investigating chromatin remodeling during cell fate transitions.
The transition from pluripotency to committed cell fates represents a fundamental process in embryonic development, requiring precise spatial and temporal coordination of gene expression programs. Epigenetic regulations, particularly post-translational modifications of histone proteins, play indispensable roles in governing these cell fate decisions by modulating chromatin architecture and accessibility [22]. During gastrulation, embryonic cells undergo profound remodeling of their transcriptional identity, transitioning from the naive epiblast state to definitive germ layers (ectoderm, mesoderm, and endoderm) through tightly regulated epigenetic mechanisms [23]. These modifications create a chromatin landscape that either permits or restricts transcription factor binding, thereby controlling lineage-specific gene expression.
The signaling dynamics that coordinate gastrulation research provide critical context for understanding epigenetic regulation. Morphogen gradients—including BMP, WNT, FGF, and Nodal signaling pathways—converge on epigenetic modifiers to establish spatially organized gene expression patterns [23]. This interplay between extracellular signaling and intracellular epigenetic machinery enables the precise temporal control of developmental genes, particularly during ZGA when the embryonic genome assumes transcriptional control from maternally-derived factors [22] [24]. The investigation of these processes has been revolutionized by advanced low-input epigenomic profiling technologies that enable comprehensive mapping of histone modification dynamics throughout critical developmental transitions.
Molecular Dynamics of Histone Modifications During Cell Fate Transitions
Histone Modification Reprogramming During Zygotic Genome Activation
ZGA represents a critical developmental milestone characterized by rapid, simultaneous activation of previously silenced chromatin regions. Recent investigations in vertebrate models have revealed distinct regulatory requirements for different gene classes during this process. Table 1 summarizes the differential dependence of gene categories on specific histone modifications during ZGA.
Table 1: Gene-Specific Histone Modification Requirements During Zygotic Genome Activation
| Gene Category | Key Histone Modifications | Dependent Writers/Enzymes | Functional Role in ZGA |
|---|---|---|---|
| Developmental Genes | H3K27ac, H3.3S31ph | CBP/P300, BRD4 | Essential for activation; requires enhanced CBP/P300 activity [24] |
| Housekeeping Genes | H3K9ac, H4K16ac, H3K14ac | Non-CBP/P300 HATs | CBP/P300-independent activation [24] |
| Primed Developmental Loci | H3K4me3 (broad domains) | KDM5B (demethylase) | Prevents premature differentiation; removal required for ZGA [22] |
Research in teleost embryos demonstrates that developmental genes and housekeeping genes are distinctly regulated during ZGA. CBP/P300 histone acetyltransferase activity is specifically required for developmental gene activation but dispensable for housekeeping gene expression [24]. This specialization ensures precise temporal control of developmental programs while maintaining cellular homeostasis. The temporal accumulation of H3.3S31ph significantly enhances CBP/P300 activity specifically at ZGA, ensuring proper activation of developmental genes [24].
Histone Modification Dynamics in Mammalian Preimplantation Development
Mouse preimplantation development exhibits sophisticated histone modification dynamics that differ significantly from non-mammalian vertebrates. Table 2 quantifies the dynamics of key histone modifications during early embryonic development.
Table 2: Quantitative Dynamics of Histone Modifications in Mouse Preimplantation Development
| Histone Modification | Oocyte/MII Stage | Zygote Stage | 2-Cell Stage | Blastocyst Stage | Functional Significance |
|---|---|---|---|---|---|
| H3K4me3 | Broad domains [22] | Inheritance of broad domains [22] | Erasure of broad domains [22] | Canonical sharp peaks [22] | Transition required for ZGA and developmental competence [22] |
| H3K27me3 | Present at developmental loci [22] | Global erasure in paternal alleles [22] | Minimal promoter presence [22] | Re-established in lineage-specific patterns [22] | Prevents premature differentiation [22] |
| H3K27ac | Limited enrichment [24] | - | Progressive accumulation [24] | Enhancer-specific patterning [25] | Marks active enhancers and promoters [24] |
A notable feature of mammalian oocytes and zygotes is the presence of noncanonical H3K4me3 existing as broad domains, which are negatively correlated with DNA methylation [22]. These broad domains are inherited after fertilization but are actively removed by the late two-cell stage, a process requiring the activation of zygotic transcription [22]. The removal of these broad domains is essential for ZGA and full developmental potential, as artificial maintenance through knockdown of H3K4me3 demethylases (particularly KDM5B) disrupts lineage differentiation at the blastocyst stage [22].
Chromatin State Remodeling During Human Pluripotent Stem Cell Differentiation
The differentiation of human pluripotent stem cells (hPSCs) provides an accessible model for investigating histone modification dynamics during cell fate commitment. During definitive endoderm differentiation, key developmental genes are transcribed before cell division, while chromatin accessibility analyses reveal early inhibition of alternative cell fates [25]. Enhancers are rapidly established and decommissioned between different cell divisions, demonstrating the dynamic nature of the epigenetic landscape during lineage commitment [25].
Histone modification patterns change progressively during this process, with activating marks (H3K4me3, H3K27ac) increasing at lineage-specific genes while repressive marks (H3K27me3) are deposited at genes associated with alternative fates [25]. The acquisition of these histone modifications occurs in a cell cycle-dependent manner, with early G1 phase representing a particularly permissive state for the initiation of differentiation [25].
Experimental Methodologies for Profiling Histone Modification Dynamics
Chromatin Immunoprecipitation Followed by Sequencing (ChIP-seq)
Chromatin Immunoprecipitation followed by Sequencing (ChIP-seq) represents the gold standard method for genome-wide mapping of histone modifications. The protocol involves specific crosslinking, immunoprecipitation, and sequencing steps that must be optimized for different biological contexts.
Table 3: Key Experimental Parameters for Histone Modification Profiling
| Method | Resolution | Sample Input Requirements | Key Applications | Technical Considerations |
|---|---|---|---|---|
| ChIP-seq | 200-500 bp | 10,000-500,000 cells (standard); 100-1,000 cells (low-input) | Genome-wide mapping of histone modifications [26] | Antibody specificity critical; crosslinking optimization required [26] |
| CUT&RUN | ~150 bp | 1,000-50,000 cells | High-resolution mapping with lower background [26] | No crosslinking; requires permeabilization [26] |
| ATAC-seq | Single-nucleotide | 500-50,000 cells | Chromatin accessibility profiling [25] | Requires viable nuclei; sensitive to mitochondrial DNA [25] |
Detailed ChIP-seq Protocol:
- Crosslinking: Treat cells with 1% formaldehyde for 8-10 minutes at room temperature to fix protein-DNA interactions.
- Cell Lysis and Chromatin Shearing: Lyse cells and fragment chromatin to 200-500 bp using sonication (15-20 cycles of 30-second pulses).
- Immunoprecipitation: Incubate fragmented chromatin with specific histone modification antibodies (e.g., anti-H3K4me3, anti-H3K27ac) overnight at 4°C with rotation.
- Washing and Elution: Wash beads sequentially with low-salt, high-salt, and LiCl buffers, then elute chromatin from beads.
- Reverse Crosslinking and Purification: Incubate eluates at 65°C overnight with proteinase K, then purify DNA using silica membrane columns.
- Library Preparation and Sequencing: Prepare sequencing libraries using compatible kits and sequence on appropriate platforms (Illumina recommended).
For low-input applications (such as preimplantation embryos), adapted protocols utilizing carrier chromatin or specialized library preparation methods are essential [22]. Recent advances have enabled the application of ChIP-seq to single cells, though this remains technically challenging for histone modifications compared to transcription factors.
Multi-omics Integration Approaches
Comprehensive understanding of epigenetic coordination requires integration of multiple data types. Simultaneous profiling of histone modifications, chromatin accessibility, DNA methylation, and transcriptome provides unprecedented insights into the regulatory logic of cell fate commitment. In synchronized hPSC differentiation systems, combined RNA-seq, ATAC-seq, and histone modification ChIP-seq (H3K4me3, H3K27me3, H3K27ac, H3K4me1, H3K36me3) have revealed coordinated changes occurring between consecutive cell divisions [25].
Experimental workflows for multi-omics profiling typically involve:
- Cell Synchronization: Using chemical inhibitors or FUCCI reporter systems to obtain populations at specific cell cycle stages [25].
- Parallel Sample Processing: Dividing samples for simultaneous DNA, RNA, and chromatin analysis.
- Data Integration: Computational approaches to correlate histone modification patterns with gene expression changes and chromatin accessibility.
The Scientist's Toolkit: Essential Research Reagents and Solutions
Table 4: Essential Research Reagents for Investigating Histone Modification Dynamics
| Reagent Category | Specific Examples | Functional Application | Key Considerations |
|---|---|---|---|
| Histone Acetyltransferase Inhibitors | A485, SGC-CBP30 [24] | Inhibition of CBP/P300 activity to investigate H3K27ac function [24] | A485 targets HAT domain; SGC-CBP30 targets bromodomain [24] |
| Histone Demethylase Inhibitors | KDM5B inhibitors [22] | Maintenance of broad H3K4me3 domains to study ZGA regulation [22] | Disrupts transition from noncanonical to canonical H3K4me3 [22] |
| Cell Cycle Synchronization Agents | Nocodazole, RO-3306 [25] | Cell cycle phase-specific analysis of epigenetic dynamics [25] | Nocodazole blocks mitosis; RO-3306 blocks G2/M transition [25] |
| Epigenomic Profiling Kits | Commercial ChIP-seq kits, ATAC-seq kits [26] | Standardized protocols for histone modification mapping [26] | Kit performance varies by application; validation required [26] |
| Pluripotency and Differentiation Media | Defined hPSC media, endoderm differentiation kits [25] | Controlled differentiation for epigenetic studies [25] | Serum-free formulations enhance reproducibility [25] |
| Europium bromide (EuBr3) | Europium bromide (EuBr3), CAS:13759-88-1, MF:Br3Eu, MW:391.68 g/mol | Chemical Reagent | Bench Chemicals |
| 2-Isopropylcyclopentanone | 2-Isopropylcyclopentanone, CAS:14845-55-7, MF:C8H14O, MW:126.2 g/mol | Chemical Reagent | Bench Chemicals |
Signaling Dynamics and Epigenetic Coordination in Gastrulation
The signaling dynamics that characterize gastrulation intersect with epigenetic regulation through multiple mechanisms. Morphogen signaling pathways directly modify the activity of epigenetic regulators, creating a framework for spatial patterning of the embryo. The following diagram illustrates the integration of signaling pathways with epigenetic modifications during cell fate commitment:
Signaling and Epigenetic Coordination in Gastrulation
Key signaling pathways operational during gastrulation include:
- BMP Signaling: Promotes ventral mesodermal fates and modulates histone acetylation through SMAD effectors.
- WNT/β-catenin Signaling: Regulates brachyury and posterior mesodermal genes through interaction with CBP/P300 HAT complexes.
- FGF Signaling: Supports mesodermal commitment and influences histone methylation patterns.
- Nodal/Activin Signaling: Directs endodermal and mesodermal fates through SMAD2/3-mediated recruitment of epigenetic regulators.
These signaling pathways converge on lineage-specific transcription factors that subsequently recruit epigenetic modifiers to establish appropriate chromatin states. For example, during endoderm differentiation, transcription factors such as EOMES, GATA6, SOX17, and FOXA2 recruit chromatin remodeling complexes to activate endodermal genes while repressing alternative lineages [25]. The p38/MAPK signaling pathway controls Activator protein-1 members that are necessary for inducing endoderm while blocking cell fate shifting toward mesoderm [25].
The following diagram illustrates the experimental workflow for investigating histone modification dynamics in cell fate commitment:
Experimental Workflow for Epigenetic Analysis
The coordination of histone modification dynamics represents a fundamental mechanism underpinning cell fate commitment during embryonic development. The integration of signaling pathways with epigenetic regulation creates a robust system for spatial and temporal control of gene expression programs essential for proper embryogenesis. Advances in low-input epigenomic technologies have revealed unprecedented details of these processes, demonstrating distinct regulatory requirements for different gene classes and developmental stages. The continued refinement of experimental approaches—particularly multi-omics integration and single-cell analyses—will further elucidate the complex epigenetic coordination governing cell fate decisions. These insights not only enhance our understanding of normal development but also provide critical frameworks for manipulating cell identities in regenerative medicine and disease modeling.
Advanced Tools and Models: From Live Imaging to Synthetic Embryos for Decoding Gastrulation
Gastrulation is a fundamental process in embryonic development, during which the simple blastula undergoes a complex reorganization to form the three primary germ layers. While the biochemical signaling dynamics that guide cell fate and movement during this process have been extensively studied, a complete understanding requires equal consideration of the physical forces and mechanical properties that enable morphogenesis. The role and importance of mechanical properties of cells and tissues in cellular function, development and disease has widely been acknowledged, however standard techniques currently used to assess them exhibit intrinsic limitations [27]. Techniques such as atomic force microscopy (AFM) and optical tweezers, while valuable, typically require physical contact with samples or are limited to surface measurements, making them unsuitable for probing the dynamic, three-dimensional mechanical changes occurring within intact, living embryos [27] [28].
In recent years, Brillouin microscopy has emerged as a non-destructive, label- and contact-free method that can probe the viscoelastic properties of biological samples with diffraction-limited resolution in 3D [27]. This all-optical technique enables mechanical characterization without disturbing the delicate physiological processes of development, providing a powerful new tool for investigating the role of biomechanics in gastrulation. By integrating mechanical data with information from biochemical signaling pathways, researchers can develop a more holistic understanding of how embryos transform from simple spherical structures into complex organisms with multiple tissue layers and axes.
This technical guide explores the principles, implementations, and applications of Brillouin microscopy specifically within the context of embryonic mechanobiology, detailing how this technology is revolutionizing our ability to map mechanical property dynamics during critical developmental events such as gastrulation.
Technical Foundations of Brillouin Microscopy
Core Physical Principles
Brillouin microscopy is based on the physical phenomenon of Brillouin light scattering (BLS), an inelastic scattering process that occurs when light interacts with thermally-driven acoustic vibrations or phonons within a material [28]. As photons propagate through a medium, a minute fraction undergoes energy exchange with the material's intrinsic density fluctuations. This interaction produces scattered light with a slightly shifted frequency compared to the incident light.
The key measurable parameters in Brillouin spectroscopy are:
- Brillouin shift (νB): The frequency shift between incident and scattered light, typically in the gigahertz (GHz) range for biological materials
- Brillouin linewidth (ΓB): The spectral width of the Brillouin peak, related to the viscous damping of acoustic waves
The Brillouin shift relates to the longitudinal modulus M' of the material through the equation:
[
ν_B = \frac{2n}{λ} \sqrt{\frac{M'}{Ï}}
]
where n is the refractive index, λ is the wavelength of the incident light, and Ï is the mass density [28]. The longitudinal modulus represents the material's resistance to compression under confined conditions and is typically 2-3 orders of magnitude larger than the more commonly referenced Young's modulus [17].
Figure 1: Fundamental principle of Brillouin light scattering. Incident light interacts with thermal acoustic vibrations in the sample, producing elastically scattered Rayleigh light and inelastically scattered Brillouin components (Stokes and Anti-Stokes) with characteristic frequency shifts.
Technical Implementations and Advancements
Brillouin microscopy has evolved through several technological generations, each addressing key limitations in speed, resolution, or phototoxicity:
Confocal Brillouin Microscopy
The earliest implementations were based on confocal scanning approaches, where a single spatial point is measured at a time [27]. While providing valuable mechanical information with diffraction-limited resolution, this method suffers from slow acquisition speeds (typically minutes to hours for a 3D volume) and relatively high illumination dosages that can cause photodamage in living samples [29].
Line-Scan Brillouin Microscopy (LSBM)
To address speed limitations, line-scan Brillouin microscopy was developed, enabling parallel acquisition of hundreds of points simultaneously [29]. This approach reduces acquisition times by approximately two orders of magnitude while simultaneously decreasing phototoxicity through the use of near-infrared (780 nm) illumination instead of the more common 532 nm lasers [29]. LSBM can be implemented in two primary configurations:
- Orthogonal-line (O-LSBM): Illumination and detection axes are separated in a 90° configuration, similar to light-sheet microscopy, minimizing total illumination dosage for volumetric imaging
- Epi-line (E-LSBM): Sample is illuminated by a focused line in a 180° backscattered geometry, mitigating effects of scattering and optical aberrations [29]
Fourier-Transform Brillouin Microscopy (FTBM)
The most recent advancement is Fourier-transform Brillouin microscopy, which enables full-field 2D spectral imaging by exploiting a custom-built imaging Fourier-transform spectrometer and the symmetric properties of the Brillouin spectrum [30]. This approach can acquire up to 40,000 spectra per second, representing an approximately three-orders-of-magnitude improvement in speed compared to standard confocal methods while maintaining high spatial resolution [30].
Table 1: Comparison of Brillouin Microscopy Modalities
| Parameter | Confocal Brillouin | Line-Scan Brillouin (LSBM) | Fourier-Transform Brillouin (FTBM) |
|---|---|---|---|
| Acquisition Method | Single-point scanning | Parallel line acquisition | Full-field 2D imaging |
| Typical Acquisition Time | Minutes to hours for 3D volumes | ~2 minutes for 3D volumes | Up to 40,000 spectra/second |
| Spatial Resolution | Diffraction-limited | ~1.5 μm laterally | Diffraction-limited |
| Laser Wavelength | Typically 532 nm | 780 nm (reduced phototoxicity) | Configurable |
| Key Advantages | High signal-to-noise | Balanced speed and viability | Extreme speed for 2D imaging |
| Limitations | Slow, potentially phototoxic | Limited to 1D multiplexing | New technology, limited adoption |
Application to Embryonic Development: Mapping Gastrulation Mechanics
Technical Workflow for Live Embryo Imaging
The application of Brillouin microscopy to living embryos requires careful consideration of sample preparation, environmental control, and data processing. The following workflow has been successfully implemented for studying Drosophila, ascidian, and mouse embryogenesis [29]:
Figure 2: Experimental workflow for Brillouin microscopy of live embryos, showing key steps from sample preparation through data analysis.
Essential Research Reagents and Equipment
Table 2: Research Reagent Solutions for Brillouin Microscopy in Developmental Biology
| Reagent/Equipment | Function/Purpose | Example Specifications |
|---|---|---|
| Narrowband Diode Laser | Excitation source for Brillouin scattering | 780 nm wavelength, 50 kHz linewidth, frequency stabilization [29] |
| High-NA Objectives | Optimal light collection and spatial resolution | NA 0.8, water immersion [29] |
| Virtually Imaged Phase Array (VIPA) | High-resolution spectral analysis of scattered light | Spectral resolution <0.5 GHz [27] |
| Atomic Gas Cell Filter | Suppression of elastically scattered Rayleigh light | ~80 dB background suppression [29] |
| Environmental Chamber | Maintenance of embryo viability during imaging | Temperature, COâ‚‚, Oâ‚‚ control [29] |
| Fluorescent Membrane Markers | Correlation of mechanical properties with cellular structures | Genetically encoded fluorescent proteins [29] |
| GPU-Accelerated Analysis Software | Real-time processing of Brillouin spectra | >1000× speed enhancement vs CPU processing [29] |
Case Study: Mechanical Dynamics During Drosophila Gastrulation
Drosophila melanogaster embryogenesis provides an excellent model for studying the mechanical aspects of gastrulation due to its well-characterized development and genetic tractability. Recent Brillouin microscopy studies have revealed fascinating mechanical dynamics during key morphogenetic events:
Ventral Furrow Formation (VFF)
During VFF, mesodermal cells on the ventral side of the embryo undergo apical constriction, leading to tissue invagination. LSBM imaging has captured a transient increase in Brillouin shift (10-20 MHz) within the mesoderm, peaking at the initiation of invagination [29] [17]. This mechanical stiffening coincides with the reorganization of sub-apical microtubules, and disruption of microtubules with Colcemid reduces the Brillouin shift during VFF, suggesting microtubules contribute to this mechanical transition [17].
Posterior Midgut Invagination (PMI)
Similar to VFF, PMI involves tissue folding but with a circular contractile domain geometry. Brillouin imaging revealed a comparable increase in Brillouin shift during this process, suggesting that mechanical stiffening is a common feature of tissue folding independent of the specific geometry [29].
Table 3: Quantitative Brillouin Shift Measurements During Drosophila Gastrulation
| Developmental Process | Tissue/Cell Type | Brillouin Shift Change | Temporal Dynamics | Biological Interpretation |
|---|---|---|---|---|
| Ventral Furrow Formation | Central mesodermal cells | +10 to +20 MHz increase | Peaks at invagination initiation (~10 min) [17] | Sub-apical stiffening potentially microtubule-mediated |
| Posterior Midgut Invagination | Posterior endodermal cells | Significant increase (quantification not specified) | During invagination process [29] | Mechanical stiffening as general feature of tissue folding |
| Epithelial-Mesenchymal Transition | Internalized mesoderm | Decrease following invagination | During EMT phase [17] | Mechanical softening accompanying change in cell state |
| Dorsal-Ventral Axis | Dorsal ectoderm vs ventral mesoderm | Differential dynamics observed [17] | Throughout gastrulation | Distinct mechanical programs for different cell fates |
Integration with Biochemical Signaling in Gastrulation
The mechanical properties revealed by Brillouin microscopy do not operate in isolation but interact intimately with biochemical signaling pathways to coordinate gastrulation. Several key intersections have been identified:
Mechanochemical Feedback Loops
Mechanical changes detected by Brillouin microscopy likely participate in feedback loops with biochemical signaling pathways. For example, the transcription factor Snail, which marks mesodermal fate during Drosophila gastrulation, not only drives genetic programs but also correlates with mechanical changes observed via Brillouin microscopy [29]. Similarly, the mechanosensitive transcription factors YAP and TAZ, known to shuttle between cytoplasm and nucleus in response to mechanical cues, may be influenced by the tissue-scale mechanical dynamics occurring during gastrulation [27].
Cytoskeletal Dynamics and Mechanical Properties
The Brillouin signal changes observed during ventral furrow formation coincide with reorganization of both actin and microtubule networks [17]. While actomyosin contraction provides the driving force for apical constriction, the concomitant stiffening detected by Brillouin microscopy appears to involve microtubules, as their disruption reduces the Brillouin shift increase during VFF [17]. This suggests that microtubules may contribute to the material properties that enable or facilitate tissue folding alongside the force-generating actomyosin network.
Interplay with Extracellular Matrix Mechanics
Beyond cell-autonomous mechanics, the extracellular matrix (ECM) also contributes to the mechanical landscape of developing embryos. Brillouin microscopy has been used to characterize mechanical properties of embryonic tendons, demonstrating the ability to detect changes in ECM cross-linking and mechanical heterogeneity during development [31]. Similar principles likely apply to the basement membrane and other ECM components that influence gastrulation movements.
Methodological Considerations and Limitations
While Brillouin microscopy provides unprecedented access to the mechanical aspects of embryogenesis, several important limitations and considerations must be acknowledged:
Interpretation of Mechanical Properties
The longitudinal modulus measured by Brillouin microscopy operates at GHz frequencies, probing material responses on nanosecond timescales—far faster than most biological processes that occur on millisecond to second timescales [17]. However, empirical correlations between Brillouin measurements and quasi-static mechanical properties have been observed, typically following a power-law relationship with exponents in the range of 0.02-0.09 [17]. This implies that relative changes in quasi-static modulus are often 10 to 50 times larger than the relative changes in Brillouin modulus.
Relationship to Other Mechanical Measurements
It is crucial to recognize that Brillouin microscopy measures the longitudinal modulus (M'), which differs conceptually from the more commonly referenced Young's modulus (E) or shear modulus (G) [17] [28]. The longitudinal modulus describes the ratio of uniaxial stress to strain under confined conditions, while Young's modulus describes unconfined deformation. For biological materials, M' is typically 2-3 orders of magnitude larger than E, so values should not be directly compared.
Technical Challenges
Current technical challenges include the need for better spatial resolution for subcellular imaging, further reductions in phototoxicity for long-term live imaging, and improved methods for correlating mechanical properties with specific molecular constituents. Additionally, the interpretation of Brillouin spectra in heterogeneous biological materials remains complex, as the signal represents an average over the phonon coherence length, which can extend beyond the optical diffraction limit in some materials [28].
Brillouin microscopy has opened a new window into the mechanical world of embryonic development, providing the first spatio-temporal descriptions of how cell material properties evolve during organismal morphogenesis. The technology's ability to perform non-invasive, label-free, 3D mechanical characterization with subcellular resolution makes it uniquely suited for investigating the role of mechanics in dynamic processes like gastrulation.
Future developments will likely focus on increasing imaging speed further, improving spatial resolution, and enhancing our ability to correlate mechanical properties with specific molecular components through multimodal imaging. As these technical advances progress, Brillouin microscopy will increasingly enable researchers to dissect the complex interplay between biochemical signaling and mechanical forces that together orchestrate the remarkable transformation of a simple embryo into a complex, multilayered organism.
The integration of mechanical mapping with established molecular and genetic approaches promises a more comprehensive understanding of gastrulation—one that acknowledges both the biochemical and physical dimensions of development. By embracing this dual perspective, researchers can address fundamental questions about how mechanical properties influence cell fate decisions, tissue patterning, and morphogenetic movements during this critical developmental period.
The process of embryonic development is orchestrated by complex signaling pathways that exhibit precise spatial and temporal dynamics. Traditional methods for investigating these pathways, including genetic knockouts, chemical inhibitors, and agonist addition, have provided invaluable insights but offer limited control over the crucial dimensions of time and space [32]. These methods often act as binary switches, failing to mimic the nuanced, dynamic patterns of morphogen activity that guide cell fate decisions in developing tissues [33]. This limitation is particularly acute in the study of gastrulation, a pivotal stage in early embryogenesis where a uniform sheet of cells transforms into the structured three-dimensional body plan with distinct head/tail, front/back, and left/right axes [2].
Optogenetics, a technology that combines genetics and optics to control molecular events with light, has emerged as a powerful solution to this challenge. By fusing light-sensitive protein domains to key signaling components, researchers can now manipulate developmental pathways with unprecedented spatial resolution (on the order of micrometers) and temporal precision (from milliseconds to minutes) [34] [33]. This precise control enables the creation of synthetic, user-defined signaling patterns in live embryos and synthetic tissues, allowing for direct testing of long-standing hypotheses about how cells interpret positional information [35]. Within the context of gastrulation research, optogenetics provides a unique toolset to dissect the role of signaling dynamics, moving beyond mere signal presence or absence to understand how the precise timing, location, and intensity of signals guides the dramatic cellular rearrangements and fate decisions that form the embryonic axes [2].
Optogenetic Systems for Controlling Signaling Pathways
Optogenetic control is achieved by harnessing natural photoreceptors from plants, algae, and bacteria. These proteins undergo conformational changes, dimerization, or oligomerization in response to specific wavelengths of light. When fused to signaling proteins of interest, these light-induced changes can be used to control protein activity, localization, and interactions [34] [33].
Common Photoreceptor Systems and Their Mechanisms
The table below summarizes the key optogenetic systems used to manipulate developmental signaling pathways.
Table 1: Key Optogenetic Systems and Their Applications in Developmental Biology
| System Name | Source Organism | Wavelength | Mode of Action | Example Applications in Development |
|---|---|---|---|---|
| CRY2/CIBN | Arabidopsis thaliana | 450 nm Blue Light | Heterodimerization | Controlling Nodal signaling; Subcellular protein localization [35] [34] |
| LOV2 (AsLOV2) | Avena sativa (Oat) | 450 nm Blue Light | Jα helix unfolding, conformational change | Steric control of protein interactions; Calcium recording (SCANR) [36] [34] |
| Phy/PIF | Arabidopsis thaliana | 660 nm (Association)740 nm (Reversion) | Heterodimerization | Control of receptor tyrosine kinase (RTK) signaling [34] [33] |
| Magnets (pMag/nMag) | Engineered from VVD | 450 nm Blue Light | Heterodimerization with electrostatic attraction | Control of gene expression and genome editing [33] |
| UVR8/COP1 | Arabidopsis thaliana | 280-315 nm UVB | Heterodimerization | Nuclear translocation and control of gene expression [33] |
| BphP1/PpsR2 | Bacteria | ~750 nm Near-Infrared | Heterodimerization | Deep-tissue activation of signaling pathways [33] |
Modes of Optogenetic Control
These photoreceptors can be deployed in several strategic ways to achieve precise control over signaling molecules:
- Control of Subcellular Localization: A protein of interest (POI) fused to one light-activated dimer component (e.g., CRY2) can be recruited to a specific organelle or membrane by illuminating a second component (e.g., CIBN) anchored to that location. This allows researchers to investigate how the signaling output of a protein changes based on its location within the cell [34].
- Control of Protein-Protein Interactions and Activity: Bringing signaling receptors into proximity is a common activation mechanism. Fusing receptor components to heterodimerizing pairs like Cry2/CIB1N or PhyB/PIF allows for light-induced assembly of active receptor complexes, thereby initiating downstream signaling cascades on demand [35] [32].
- Light-Induced Asymmetric Signaling: By applying a focused beam or gradient of light to one side of a cell, an optogenetic actuator can be asymmetrically activated. This creates polarized signaling within a single cell, a powerful approach for mimicking and studying processes like cell migration and division [34].
Figure 1: Core Optogenetic Control Mechanisms. Light input triggers photoreceptors like CRY2, LOV2, or PhyB, leading to dimerization or conformational changes that control protein localization, pathway activation, or inhibition.
Application: Optogenetic Dissection of Nodal Signaling in Gastrulation
The Nodal signaling pathway, a member of the TGF-β family, is a key morphogen responsible for organizing the mesendodermal germ layers and establishing the embryonic axes during vertebrate gastrulation [35]. A recent study exemplifies the power of optogenetics to dissect this pathway with high spatiotemporal precision.
Development of the optoNodal2 System
To achieve precise control over Nodal signaling, researchers engineered an improved optogenetic reagent, optoNodal2 [35]. This system addresses key limitations of earlier versions, including problematic dark activity and slow response kinetics.
Table 2: Key Features of the optoNodal2 System for Controlling Nodal Signaling
| Feature | Description | Advantage |
|---|---|---|
| Photoreceptor Pair | Cry2/CIB1N from Arabidopsis thaliana | Rapid association (~seconds) and dissociation (~minutes) kinetics [35]. |
| Receptor Engineering | Type I (acvr1b) and Type II (acvr2b) Nodal receptors fused to Cry2 and CIB1N, respectively. | Enables light-induced heterodimerization to initiate signaling [35]. |
| Sequestration Strategy | Removal of myristoylation motif from the Type II receptor, rendering it cytosolic in the dark. | Greatly reduces background (dark) activity, improving dynamic range [35]. |
| Signaling Output | Phosphorylation of Smad2 (pSmad2), nuclear translocation, and activation of target genes (e.g., gsc, sox32). | Reports directly on pathway activity; induces downstream mesendodermal fates [35]. |
The performance of optoNodal2 was quantitatively benchmarked. In mutant zebrafish embryos lacking endogenous Nodal signaling (Mvg1), pSmad2 levels reached a maximum approximately 35 minutes after a 20-minute light impulse and returned to baseline about 50 minutes later. This represented a significant kinetic improvement over first-generation tools, where signaling continued to accumulate for over 90 minutes post-illumination [35].
Experimental Workflow for Spatial Patterning of Nodal
The following diagram and protocol detail the methodology for creating synthetic Nodal signaling patterns in zebrafish embryos.
Figure 2: Workflow for high-throughput spatial patterning of Nodal signaling in zebrafish embryos using the optoNodal2 system.
Detailed Protocol:
- Sample Preparation: Inject one-cell stage zebrafish embryos (preferably in a Nodal signaling mutant background like Mvg1 or MZoep to eliminate confounding endogenous signals) with 10-30 pg of mRNA encoding the optoNodal2 receptor components [35].
- Embryo Mounting: At the desired developmental stage (e.g., shield stage), mount up to 36 embryos in parallel on an agarose-lined dish or within a microfluidic device compatible with the illumination platform.
- Spatial Patterning: Use an ultra-widefield patterned illumination microscope [35] or a Digital Micromirror Device (DMD) system [33] to project user-defined patterns of blue light (e.g., 470 nm, ~20 μW/mm²) onto the embryos. Patterns can include gradients, stripes, or spots to mimic or perturb natural signaling landscapes.
- Fixation and Immunostaining: At specific timepoints post-illumination, fix embryos and perform immunostaining for pSmad2 to visualize the spatial pattern of Nodal signaling activity [35].
- Downstream Analysis:
- Gene Expression: Analyze the expression of Nodal target genes (e.g., gsc, sox32, ntl) via in situ hybridization or RNA sequencing to determine the functional outcome of the synthetic signal.
- Cell Behavior: Track cell internalization movements during gastrulation in response to the patterned signal [35].
- Phenotypic Rescue: Assess the ability of patterned optoNodal2 activation to rescue characteristic developmental defects (e.g., mesendodermal deficiencies) in Nodal signaling mutants [35].
Application: Integrating Mechanical Forces and BMP Signaling in Human Gastrulation Models
Gastrulation involves an intricate interplay between biochemical signaling and physical forces. A groundbreaking study using human synthetic embryo models demonstrated that biochemical signals alone are insufficient to drive symmetry breaking and requires conjunction with specific mechanical contexts [2].
Optogenetic Control of BMP4 Signaling
To investigate the role of the morphogen BMP4, researchers developed an optogenetic human embryonic stem cell (hESC) line where exposure to a specific wavelength of light permanently turns on the BMP4 gene [2]. This system provides unparalleled precision in choosing when and where the signal is activated within a colony of cells.
Key Experimental Findings:
- In unconfined, low-tension environments, light-induced BMP4 activation alone was sufficient to induce extra-embryonic cell types (e.g., amnion) but failed to generate the mesoderm and endoderm layers essential for gastrulation.
- In confined, high-tension environments (e.g., using tension-inducing hydrogels), the same BMP4 signal initiated a full gastrulation response, including the formation of mesoderm and endoderm.
- Further analysis revealed that mechanical tension, sensed by the mechanosensory protein YAP1, acts as a molecular brake on gastrulation. Nuclear YAP1 must be downregulated for the biochemical signaling cascade (BMP4, WNT, Nodal) to proceed and instruct cells to form the appropriate germ layers [2].
This research provides a powerful experimental framework for dissecting the crosstalk between tissue mechanics and biochemical signaling, a frontier in developmental biology.
The Scientist's Toolkit: Essential Reagents and Hardware
Success in optogenetic experiments relies on a suite of well-characterized reagents and specialized hardware.
Table 3: Essential Research Reagent Solutions for Developmental Optogenetics
| Item | Function | Example Variants / Systems |
|---|---|---|
| Optogenetic Actuators | Core light-sensitive proteins fused to signaling molecules to control their activity. | optoNodal2 (Cry2/CIB1N) [35], LOV-based tools [36] [34], Phy/PIF [33] |
| Cell/Embryo Lines | Genetically engineered systems expressing optogenetic actuators. | Optogenetic BMP4 hESCs [2], Zebrafish embryos injected with optoNodal2 mRNA [35] |
| Chromophore | The light-absorbing cofactor required for photoreceptor function. | All-trans-retinal (for microbial opsins) [37], Flavin Adenine Dinucleotide (FAD for Cry2) [32], Phycocyanobilin (PCB for Phy) [33] |
| Illumination Hardware | Devices to deliver light of specific wavelengths, patterns, and intensities to samples. | Ultra-widefield microscopy [35], DMD microscopes [33], LED arrays [33], LAVA boards [33] |
| Bis(cyanopropyl)dichlorosilane | Bis(cyanopropyl)dichlorosilane, CAS:1071-17-6, MF:C8H12Cl2N2Si, MW:235.18 g/mol | Chemical Reagent |
| Mannose-1,6-bisphosphate | Mannose-1,6-bisphosphate, CAS:19504-70-2, MF:C6H14O12P2, MW:340.12 g/mol | Chemical Reagent |
Optogenetics has fundamentally transformed the toolkit available for investigating developmental pathways. By enabling the precise manipulation of signaling proteins with high spatiotemporal resolution, it allows researchers to move from observing correlations to establishing causality in complex processes like gastrulation. The ability to create synthetic signaling patterns and probe the interplay between biochemical and mechanical cues has already yielded profound insights, revealing that cells must be both chemically and mechanically competent to execute developmental programs [2].
Future directions in the field are poised to expand its capabilities further. The development of novel optogenetic actuators with red-shifted spectra will improve tissue penetration and enable multi-color control of distinct pathways [38] [37]. The integration of optogenetics with live-cell biosensors will allow for closed-loop control systems, where signaling dynamics are manipulated in real-time based on readouts from the cell itself. Furthermore, the application of these tools in more complex in vitro models, such as organoids, and the continued refinement of high-throughput illumination platforms will accelerate the systematic dissection of the rules governing embryogenesis. This progress will not only deepen our understanding of fundamental biology but also pave the way for innovative strategies in regenerative medicine and the treatment of developmental disorders.
Human gastrulation represents a pivotal developmental milestone during the third week of embryonic development, when a uniform sheet of cells undergoes profound transformation to establish the three fundamental germ layers—ectoderm, mesoderm, and endoderm—that give rise to all fetal tissues. This process involves sophisticated symmetry breaking events that define the anterior-posterior, dorsal-ventral, and left-right body axes, creating a spatial blueprint determining where the head, spine, and limbs will eventually form [2]. Despite its fundamental importance, our understanding of human gastrulation remains limited due to the inaccessibility of the in vivo condition, scarcity of tissue material, and ethical constraints encapsulated in the 14-day rule for embryo research [39]. Furthermore, significant species-specific differences between humans and model organisms underscore the necessity for human-specific model systems [39].
The emergence of stem cell-based models, particularly gastruloids and embryoids, has revolutionized our ability to investigate human gastrulation in vitro. These models leverage the self-organizing capacity of human pluripotent stem cells (hPSCs) to recapitulate aspects of early human development, providing unprecedented access to a previously inaccessible window of human embryogenesis [39] [40]. Within these models, precisely orchestrated signaling dynamics drive cell fate decisions and morphological transformations, offering researchers a powerful platform to dissect the molecular mechanisms underlying normal development and its dysregulation in reproductive failures and congenital disorders [39] [41].
Stem cell-based embryo models can be broadly categorized into non-integrated and integrated systems, each with distinct capabilities and applications in gastrulation research.
Table 1: Categories of Stem Cell-Based Human Embryo Models
| Model Type | Key Characteristics | Examples | Applications |
|---|---|---|---|
| Non-integrated Models | Mimic specific aspects of development; usually lack complete extra-embryonic lineages [39] | 2D Micropatterned Colonies (Gastruloids), PASE, PTED Embryoids [39] | Studying specific lineage specification, symmetry breaking, high-throughput screening [39] [41] |
| Integrated Models | Contain both embryonic and extra-embryonic cell types; model integrated development of entire conceptus [39] | Human Embryoids, ETiX Embryoids [40] [42] | Investigating tissue-tissue crosstalk, embryo organization, developmental potential [40] |
Non-integrated Models: Focusing on Specific Aspects
The 2D micropatterned colony (gastruloid) system involves culturing hESCs on circular micropatterned surfaces coated with extracellular matrix. Upon BMP4 treatment, these colonies self-organize into radial patterns containing an ectodermal center surrounded by concentric rings of mesoderm and endoderm, with an outermost ring of extra-embryonic cells of unclear origin [39]. This highly reproducible system generates all three germ layers but lacks the three-dimensional architecture and bilateral symmetry of natural embryos [39] [41].
Post-implantation amniotic sac embryoids (PASE) represent a three-dimensional model where hPSCs placed on a soft gel bed form an amniotic sac-like structure. During PASE development, hPSCs undergo lumenogenesis, causing the amniotic cavity to open up, while the emerging extra-embryonic amnion separates from the disk-like epiblast [39]. The epiblast subsequently develops a primitive streak-like structure with cells undergoing epithelial-to-mesenchymal transition (EMT) [39].
Integrated Models: Recapitulating Whole Embryo Development
Human embryoids combine wild-type embryonic stem cells with induced extraembryonic-like cells (hypoblast-like and trophoblast-like) generated through transcription factor overexpression. These modular systems self-organize into structures that mimic multiple hallmarks of post-implantation development, including lumenogenesis, amniogenesis, primordial germ cell formation, and anterior hypoblast specification [40]. Crucially, they demonstrate functional tissue-tissue crosstalk, where extraembryonic-like cells influence epiblast differentiation [40].
The ETiX embryoid system, demonstrated in mouse, combines ES cells, trophoblast stem cells, and extraembryonic endoderm stem cells to create structures that progress through gastrulation to neurulation and early organogenesis, developing brain regions, a beating heart-like structure, neural tube, somites, and gut tube [42]. This complete embryo model develops within an extraembryonic yolk sac that initiates blood island formation [42].
Signaling Pathways Governing Gastrulation
The intricate process of gastrulation is coordinated by an interplay of conserved signaling pathways that pattern the embryonic tissues and direct cell fate decisions. These pathways form complex regulatory networks with cross-talk and feedback loops that ensure proper spatiotemporal organization of the developing embryo.
Core Signaling Pathways
Table 2: Key Signaling Pathways in Human Gastrulation
| Signaling Pathway | Key Components | Role in Gastrulation | Experimental Manipulation |
|---|---|---|---|
| BMP Signaling | BMP4, SMAD1/5/8, NOGGIN | Initiates symmetry breaking, induces primitive streak formation, promotes mesoderm and extra-embryonic differentiation [16] [43] | BMP4 supplementation (100ng/mL) [43]; Optogenetic activation [2] [16] |
| WNT/β-catenin | β-catenin, CHIR99021, Cardamonin | Regulates primitive streak formation, posterior patterning, interacts with BMP and Nodal pathways [41] [43] | CHIR99021 (GSK3 inhibitor) for activation [3]; Cardamonin for inhibition [43] |
| Nodal/Activin/TGF-β | Nodal, Activin A, SB431542 | Controls mesendoderm specification, left-right asymmetry, regulates SMAD2/3 signaling [41] [43] | Activin A (50ng/mL) for activation [43]; SB431542 (10μM) for inhibition [43] |
| Hippo Pathway | YAP/TAZ, TEAD1-4, TRULI | Translates mechanical cues into gene expression, regulates tissue growth and differentiation [16] [43] | TRULI (2.5μM) for inhibition [43] |
| FGF Signaling | FGF2, PD0325901, PD173074 | Promotes mesoderm differentiation, regulates cell migration and fate decisions [43] | FGF2 (250ng/mL) for activation [43]; PD0325901 (1μM) for inhibition [43] |
Integration of Biochemical and Mechanical Signals
Recent research has revealed that mechanical forces play an indispensable role alongside biochemical signals in regulating gastrulation. Studies using optogenetic tools to precisely activate BMP4 signaling demonstrate that chemical cues alone are insufficient to drive complete gastrulation [2] [16]. The transformation only proceeds fully when cells are under appropriate mechanical conditions, specifically confinement and tension [2] [16].
The mechanosensory protein YAP1 serves as a critical integrator of mechanical and biochemical signals. In high-tension environments, nuclear YAP1 acts as a molecular brake on gastrulation, preventing premature transformation [2] [16]. Mechanical tension via YAP1 fine-tunes downstream WNT and Nodal signaling pathways, which ultimately instruct cells toward specific tissue fates [2] [16]. This interdependence between tissue mechanics and molecular signaling represents a fundamental principle of embryonic development.
Diagram 1: Signaling integration in gastrulation. The pathway shows how mechanical forces and biochemical signals converge through YAP1 to regulate gastrulation.
Experimental Protocols and Methodologies
Generating 2D Micropatterned Gastruloids
Workflow Overview:
- Micropattern Fabrication: Create arrays of circular extracellular matrix (ECM) islands (typically 500-800μm diameter) on cell-repellent surfaces using microcontact printing or photolithography [39].
- Cell Seeding: Dissociate human embryonic stem cells to single cells and seed at optimized density (e.g., 2-3×10^6 cells/mL) to achieve confluent monolayers on each micropattern [39].
- BMP4 Induction: 24 hours after seeding, treat with BMP4 (10-50ng/mL) in N2B27 medium to initiate self-organization [39] [41].
- Fixation and Analysis: Fix cells at specific timepoints (typically 42-72 hours post-BMP4) for immunofluorescence analysis of germ layer markers [41].
Key Technical Considerations:
- Maintain consistent cell density across experiments to minimize variability in patterning outcomes [41].
- Include appropriate controls: no-BMP4 negative controls and Wnt-activating positive controls (e.g., CHIR-98014) [41].
- For high-throughput applications, automate image acquisition and analysis using custom segmentation algorithms to quantify marker expression patterns [41].
Establishing Integrated Human Embryoids
Modular Assembly Protocol:
- Lineage Preparation:
- Generate induced hypoblast-like cells by dual overexpression of GATA6 and SOX17 in RSeT human ES cells [40].
- Generate induced trophoblast-like cells by dual overexpression of GATA3 and TFAP2C in RSeT human ES cells [40].
- Maintain wild-type embryonic stem cells in RSeT medium to preserve peri-implantation pluripotency state [40].
Embryoid Assembly:
- Combine wild-type ES cells, induced hypoblast-like cells, and induced trophoblast-like cells in specific ratios (typically 3:1:1) in low-attachment plates [40].
- Culture in basal medium without exogenous signaling factors to permit tissue-driven self-organization [40].
- Monitor development over 5-8 days, with medium changes every 48 hours [40].
Quality Assessment:
Optogenetic Control of Gastrulation
Light-Inducible System for BMP4:
- Genetic Engineering: Stably integrate human BMP4 coding sequence downstream of a loxP-flanked stop cassette in hESCs using piggyBac vector system [16].
- Light Sensitivity: Render cells light-sensitive through doxycycline-induced expression of Cre recombinase fused to light-responsive domains [16].
- Spatiotemporal Control: Activate BMP4 expression with precise spatiotemporal control using specific wavelengths of light (e.g., 650nm illumination) [16].
- Mechanical Context: Apply stimulation to cells in confined micropatterns or tension-inducing hydrogels to provide appropriate mechanical environment [2] [16].
Diagram 2: Experimental workflows for gastrulation models. The chart outlines three principal methodologies for generating stem cell-based gastrulation models.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Research Reagents for Gastrulation Studies
| Reagent Category | Specific Examples | Function/Application | Key Findings Enabled |
|---|---|---|---|
| Signaling Agonists | BMP4 (100ng/mL) [43], Recombinant Wnt3a (100ng/mL) [43], Activin A (50ng/mL) [43] | Activate specific signaling pathways to direct cell fate decisions | BMP4 initiates symmetry breaking and germ layer patterning [39] [16] |
| Signaling Inhibitors | Cardamonin (20μM, Wnt inhibitor) [43], SB431542 (10μM, Nodal/Activin/TGF-β inhibitor) [43], PD0325901 (1μM, FGF inhibitor) [43] | Inhibit specific pathways to dissect their roles in gastrulation | Nodal inhibition expands epiblast population [43] |
| Mechanical Manipulation | TRULI (2.5μM, Hippo inhibitor) [43], Synthetic hydrogels, Micropatterned substrates | Modulate mechanical environment and tissue tension | Demonstrated mechanical force requirement for gastrulation [2] [16] |
| Optogenetic Tools | Light-inducible BMP4 system [16], Cyan light-activated Cre recombinase | Spatiotemporal control of signaling pathway activation | Revealed interdependence of mechanics and BMP signaling [2] [16] |
| Cell Lines | RSeT hESCs (peri-implantation-like) [40], Inducible GATA6-SOX17 hESCs [40], Inducible GATA3-TFAP2C hESCs [40] | Provide starting material with specific pluripotency states | RSeT cells optimal for generating integrated embryoids [40] |
| Analysis Tools | Germ layer markers (SOX2, BRA, GATA3) [41], Custom image segmentation algorithms [41], Mathematical modeling frameworks [16] | Quantify patterning outcomes and model signaling dynamics | Identified cell density and SOX2 stability as key variance sources [41] |
| 2,5-Bis(4-pyridyl)-1,3,4-thiadiazole | 2,5-Bis(4-pyridyl)-1,3,4-thiadiazole, CAS:15311-09-8, MF:C12H8N4S, MW:240.29 g/mol | Chemical Reagent | Bench Chemicals |
| 2,4,4,6-Tetramethyl-1,3-dioxane | 2,4,4,6-Tetramethyl-1,3-dioxane, CAS:5182-37-6, MF:C11H11N3OS, MW:233.29 g/mol | Chemical Reagent | Bench Chemicals |
Applications and Research Implications
Modeling Developmental Disorders and Pregnancy Loss
Human gastrulation represents a critical vulnerability window during which substantial pregnancy failure occurs, with an estimated 30% of pregnancies failing at this stage [41]. Gastruloids and embryoids provide a unique platform to investigate the mechanisms underlying these failures, enabling researchers to identify specific failure modes and their molecular causes. By exposing these models to teratogenic compounds, researchers can map the morphospace of abnormal development and identify critical parameters whose perturbation leads to specific congenital malformations [41].
High-Throughput Teratogenicity Screening
The 2D gastruloid system offers particular advantages for high-throughput applications due to its reproducibility and scalability. Recent studies have demonstrated the utility of this platform for systematic drug screening, testing 210 compounds with known effects on stem cell signaling pathways [41]. By combining high-throughput perturbations with mathematical modeling, researchers have created predictive frameworks for identifying human-specific teratogens that might be missed in traditional animal models [41]. This approach addresses a significant limitation in current teratogenicity assessment, as conventional models like mice and rats are notoriously resistant to certain human teratogens such as thalidomide [41].
Illuminating Human-Specific Developmental Features
Comparative analyses between human embryo models and those from other species have revealed important human-specific aspects of gastrulation. For instance, during human embryogenesis, the epiblast-derived amnion forms ahead of primitive streak development, whereas in rodents, amnion genesis follows extraembryonic mesoderm formation from the primitive streak [39]. These differences in developmental timing and organization underscore the importance of human-specific models for understanding our unique biology and the evolutionary diversification of developmental mechanisms.
Future Directions and Concluding Perspectives
The field of stem cell-based embryo modeling continues to advance rapidly, with ongoing efforts focused on increasing the fidelity and complexity of these systems. Future developments will likely include improved vascularization, integration of immune cells, and extension of developmental competence to later stages of organogenesis. The combination of these experimental models with advanced computational approaches, including quantitative mathematical modeling and artificial intelligence-based pattern recognition, will further enhance their predictive power and research utility [16] [41].
As the field progresses, important ethical considerations remain paramount. The International Society for Stem Cell Research has categorized attempts to transfer human stem cell-based embryo models to uteruses as prohibited activities [39]. The scientific community continues to engage in thoughtful dialogue regarding the ethical dimensions of this research, balancing the tremendous potential for understanding human development and reducing reproductive failure against important ethical considerations.
In conclusion, gastruloids and embryoids represent transformative tools that have opened unprecedented windows into human gastrulation. By reconstructing the complex signaling dynamics that guide this fundamental process, these models provide insights that were previously inaccessible, advancing both basic developmental biology and clinical applications in reproductive medicine and teratology screening.
Gastrulation represents a pivotal stage in embryonic development, during which a homogeneous layer of pluripotent epiblast cells undergoes dramatic reorganization to form the three primary germ layers—ectoderm, mesoderm, and endoderm—that establish the basic body plan [44] [45]. This process is tightly controlled by a complex interplay of signaling dynamics and gene regulatory networks that gradually restrict developmental potential. Single-cell multi-omics technologies have revolutionized our ability to dissect these molecular events at unprecedented resolution, moving beyond bulk tissue analysis to reveal the cellular heterogeneity and epigenetic reprogramming that underlies cell fate decisions [46] [47]. By simultaneously profiling multiple molecular layers within individual cells, researchers can now delineate how signaling dynamics during gastrulation are translated into transcriptional and epigenetic changes that drive lineage specification.
The integration of stem cell technology with advanced molecular profiling tools has provided unprecedented insights into early lineage specification and the morphogenetic events that shape mammalian development [44]. These approaches are particularly valuable for studying human development, which presents significant ethical and technical challenges. Engineered models of peri-gastrulation, including blastoids, gastruloids, and somitoids, now enable the investigation of signaling dynamics and molecular mechanisms in experimentally accessible systems that mimic key aspects of embryonic development [6] [44]. Within this context, single-cell multi-omics serves as an essential tool for validating these models and elucidating the precise molecular relationships between signaling pathways, epigenetic states, and transcriptional outputs during germ layer formation.
Technological Foundations of Single-Cell Multi-Omics
Core Methodological Principles
Single-cell multi-omics technologies characterize cell states and activities by simultaneously integrating various single-modality omics methods that profile the transcriptome, genome, epigenome, epitranscriptome, proteome, metabolome, and other emerging omics modalities [47]. The fundamental workflow begins with single-cell isolation, typically achieved through fluorescence-activated cell sorting (FACS), magnetic-activated cell sorting (MACS), or various microfluidic technologies [46]. Microfluidic devices have revolutionized single-cell analysis by enabling high-throughput processing of tens of thousands of single cells through droplet-based systems or nanowells that provide individual compartments for single cells [46] [48].
A crucial step in single-cell sequencing workflows is cell barcoding, which allows libraries from multiple individual cells to be sequenced together in a single pool while preserving cellular identity [46]. In plate-based techniques, the cell barcode is typically added to the final PCR step before sequencing, whereas microfluidics-based barcoding methods incorporate cell barcodes earlier in the protocol, often processing the entire pool of libraries in a single tube [46]. The incorporation of unique molecular identifiers (UMIs) has been instrumental in advancing these technologies by enabling accurate quantification of original molecule abundance before amplification and allowing researchers to detect and correct artifacts introduced during the aggressive amplification process required for single-cell analysis [48].
Multi-Omics Integration Platforms
Numerous single-cell multi-omics techniques have been developed into high-throughput, routinely accessible platforms that delineate precise relationships among different layers of the central dogma [48]. These technologies have evolved significantly over the past decade, with improvements in throughput, resolution, modality integration, and accuracy [47]. The table below summarizes representative single-cell multi-omics methods and their applications:
Table 1: Evolution of Single-Cell Multi-Omics Technologies
| Year | Method | Technology | Data Types | Resolution | Reference |
|---|---|---|---|---|---|
| 2015 | G&T-seq | Plate-based sequencing | DNA, mRNA | Single cell | [47] |
| 2016 | scTrio-seq | Pipette cell-picking sequencing | DNA, RNA, DNA methylation | Single cell | [47] |
| 2017 | CITE-seq | Droplet-based microfluidics | mRNA, protein | Single cell | [47] |
| 2019 | scNOMe-seq | Plate-based sequencing | DNA methylation, chromatin accessibility | Single cell | [47] |
| 2019 | SNARE-seq | Droplet-based microfluidics | Accessible chromatin, mRNA | Single cell | [47] |
| 2021 | ASAP-seq | Droplet-based microfluidics | Accessible chromatin, mRNA, proteins, mitochondrial DNA | Single cell | [47] |
| 2022 | NEAT-seq | Droplet-based microfluidics | Intracellular proteins, accessible chromatin, mRNA | Single cell | [47] |
| 2022 | Spatial ATAC-seq | Microchannel-based microfluidics | Chromatin accessibility | Spatial, 10-50 μm | [47] |
Recent advancements have extended multi-omics profiling to spatial contexts, enabling the mapping of gene expression and chromatin accessibility within the tissue architecture [47]. Methods such as DBiT-seq (deterministic barcoding in tissue for spatial omics sequencing) enable simultaneous mapping of mRNA and proteins at spatial resolutions of 10-50 μm, while Spatial ATAC-RNA-seq allows parallel assessment of chromatin accessibility and transcriptome in the same tissue section [47]. These spatial multi-omics approaches are particularly valuable for gastrulation research, where the positional information of cells is critical for understanding patterning and morphogenetic events.
Experimental Design and Workflow Integration
Strategic Planning for Gastrulation Studies
When designing single-cell multi-omics experiments for gastrulation research, careful consideration must be given to the biological question, appropriate model systems, and technology selection. For studies focusing on signaling dynamics during early lineage specification, a time-series approach capturing multiple developmental stages is essential to resolve transitional states [45]. Research questions aimed at understanding the molecular basis of cell fate decisions might employ transcriptome-epigenome integration, while investigations of signaling pathway activation might benefit from combining transcriptome with proteome or phosphoproteome profiling.
The selection of an appropriate model system depends on the specific aspect of gastrulation under investigation. For pre-gastrulation events, blastoid models derived from naive pluripotent stem cells can mimic blastocyst formation and implantation [44]. For gastrulation proper, 2D micropatterned systems or 3D gastruloids provide insights into germ layer specification and tissue organization [6] [44]. Post-gastrulation events such as somitogenesis and axial elongation can be studied using somitoid models [44]. Each system offers distinct advantages and limitations in terms of scalability, physiological relevance, and compatibility with multi-omics protocols.
Integrated Multi-Omics Workflow
A typical integrated single-cell multi-omics workflow for gastrulation research involves several key stages, from sample preparation through data integration and interpretation. The following diagram illustrates a generalized workflow:
Diagram 1: Integrated Multi-omics Workflow
This workflow begins with sample preparation from appropriate model systems, followed by single-cell isolation using FACS or microfluidic platforms. Multi-omics profiling then simultaneously captures multiple molecular modalities, after which libraries are prepared and sequenced. The data analysis phase involves processing and quality control, integration of different modalities, cell clustering and annotation, regulatory network analysis, and finally biological interpretation and validation.
Multi-Omics Applications in Gastrulation Research
Decoding Epigenetic Dynamics During Lineage Specification
Single-cell multi-omics approaches have revealed extensive epigenetic reprogramming during mouse gastrulation, with distinct histone modification dynamics accompanying the specification of each germ layer [45]. A recent study utilizing single-cell ChIP-seq for H3K27ac and H3K4me1 in mouse embryos across six developmental stages (from Pre-Primitive Streak to Early Headfold stages) demonstrated that significant epigenetic priming is evident before morphological signs of lineage commitment [45]. The research found germ layer-specific subpopulations defined by H3K27ac signals emerging as early as the Pre-Primitive Streak stage, indicating effective epigenetic priming for lineage specification before gastrulation initiation.
The study further revealed asynchronous cell fate commitment of each germ layer at distinct histone modification levels [45]. Principal components analysis of H3K27ac and H3K4me1 signals showed different segregation patterns: at the H3K27ac level, ectoderm clusters located closer to mesenchyme-to-mesoderm, while separating from endodermal lineages across all stages [45]. Interestingly, highly similar H3K27ac patterns were observed between neural ectoderm and epiblast cells at Pre-Primitive Streak stages, suggesting that neural ectoderm fate was already primed at this early stage. In contrast, germ layer-specific differences appeared more pronounced in H3K4me1 patterns, indicating distinct regulatory functions for these histone marks during lineage commitment.
Resolving Temporal Relationships Between Signaling, Epigenetics, and Transcription
Integrated analysis of single-cell transcriptome and epigenome data has unveiled a "time lag" transition pattern between enhancer activation and gene expression during germ-layer specification [45]. This finding suggests that epigenetic changes in enhancer activity precede transcriptional activation of associated genes, providing a molecular mechanism for the gradual restriction of developmental potential during gastrulation. By utilizing H3K27ac and H3K4me1 co-marked active enhancers, researchers have constructed gene regulatory networks centered on pivotal transcription factors, highlighting potential critical regulators such as Cdkn1c in mesoderm lineage specification [45].
The application of scNMT-seq (single-cell nucleosome, methylation, and transcription sequencing) to study mouse gastrulation revealed that epigenetic states in DNA methylation and chromatin accessibility in cells fated to ectoderm are already established in the early epiblast, whereas cells committed to mesoderm and endoderm undergo extensive epigenetic reprogramming [45] [46]. This differential epigenetic dynamics across germ layers underscores the distinct regulatory strategies employed during lineage specification and reflects the influence of signaling dynamics on epigenetic states.
Detailed Experimental Protocols
Single-Cell Multi-Omics Profiling of Gastrulation Models
This protocol describes an integrated approach for simultaneous profiling of transcriptome and chromatin accessibility in gastrulation models using the 10x Genomics Multiome ATAC + Gene Expression platform, adapted from methodologies described in the search results [49] [45].
Sample Preparation and Nuclei Isolation
Begin with gastruloids or embryonic tissues at appropriate developmental stages. For dissociation, wash samples with PBS and incubate in Accutase or TrypLE at 37°C for 10-15 minutes with gentle pipetting to dissociate into single cells. Quench the dissociation reagent with complete medium, then filter through a 40μm flow-through filter. Centrifuge at 300-500g for 5 minutes and resuspend in cold PBS with 0.04% BSA. For nuclei isolation, centrifuge the cell suspension and resuspend the pellet in cold lysis buffer (10mM Tris-HCl, 10mM NaCl, 3mM MgCl2, 0.1% Tween-20, 0.1% Nonidet P-40, 0.01% Digitonin, 1% BSA in nuclease-free water) for 3-5 minutes on ice. Immediately dilute with wash buffer (without detergents) and filter through a 40μm flow-through filter. Count nuclei using a hemocytometer and adjust concentration to 1,000-10,000 nuclei/μL, keeping samples on ice.
Multiome ATAC + Gene Expression Library Preparation
Proceed with the transposition reaction using the 10x Genomics Nuclei Buffer and Tn5 Transposase. Incubate at 37°C for 60 minutes with mild shaking. After transposition, add the Stop Buffer and proceed immediately to partitioning. Load the transposed nuclei into the 10x Genomics Chromium Chip along with the Master Mix and Partitioning Oil. After partitioning, recover the barcoded gel beads-in-emulsion (GEMs) and perform the GEM incubation (53°C for 45 minutes). Break the GEMs and clean up the post-ATAC and post-GEX products separately. Amplify the ATAC library using custom primers and the following PCR program: 98°C for 45s; 10-14 cycles of (98°C for 20s, 67°C for 30s, 72°C for 60s); 72°C for 60s. Amplify the GEX library using: 98°C for 45s; 11-14 cycles of (98°C for 20s, 67°C for 30s, 72°C for 60s); 72°C for 60s. Perform double-sided size selection on both libraries using SPRIselect beads and quantify using Qubit and Bioanalyzer.
Single-Cell ChIP-seq for Histone Modifications During Gastrulation
This protocol describes the CoBATCH method for single-cell ChIP-seq of histone modifications, as applied to mouse gastrulation studies [45].
Cell Fixation and Barcoding
Harvest gastruloids or embryonic tissues and dissociate into single cells as described in 5.1.1. Fix cells with 1% formaldehyde for 10 minutes at room temperature, then quench with 125mM glycine for 5 minutes. Wash twice with cold PBS and resuspend in cell lysis buffer (10mM Tris-HCl pH 7.5, 10mM NaCl, 3mM MgCl2, 0.1% Tween-20) for 10 minutes on ice. Centrifuge and resuspend the nuclei in Rxn-X buffer (50mM Tris-HCl pH 7.5, 10mM MgCl2, 100mM NaCl, 0.1mg/mL BSA, 1mM DTT). Add protein A-Tn5 transposase preloaded with mosaic ends and specific antibodies against histone modifications (e.g., H3K27ac, H3K4me1). Incubate at 4°C for 2 hours with gentle rotation to allow antibody binding. Add MgCl2 to a final concentration of 10mM and incubate at 37°C for 1 hour to perform tagmentation. Stop the reaction with 10mM EDTA and 1% SDS, then incubate at 40°C for 1 hour to reverse cross-linking.
Library Preparation and Sequencing
Purify DNA using SPRIselect beads and elute in TE buffer. Amplify libraries using i5 and i7 indexing primers with the following PCR program: 72°C for 5min; 98°C for 30s; 12-15 cycles of (98°C for 10s, 63°C for 30s, 72°C for 60s); 72°C for 5min. Perform double-sided size selection (0.5x and 1.0x SPRIselect) to remove primer dimers and large fragments. Quality control using Qubit and Bioanalyzer/Tapestation. Pool libraries at appropriate molar ratios and sequence on Illumina platforms (NovaSeq 6000 recommended) with paired-end reads (2×150bp) to achieve sufficient coverage.
Signaling Pathways in Gastrulation: A Multi-Omics Perspective
Key Signaling Pathways and Their Epigenetic Regulation
Gastrulation is coordinated by a complex interplay of signaling pathways that guide cell fate decisions and morphogenetic movements. Single-cell multi-omics approaches have been particularly instrumental in elucidating how these signaling dynamics are translated into epigenetic and transcriptional changes. The following diagram illustrates the major signaling pathways involved in gastrulation and their coordination:
Diagram 2: Signaling to Cell Fate Regulation
The BMP, WNT, and Nodal pathways play particularly crucial roles in germ layer patterning. BMP signaling promotes epidermal differentiation at high levels and mesodermal differentiation at lower levels, primarily through SMAD-mediated regulation of target genes [44]. WNT/β-catenin signaling is essential for primitive streak formation and mesendodermal specification, with different concentration thresholds activating distinct target genes [44]. Nodal signaling, a member of the TGF-β superfamily, is critical for mesendodermal patterning and left-right asymmetry establishment, acting through SMAD2/3 transcription factors [44].
Single-cell multi-omics analyses have revealed that these signaling pathways induce specific epigenetic changes that reinforce lineage commitment. For instance, WNT signaling promotes the opening of chromatin regions near mesodermal genes while facilitating the closure of ectodermal gene loci [45]. Similarly, BMP signaling induces H3K27ac deposition at enhancers associated with epidermal genes while repressing histone modifications at neural genes [45]. These epigenetic changes create a permissive environment for the activation of lineage-specific transcriptional programs while restricting alternative fate potentials.
Multi-Omics Insights into Signaling Gradient Interpretation
A key question in gastrulation research concerns how cells interpret morphogen gradients to adopt specific positional identities along the embryonic axes. Single-cell multi-omics approaches have provided unprecedented insights into this process by simultaneously capturing signaling pathway activity, epigenetic states, and transcriptional outputs in individual cells. Studies using micropatterned human pluripotent stem cell colonies exposed to BMP4 gradients have revealed that distinct concentration thresholds trigger different epigenetic and transcriptional responses [44].
Integration of scRNA-seq and scATAC-seq data from gastrulation models has demonstrated that cells exposed to different morphogen concentrations exhibit distinct chromatin accessibility profiles at key developmental loci before the full activation of lineage-specific genes [45]. This suggests that epigenetic priming represents an early response to signaling gradients, potentially establishing competence for subsequent lineage commitment. Furthermore, multi-omics analyses have revealed that the same signaling pathway can induce different epigenetic changes depending on cellular context and history, explaining how identical signals can elicit distinct outcomes in different regions of the embryo.
Essential Research Reagents and Computational Tools
Research Reagent Solutions for Gastrulation Multi-Omics
Table 2: Essential Research Reagents for Single-Cell Multi-Omics Studies
| Category | Specific Reagents | Function | Application Notes |
|---|---|---|---|
| Cell Dissociation | Accutase, TrypLE Select, Collagenase IV | Tissue dissociation into single cells | Gentle enzymes preserve cell viability and surface epitopes; concentration and timing critical for embryonic tissues |
| Nuclei Isolation | Dounce homogenizers, IGEPAL, Tween-20, Digitonin | Nuclei extraction and purification | Optimized lysis conditions essential for preserving nuclear integrity and chromatin accessibility |
| Epigenetic Profiling | Tn5 transposase, Protein A-Tn5 conjugates, Histone modification antibodies | Tagmentation and immunoprecipitation | Antibody validation critical for ChIP-seq; Tn5 concentration optimization needed for different sample types |
| Single-Cell Barcoding | 10x Genomics Chromium chips, MULTI-seq barcodes, Cell hashing antibodies | Cell multiplexing and sample indexing | Enables sample pooling, reduces batch effects, and decreases per-sample costs |
| Library Preparation | SPRIselect beads, Custom primers, PCR enzymes | Library amplification and size selection | Double-sided size selection improves library complexity and removes adapter dimers |
| Quality Control | Bioanalyzer/Pico chips, Qubit dsDNA HS assay, DAPI/Propidium Iodide | Assessment of nucleic acid quality and quantity | Critical for determining input quality and optimizing sequencing depth |
Computational Tools for Multi-Omics Data Integration
The analysis of single-cell multi-omics data requires specialized computational tools that can handle the complexity and scale of these datasets. For processing raw sequencing data, Cell Ranger (10x Genomics) and Cell Ranger ATAC provide standardized pipelines for demultiplexing, barcode processing, and alignment [49]. For quality control and initial analysis, Seurat and ArchR offer comprehensive frameworks for filtering cells, dimensionality reduction, clustering, and visualization [49] [45]. The integration of different omics modalities can be achieved using tools such as Harmony for batch correction, Signac for integrated analysis of scRNA-seq and scATAC-seq data, and MOFA+ for multi-omics factor analysis that identifies latent factors across data modalities [49].
For biological interpretation, tools like CellChat enable inference of cell-cell communication networks from scRNA-seq data, while GREAT facilitates functional annotation of cis-regulatory elements identified through epigenomic profiling [49] [45]. Additionally, pySCENIC allows reconstruction of gene regulatory networks by combining transcription factor binding motifs with gene expression data, providing insights into the regulatory logic underlying cell fate decisions during gastrulation [45].
Technical Considerations and Limitations
Methodological Challenges and Optimization Strategies
Despite the powerful insights provided by single-cell multi-omics approaches, several technical challenges must be addressed in experimental design and data interpretation. The table below summarizes key considerations and recommended optimization strategies:
Table 3: Technical Challenges and Optimization in Single-Cell Multi-Omics
| Challenge | Impact on Data Quality | Optimization Strategies |
|---|---|---|
| Cell Viability and Integrity | Poor viability increases background noise and technical artifacts | Maintain strict temperature control during dissociation; use viability dyes for selection; minimize processing time |
| Nuclear Integrity | Compromised nuclei yield poor chromatin accessibility data | Optimize lysis conditions; validate nuclei morphology by microscopy; use sucrose gradients for purification |
| Amplification Bias | Uneven coverage, allele dropout, false positives | Incorporate UMIs; use unique polymerases (e.g., phi29 for WGA); optimize cycle number; employ spike-in controls |
| Multiplet Rate | Misassignment of molecular signatures to single cells | Optimize cell concentration loading; employ doublet detection algorithms; use species-mixing controls |
| Batch Effects | Technical variation obscures biological signals | Include biological replicates; use sample multiplexing; apply batch correction algorithms |
| Sparse Data | Limited detection of low-abundance molecules | Increase sequencing depth; employ targeted approaches; use imputation algorithms cautiously |
Integration with Functional Validation
While single-cell multi-omics provides comprehensive molecular maps, functional validation remains essential for establishing causal relationships. Perturbation approaches such as CRISPR-Cas9 screens, inducible expression systems, and small molecule inhibitors can test hypotheses generated from observational multi-omics data [47]. For gastrulation research, engineered model systems offer particularly valuable platforms for functional validation, as they enable precise manipulation of signaling pathways and transcription factors in a controlled environment [44].
Live imaging approaches complement single-cell multi-omics by providing dynamic information about cell behaviors and morphogenetic movements. Combining lineage tracing with multi-omics profiling enables direct correlation of lineage relationships with molecular states, revealing how epigenetic and transcriptional changes are inherited or modified during cell division and differentiation [47] [44]. This integrated approach provides a more complete understanding of how signaling dynamics are translated into cell fate decisions during gastrulation.
Future Perspectives and Concluding Remarks
Single-cell multi-omics technologies have fundamentally transformed our ability to dissect the molecular mechanisms underlying gastrulation, providing unprecedented resolution of the epigenetic and transcriptional landscapes that guide cell fate decisions. These approaches have revealed the extensive cellular heterogeneity within seemingly homogeneous cell populations, uncovered previously unrecognized transitional states, and elucidated the complex regulatory networks that translate signaling dynamics into lineage commitment. The integration of multi-omics data with engineered model systems has been particularly powerful, enabling the validation of these models while providing insights into human-specific aspects of development that are difficult to study in vivo.
Looking forward, several emerging technologies promise to further advance gastrulation research. Spatial multi-omics approaches will enable the direct correlation of molecular profiles with positional information within embryonic structures, revealing how spatial organization influences and is influenced by signaling dynamics [47]. Long-read sequencing technologies will improve the detection of isoform diversity and structural variants, while emerging methods for profiling additional modalities such as protein post-translational modifications, metabolic states, and chromatin conformation will provide more comprehensive views of cellular states [48]. Computational methods for multi-omics data integration and causal inference will also continue to evolve, enabling more accurate reconstruction of regulatory networks and predictive models of cell fate decisions.
In conclusion, single-cell multi-omics approaches have established a new paradigm for studying gastrulation, moving beyond descriptive characterization to mechanistic understanding of how signaling dynamics coordinate epigenetic reprogramming and transcriptional activation to guide the formation of the basic body plan. As these technologies continue to mature and become more accessible, they will undoubtedly yield further insights into the fundamental principles of embryonic development and the etiology of developmental disorders.
The precise signaling dynamics that coordinate human gastrulation are a critical focus of developmental biology research. Beyond fundamental understanding, these processes hold the key to evaluating drug-induced developmental abnormalities, known as teratogenicity. Traditional animal models often fail to predict human-specific outcomes, creating a significant bottleneck in pharmaceutical safety assessment [43]. The emergence of synthetic embryo platforms, particularly those incorporating optogenetic control and defined mechanical environments, now enables unprecedented high-throughput investigation of these crucial early developmental events [2]. This whitepaper details methodologies leveraging these advanced platforms to screen for compound teratogenicity by directly interrogating the signaling cascades governing gastrulation.
Signaling Pathways Governing Gastrulation and Susceptibility to Teratogenic Disruption
Successful gastrulation requires the meticulous spatiotemporal coordination of multiple conserved signaling pathways. Disruption at any key node can cause severe developmental defects, making these pathways prime targets for teratogenicity screening.
Hippo Signaling Pathway: Regulating TE Differentiation: The Hippo pathway is a highly conserved kinase cascade that controls the first lineage specification in the preimplantation embryo, determining the fate of the trophectoderm (TE) versus the inner cell mass (ICM). Its core components include MST1/2, LATS1/2, and the transcriptional coactivators YAP/TAZ. When active, the kinase cascade phosphorylates YAP/TAZ, sequestering them in the cytoplasm. When inhibited, dephosphorylated YAP/TAZ translocate to the nucleus and complex with TEAD transcription factors to activate TE-specific genes like CDX2 [43]. This pathway is mechanically sensitive; in outer, polarized cells, apical polarity complexes suppress the Hippo pathway, allowing YAP/TAZ nuclear localization and TE specification [43]. Teratogens that disrupt cell polarity or mechanics can therefore cause lineage mis-specification.
Wnt/β-Catenin Pathway: A Key Regulator of Axis Patterning: The Wnt/β-catenin pathway is fundamental for establishing the body axes during gastrulation. In a new study, researchers found that mechanical tension, mediated by YAP1, fine-tunes the downstream biochemical signaling pathways mediated by WNT (and Nodal) to instruct cell fate decisions [2]. This indicates that proper gastrulation requires an interplay between mechanical forces and Wnt signaling.
BMP4 Signaling and Mechanical Competence in Symmetry Breaking: BMP4 is a well-known morphogen that influences cell behavior and regulates embryonic development. Recent research using optogenetic tools reveals that BMP4 signaling alone is insufficient to drive complete gastrulation. In unconfined, low-tension environments, BMP4 activation generated extra-embryonic cell types but failed to produce the mesoderm and endoderm layers that build the body's organs [2]. Gastrulation's missing layers only formed when BMP4 was activated in confined cell colonies or tension-inducing hydrogels [2]. This demonstrates that cells must be both chemically prepared (via BMP4) and physically primed (via mechanical tension) for gastrulation to occur. The mechanosensory protein YAP1 acts as a molecular brake, preventing gastrulation from occurring too soon [2].
Table 1: Key Signaling Pathways in Human Preimplantation Development and Teratogenic Vulnerability
| Pathway | Core Components | Primary Role in Gastrulation | Potential Teratogenic Mechanism |
|---|---|---|---|
| Hippo | MST1/2, LATS1/2, YAP/TAZ, TEAD1-4 [43] | First lineage specification (ICM vs. TE); Mechanosensing [43] [2] | Disruption of cell polarity/mechanics causing lineage mis-specification [43] |
| Wnt/β-catenin | Wnt ligands, β-catenin, TCF/LEF | Axis patterning, Cell fate determination [2] | Ectopic activation/inhibition disrupting axial patterning [43] |
| TGF-β/Nodal | Nodal, Activin, TGF-β, Smads | Mesendoderm specification, Primitive streak induction [43] | Aberrant primitive streak formation |
| BMP | BMP4, BMP receptors, Smads | Dorsoventral patterning, Epithelial-mesenchymal transition [2] | Disrupted symmetry breaking and axis formation [2] |
| FGF | FGF ligands, FGFR, RAS-MAPK | ICM lineage segregation (EPI vs. PrE) [43] | Altered balance between epiblast and primitive endoderm [43] |
Synthetic Embryo Platforms: Engineered Systems for High-Throughput Screening
Synthetic embryo platforms, such as gastruloids and optogenetically controlled models, provide a scalable and ethically manageable system for teratogenicity testing. These systems recapitulate key aspects of early human development, including symmetry breaking, lineage specification, and the response to morphogen gradients, without the use of natural human embryos [2].
A pivotal advancement is the development of a light-based "remote-control" synthetic embryo system. This platform uses human embryonic stem cells (hESCs) engineered to express a light-activated switch for key developmental genes like BMP4 [2]. This optogenetic tool allows researchers to trigger specific signaling pathways with exceptional spatial and temporal precision, enabling the direct testing of how tissue geometry and mechanical stress influence developmental outcomes [2].
These platforms are built on microchips, allowing for high-throughput experimentation and the parallel testing of multiple compounds or conditions [2]. The integration of a mathematical "digital twin" that simulates the interplay between biochemical signals and physical forces further enhances the predictive power of these systems [2].
Experimental Protocols for Teratogenicity Assessment
Core Platform Setup and Workflow
The following Dot language code defines the experimental workflow for high-throughput teratogenicity screening using synthetic embryo platforms.
Title: HTS Teratogenicity Screening Workflow
Protocol 1: Establishing Optogenetically Controlled Synthetic Embryos
- Cell Line Preparation: Utilize an established human pluripotent stem cell (hPSC) line (e.g., H9) engineered to express a light-activated switch (e.g., Opto-BMP4) for inducible control of the gene of interest [2].
- Micropatterned Surface Seeding: Seed the optogenetic hPSCs onto micropatterned cell culture substrates (e.g., Cytoo chips or similar) that define tissue geometry and induce reproducible mechanical confinement [2].
- Coat substrates with extracellular matrix (e.g., Matrigel or Laminin-521).
- Seed a single-cell suspension at a defined density to form uniformly sized colonies.
- Pre-culture Maintenance: Culture cells in a defined, feeder-free medium for 24-48 hours to allow colony formation and stabilization under minimal morphogen conditions.
Protocol 2: High-Throughput Compound Screening and Pathway Challenge
- Compound Library Application: Using an automated liquid handler, transfer a library of test compounds (or vehicle controls) to the assay plates. Incubate for a predefined period (e.g., 24 hours) to expose the synthetic embryos to potential teratogens [2].
- Optogenetic Pathway Activation: Expose the plates to a specific wavelength of light (e.g., 650 nm for Cy5- or phytochrome-based systems) to uniformly activate the target pathway (e.g., BMP4) across all wells. The precise timing and duration of light exposure should be optimized for the specific pathway [2].
- Tension Modulation (Optional): For compounds suspected of affecting mechanosensing, a subset of plates can be cultured in tension-inducing hydrogels (e.g., fibrin or PEG-based gels) to test the interplay between compound effect and mechanical context [2].
Endpoint Analysis and Data Collection
The following Dot language code visualizes the key signaling pathways and their interactions that are analyzed to assess teratogenicity.
Title: Core Gastrulation Signaling Network
Protocol 3: High-Content Imaging and Molecular Phenotyping
- Fixation and Staining: At the experimental endpoint (e.g., 72 hours post-induction), fix cells and perform immunofluorescence staining for key lineage and polarity markers.
- Automated Image Acquisition: Use a high-content imaging system (e.g., ImageXpress Micro Confocal) to automatically acquire high-resolution images (20x or 40x objective) from each well of the screening plate.
- Image Analysis Pipeline:
- Segmentation: Identify individual cell nuclei (DAPI) and colony boundaries.
- Intensity Quantification: Measure fluorescence intensity of lineage markers in segmented regions.
- Spatial Analysis: Calculate the spatial distribution of markers (e.g., radial profile analysis) to detect aberrant patterning.
- Morphometric Analysis: Quantify colony morphology features (e.g., circularity, aspect ratio).
Data Analysis and Teratogenicity Scoring
Quantitative data from high-content imaging is aggregated to generate a teratogenicity risk score for each test compound. The following table summarizes the key quantitative benchmarks derived from studies modulating specific pathways, serving as a baseline for identifying compound-induced deviations [43].
Table 2: Quantitative Effects of Pathway Modulation on Blastocyst Development and Lineage Specification
| Small Molecule / Treatment | Target Pathway | A./I. | Blastocyst Development Rate (Control) | ICM Marker | TE Marker | PrE Marker | Reference |
|---|---|---|---|---|---|---|---|
| CRT0276121 | Hippo | A. | 25% (83%) | → | ↓ | - | [43] |
| TRULI | Hippo | I. | 100% (100%) | ↑ | ↓ | - | [43] |
| 1-Azakenpaullone | Wnt/β-catenin | A. | 70% (86%) | → | ↓ | - | [43] |
| Cardamonin | Wnt/β-catenin | I. | 46% (75%) | → | ↓ | - | [43] |
| PD0325901 + PD173074 | FGF | I. | - | → | - | → | [43] |
| FGF2 | FGF | A. | - | ↓ | - | ↑ | [43] |
| SB431542 | TGF-β/Activin/Nodal | I. | 25% (28%) | ↑ | - | → | [43] |
| BMP4 | BMP | A. | 17.4% (61.5%) | → | → | → | [43] |
Teratogenicity Scoring Algorithm:
- Primary Hits: Identify compounds that cause a statistically significant (p < 0.01) deviation in blastocyst development rate or a >50% change in lineage marker expression compared to vehicle controls.
- Pathway Fingerprinting: Classify hits based on the specific pattern of lineage marker disruption, which can indicate the pathway affected (e.g., a compound that suppresses CDX2 and increases NANOG may be a Hippo pathway activator).
- Mechanistic Confirmation: Use secondary assays (e.g., qPCR for pathway target genes, Western blot for phospho-proteins) on hit compounds to confirm the hypothesized mechanism of action.
The Scientist's Toolkit: Essential Reagents and Materials
Table 3: Research Reagent Solutions for Synthetic Embryo Teratogenicity Screening
| Item | Function / Application | Example / Notes |
|---|---|---|
| Opto-hPSC Line | Engineered cell line for light-controlled gene expression. | hPSCs with Opto-BMP4 construct for precise, remote-controlled pathway activation [2]. |
| Micropatterned Plates | Defines colony geometry and induces reproducible mechanical confinement. | Cytoochips or similar; crucial for standardizing the mechanical microenvironment [2]. |
| Tension-Inducing Hydrogels | Provides a tunable 3D mechanical environment. | Fibrin or PEG-based hydrogels; used to test the role of mechanosensing in compound toxicity [2]. |
| Pathway Agonists/Antagonists | Positive and negative controls for pathway modulation. | TRULI (Hippo inhibitor), Cardamonin (Wnt inhibitor), FGF2 (FGF activator), SB431542 (Nodal inhibitor) [43]. |
| Validated Antibodies | Immunofluorescence staining for lineage and polarity markers. | Anti-NANOG (ICM), Anti-CDX2 (TE), Anti-GATA6 (PrE), Anti-YAP/TAZ (mechanosensing) [43] [2]. |
| High-Content Imager | Automated, high-throughput image acquisition. | Systems like ImageXpress Micro Confocal for consistent, hands-off data collection across 96/384-well plates. |
| "Digital Twin" Model | Mathematical simulation of signaling and mechanics. | Computer model predicting how biochemical signals and mechanical tension interact to direct development [2]. |
| trans-3,5-Diethyl-1,2,4-trithiolane | trans-3,5-Diethyl-1,2,4-trithiolane, CAS:38348-26-4, MF:C6H12S3, MW:180.4 g/mol | Chemical Reagent |
Overcoming Challenges: Optimizing Models and Interpreting Complex Signaling Data
Addressing Ethical and Technical Barriers in Human Gastrulation Research
Human gastrulation is a pivotal yet elusive stage in early embryonic development, occurring approximately two weeks after fertilization. During this process, a uniform sheet of cells transforms into the foundational blueprint of the human body, establishing the head-tail, ventral-dorsal, and right-left axes that guide all subsequent tissue and organ formation [2]. Despite its fundamental importance, research into human gastrulation faces significant ethical constraints and technical limitations, as this developmental milestone occurs too early and deeply within the uterus for direct observation or experimentation [2].
The study of gastrulation mechanisms represents a critical frontier in developmental biology with profound implications for regenerative medicine and fertility therapies. Recent advances have revealed that this process is guided by a precise interplay between biochemical signals and physical forces, moving beyond the traditional paradigm that focused primarily on molecular signaling pathways [2] [16]. This whitepaper examines the current landscape of human gastrulation research within the context of signaling dynamics, addressing both the ethical frameworks that enable responsible investigation and the technical innovations that are opening new windows into this transformative developmental stage.
Ethical Barriers and Solutions
Primary Ethical Constraints
The 14-day rule, an international ethical guideline that limits embryo research to the first two weeks of development, presents a fundamental constraint on gastrulation studies. This boundary was established because gastrulation represents the emergence of the embryonic body plan and the point at which twinning can no longer occur. While this regulation prevents direct investigation of later developmental stages, it simultaneously motivates the development of alternative model systems that can ethically recapitulate key developmental events.
The challenge of obtaining human embryo specimens further compounds these ethical limitations. Donations for research purposes are scarce, and the logistical complexities of collection, preservation, and experimentation create substantial barriers to comprehensive studies. These constraints have necessitated the development of innovative model systems that can bypass these ethical concerns while providing scientifically valid insights into human development.
Ethical Research Models
Table 1: Alternative Models for Gastrulation Research
| Model System | Description | Advantages | Limitations |
|---|---|---|---|
| Synthetic Embryoids | Light-controlled human embryonic stem cells that self-organize | Enables study of symmetry breaking; avoids embryo use [2] | May not fully replicate in vivo complexity |
| 2D Gastruloids | Human embryonic stem cells grown on micropatterned substrates | Recapitulates some aspects of germ layer formation [16] | Limited to two-dimensional organization |
| Animal Models | Studies in Drosophila, quail, and other species | Provides evolutionary insights; fewer ethical restrictions [50] | Species-specific differences exist |
The emergence of synthetic embryo models represents a paradigm shift in gastrulation research. These systems, derived from human pluripotent stem cells, can self-organize and mimic key developmental events without the ethical concerns associated with natural embryos. As one researcher noted: "We can now generate self-organization and different cell types, just by shining light on it. This allowed us to make a major discovery about the role of mechanical forces in embryonic development" [2]. These models are particularly valuable for studying the symmetry breaking process that initiates gastrulation, a phenomenon that was previously inaccessible to direct observation.
Technical Challenges and Advanced Methodologies
Limitations of Traditional Approaches
Conventional methods for studying embryogenesis have faced significant technical hurdles. Animal models, while invaluable for establishing fundamental principles, exhibit critical species-specific differences that limit their applicability to human development. For instance, studies in Drosophila have revealed evolutionary conservation in anterior-posterior patterning genes, but the regulatory DNA sequences controlling these genes have diverged over 40 million years of evolution [50]. Similarly, research in frog and chick embryos suggested that biochemical signaling alone was insufficient to explain gastrulation, hinting at the involvement of physical forces that were difficult to measure with traditional techniques [2].
The inaccessibility of human embryos during implantation has represented perhaps the most significant technical barrier. As gastrulation occurs after the embryo has embedded in the uterine wall, direct observation has been impossible, forcing researchers to rely on static histological sections or inference from model organisms. This limitation has obscured the dynamic, coordinated cellular movements and signaling events that characterize this critical developmental window.
Optogenetic Control of Development
Recent breakthroughs in optogenetic tools have revolutionized our ability to study gastrulation with unprecedented precision. Researchers have developed a light-inducible system that allows remote control of key developmental genes in human embryonic stem cells. By inserting the human BMP4 coding sequence downstream of a loxP-flanked stop cassette in a piggyBac vector, scientists created cells that express this critical morphogen in response to specific light wavelengths [16].
This optogenetic approach enables spatiotemporal precision in activating developmental pathways, allowing researchers to test how tissue geometry and mechanical stress influence embryonic patterning. When BMP4 signaling was triggered in unconfined, low-tension environments, gastrulation failed to fully progress, demonstrating that morphogens alone are insufficient to drive this process [2]. Only when mechanical tension was applied through confinement or tension-inducing hydrogels did the complete sequence of germ layer formation occur.
Mechanical Force Integration
The role of physical forces in gastrulation has emerged as a crucial factor that was previously underappreciated. Research has revealed that mechanical tension, mediated through the mechanosensory protein YAP1, fine-tunes downstream biochemical signaling pathways including WNT and Nodal, which instruct cells on their developmental fates [2]. Nuclear YAP1 appears to act as a molecular brake on gastrulation, preventing these transformations from occurring prematurely [2].
These findings suggest that gastrulation requires both chemical competence and physical priming – cells must be both biochemically prepared and mechanically positioned to undergo proper differentiation. The concept of mechanical competence represents a new framework for understanding how embryos progress through developmental milestones, potentially involving a "mechanical organizer" that complements classical signaling centers [2].
Signaling Pathways in Gastrulation
Core Signaling Networks
Multiple evolutionarily conserved signaling pathways coordinate the complex process of gastrulation. The BMP4 pathway serves as a primary initiator, activating downstream cascades that establish the fundamental body plan. Following BMP4 activation, WNT and NODAL signaling pathways are induced, controlling the onset of gastrulation both in animal models and in human embryonic stem cells [16]. These pathways interact in a precise spatiotemporal sequence to guide symmetry breaking and germ layer specification.
The Hippo signaling pathway represents another crucial regulator, particularly through its effectors YAP and TAZ. This pathway is highly conserved and centers on a serine/threonine kinase core that negatively regulates YAP/TAZ activity. During development, Hippo signaling helps determine cell fate decisions between the inner cell mass and trophectoderm, with implications for later embryonic patterning [43].
Pathway Crosstalk and Integration
The interplay between these signaling pathways creates a robust regulatory network that guides gastrulation. Research using synthetic embryo models has demonstrated that mechanical signals are crucial regulators of the BMP4 signaling cascade, which in turn regulates WNT and YAP activity, shaping the fate patterns of gastrulation [16]. This crosstalk ensures that developmental transitions occur at the appropriate time and location, integrating both biochemical and biophysical cues.
Figure 1: Signaling Pathway Crosstalk in Gastrulation. Mechanical forces and biochemical signals integrate to regulate germ layer formation.
Experimental Protocols and Workflows
Optogenetic Gastrulation Assay
The development of optogenetic tools has enabled precise experimental control over gastrulation events. The following protocol outlines the key steps for establishing a light-inducible system to study symmetry breaking:
Cell Line Preparation
- Engineer human embryonic stem cells (RUES2 NIH hESC-09-0013) to contain the human BMP4 coding sequence downstream of a loxP-flanked stop cassette in a piggyBac vector [16].
- Introduce a light-sensitive element that enables controlled gene expression via light-induced loxP recombination.
- Treat cells with DOX to confer light sensitivity by inducing Cre-ER expression.
Experimental Setup
- Culture optogenetic-programmed hESCs under defined conditions in geometrical confinement on micropatterns.
- Activate BMP4 signaling using specific wavelengths of light with precise spatial and temporal control.
- Apply mechanical confinement through micropatterned substrates or tension-inducing hydrogels to specific cell regions.
- Fix cells at predetermined time points for immunostaining and RNA sequencing analysis.
Analysis Methods
- Perform immunofluorescence staining for key developmental markers including BRA (mesoderm), GATA6 (primitive endoderm), SOX17 (definitive endoderm), and phospho-SMAD1/5 (BMP signaling activity) [16].
- Process z-stack images using specialized software to segment individual nuclei and generate pointcloud files containing 3D coordinates and fluorescence levels.
- Use mathematical modeling to simulate biochemical signal movement and interaction with physical forces.
- Compare experimental results with computational predictions to validate hypotheses.
Quantitative Analysis of Gene Expression
Advanced imaging and computational methods enable precise quantification of gene expression patterns during gastrulation:
Embryo Processing
- Collect and fix embryos at specific developmental stages using formaldehyde-based fixation protocols.
- Perform in situ hybridization with species-specific RNA probes for key patterning genes.
- Use sequential detection with horseradish-peroxidase conjugated antibodies and fluorescent tyramide amplification.
- Stain nuclei with Sytox Green to enable cellular resolution analysis.
Image Analysis Pipeline
- Acquire z-stacks of embryos using confocal microscopy with plan-apochromat objectives.
- Process images using specialized software to unmix channels and segment individual nuclei.
- Generate pointcloud files containing 3D coordinates and fluorescence levels for each nucleus.
- Create morphological models that contain average nuclear positions and expression patterns.
- Align individual embryos to templates using rigid-body transformation and non-rigid warping.
- Compute expression values by averaging measurements across corresponding nuclei after spatial registration [50].
Figure 2: Experimental Workflow for Gastrulation Research. The integrated approach combines stem cell biology, optogenetics, and computational modeling.
Research Reagent Solutions
Table 2: Essential Research Reagents for Gastrulation Studies
| Reagent/Cell Line | Function | Application in Gastrulation Research |
|---|---|---|
| RUES2 hESCs | Human embryonic stem cell line | Base cell line for optogenetic engineering and differentiation studies [16] |
| Light-Inducible BMP4 System | Optogenetic tool for spatial control of BMP4 | Enables precise activation of gastrulation initiation signals [2] [16] |
| Anti-BRA Antibody | Mesoderm marker identification | Detects emergence of mesodermal lineages during symmetry breaking [16] |
| Anti-phospho-SMAD1/5 | BMP pathway activity reporter | Measures activation of BMP signaling cascade in response to induction [16] |
| TRULI (2.5 μM) | Hippo pathway inhibitor | Modulates YAP/TAZ activity to test mechanical force hypotheses [43] |
| SB431542 (10 μM) | TGF-β/Activin/Nodal inhibitor | Tests requirement of Nodal signaling in germ layer specification [43] |
| Micropatterned Substrates | Geometric confinement system | Applies controlled mechanical tension to specific cell regions [2] |
Quantitative Data and Mathematical Modeling
Key Experimental Findings
Table 3: Quantitative Effects of Pathway Modulation on Blastocyst Development
| Treatment | Target Pathway | Concentration | Blastocyst Development Rate (Control) | ICM Marker | TE Marker |
|---|---|---|---|---|---|
| CRT0276121 | Hippo activator | 1.5 μM | 25% (83%) | → | ↓ [43] |
| TRULI | Hippo inhibitor | 2.5 μM | 100% (100%) | ↑ | ↓ [43] |
| 1-Azakenpaullone | Wnt/β-catenin activator | 20 μM | 70% (86%) | → | ↓ [43] |
| Cardamonin | Wnt/β-catenin inhibitor | 20 μM | 46% (75%) | → | ↓ [43] |
| BMP4 | BMP activator | 100 ng/mL | 17.4% (61.5%) | → | → [43] |
Mathematical modeling has become an indispensable tool for understanding gastrulation dynamics. Researchers have developed "digital twin" embryo simulations that show how biochemical signals like BMP4, WNT, and NODAL move through tissues and interact with physical forces [2]. These computational models, built using Python and Jupyter notebooks, incorporate actual measurements of mechanical tension to predict how signaling patterns and tissue organization lead to specific cell layers [16].
The simulations closely match experimental observations, demonstrating that both biochemical signals and mechanical tension must work together for proper embryological signaling cascade self-organization. This integrated approach provides a quantitative framework for understanding how the embryo changes during early development, enabling researchers to test hypotheses in silico before conducting wet-lab experiments.
Future Directions and Clinical Applications
Emerging Research Frontiers
The field of gastrulation research is rapidly evolving, with several promising directions emerging. The concept of a mechanical organizer – a force-based counterpart to classical signaling centers – represents a provocative idea that could prove transformative for understanding embryonic patterning [2]. This hypothetical structure would complement known biochemical organizers by satisfying specific physical conditions required for developmental progression.
Advanced microfluidic systems are being developed to create more physiologically relevant culture environments for embryonic development studies. These platforms enable precise control over biochemical and mechanical cues, allowing researchers to recreate complex in vivo conditions in vitro. The integration of multi-omics approaches – including single-cell transcriptomics, proteomics, and metabolomics – promises to unveil unprecedented detail about the molecular changes occurring during gastrulation.
Clinical Implications
Understanding gastrulation has profound implications for regenerative medicine and reproductive health. Insights from basic research are already informing strategies to refine stem-cell therapies that could be activated on demand to replace damaged tissues or organs [2]. Additionally, this work illuminates why early pregnancies sometimes fail, potentially leading to interventions that could support successful gestation.
As one researcher noted: "When we improve our understanding of the underlying rules of embryogenesis, we can use that information to give people the best opportunities for building future families" [2]. The potential to enhance assisted reproductive technology outcomes represents a particularly promising clinical application, with blastocyst quality being a key factor affecting implantation and clinical pregnancy rates [43].
The study of human gastrulation has transitioned from an inaccessible biological mystery to a tractable research frontier through the strategic development of ethical model systems and advanced technologies. The integration of optogenetic control, mechanical force measurements, and computational modeling has revealed the profound interdependence between biochemical signaling and physical forces during this critical developmental window. These advances have not only addressed longstanding ethical and technical barriers but have also fundamentally transformed our understanding of how human life emerges from a seemingly uniform cellular sheet.
As research continues to unravel the complex signaling dynamics of gastrulation, the potential applications in regenerative medicine and fertility treatments grow increasingly tangible. The ongoing refinement of synthetic embryo models, combined with increasingly sophisticated analytical approaches, promises to further illuminate the molecular and mechanical conversations that guide the formation of human body plan – conversations that represent our own developmental origins.
The process of gastrulation represents a pivotal milestone in early embryogenesis, where a homogeneous sheet of embryonic cells undergoes symmetry breaking to establish the foundational body axes and the primordia of all future organs [16]. This complex transformation is orchestrated by an evolutionarily conserved set of signaling pathways—including BMP, Nodal, Wnt, FGF, and others—that balance tissue growth, differentiation, and morphogenesis through precise spatiotemporal dynamics [51]. Despite their fundamental importance, distinguishing the specific contributions of individual signaling pathways has presented a persistent challenge to developmental biologists, as reduction or abolition of their activity often leads to overlapping phenotypic defects that are difficult to disentangle using conventional approaches [51].
The core of this challenge lies in the inherent complexity of developmental systems. For instance, both Nodal and Sonic hedgehog (Shh) mutants can exhibit cyclopia, though the underlying mechanisms differ substantially—Nodal mutants display this defect due to an early lack of mesoderm, while Shh mutants develop cyclopia from later midline patterning defects [51]. Similarly, while misregulation of BMP, Wnt, RA, FGF, and PCP signaling pathways leads to specific defects, all these mutants also share common malformations such as shortened tails [51]. These overlapping manifestations complicate the linkage of phenotypic observations to specific signaling mechanisms, necessitating advanced approaches that can decode phenotypic complexity with greater precision and temporal resolution than human observation alone can provide.
Within the context of gastrulation research, recent advances have revealed that mechanical forces and tissue geometry play previously underappreciated roles in modulating signaling pathway activity [16] [2]. The emerging paradigm suggests that biochemical signals alone are insufficient to drive proper gastrulation; instead, cells must be both chemically prepared and physically primed through the integration of morphogen signaling and mechanical tension [2]. This intersection of physical and biochemical signaling further compounds the challenge of phenotypic interpretation, demanding new methodological frameworks capable of dissecting these multifaceted interactions.
The Nature of the Problem: Overlapping Phenotypes in Signaling Mutants
Characteristic Phenotypic Overlaps in Early Development
The challenge of distinguishing signaling mutants manifests most prominently in the recurrent phenotypic motifs observed across different pathway perturbations. Research has systematically documented that multiple pathway disruptions converge on similar morphological outcomes, creating a classification problem that often confounds even experienced developmental biologists [51]. For example, in zebrafish models, which serve as a fundamental system for vertebrate developmental studies, loss-of-function phenotypes for seven major signaling pathways (BMP, RA, Wnt, FGF, Nodal, Shh, and PCP) demonstrate significant overlap in their external morphological presentations [51].
The tail defect paradigm illustrates this challenge particularly well. While each signaling pathway controls distinct aspects of axis patterning and tissue specification, disruption of any of five different pathways (BMP, Wnt, RA, FGF, and PCP) results in malformed, shortened tails [51]. This common phenotypic outcome emerges despite the fact that each pathway operates through different molecular mechanisms and acts at slightly different developmental timepoints. The convergence of these diverse signaling disruptions on similar morphological outcomes suggests the existence of bottleneck processes in development that are particularly vulnerable to perturbation across multiple signaling axes.
Limitations of Traditional Phenotypic Analysis
Conventional approaches to phenotypic analysis in developmental biology have primarily relied on manual assessment by trained experts who identify defects based on morphological landmarks and temporal progression. While this approach has yielded fundamental insights, it faces inherent limitations in resolving complex phenotypic overlaps. Systematic evaluations have demonstrated that non-expert assessors correctly identify signaling defects with only approximately 53% accuracy, while even experts achieve approximately 79% accuracy under controlled conditions [51].
The temporal dimension of phenotypic emergence presents particular challenges for traditional analysis. Signaling pathway disruptions often initiate molecular changes long before morphological manifestations become apparent to human observers. Additionally, many critical phenotypic transitions occur during narrow developmental windows that are easily missed without continuous monitoring. The subjective nature of human assessment also introduces consistency challenges, particularly when classifying phenotypes that fall along a spectrum rather than into discrete categories.
Advanced Approaches for Phenotype Resolution
Deep Learning-Based Phenotyping Systems
EmbryoNet represents a transformative approach to phenotypic classification, employing a deep convolutional neural network (CNN) specifically trained to identify and distinguish signaling defects in early development [51]. This system addresses the limitations of human assessment through automated, high-throughput phenotyping that can detect subtle morphological patterns invisible to the human eye. The architecture incorporates a modified ResNet18 CNN that includes timestamps of developmental images, enabling the model to interpret phenotypes within their proper developmental context [51].
The performance advantages of EmbryoNet are substantial. In controlled comparisons, the system classified embryonic phenotypes with 91% accuracy, significantly outperforming both non-expert teams (53% accuracy) and expert developmental biologists (79% accuracy) [51]. Perhaps more importantly, EmbryoNet detected phenotypic deviations much earlier in development than human assessors could identify them, providing a crucial temporal advantage for experimental interventions. The system's classification capabilities extend across the seven major vertebrate signaling pathways, successfully distinguishing between mutants with overlapping morphological presentations such as Nodal and Shh pathway disruptions [51].
Integrating Mechanical and Biochemical Signaling Data
Recent research has revealed that mechanical forces play an essential role in modulating signaling pathway activity during gastrulation, suggesting that comprehensive phenotypic analysis must account for both biochemical and biophysical parameters [16] [2]. Studies using optogenetic tools to activate BMP4 with spatiotemporal precision demonstrated that biochemical signaling alone is insufficient to initiate gastrulation; proper transformation requires the additional presence of specific mechanical conditions at the cellular level [2].
The molecular integration of mechanical and biochemical signaling occurs through mechanosensory proteins such as YAP1, which fine-tunes downstream biochemical signaling pathways mediated by WNT and Nodal [2]. Nuclear YAP1 appears to act as a molecular brake on gastrulation, preventing these transformations from occurring prematurely until appropriate mechanical conditions are met [2]. This interdependence creates a signaling cascade where cells must be both chemically prepared and physically primed to undergo developmental transitions, explaining why traditional approaches focusing solely on biochemical signaling have often failed to resolve phenotypic complexity.
Table 1: Quantitative Performance Comparison of Phenotyping Methods
| Method | Accuracy | Processing Speed | Early Detection Capability | Multi-Pathway Discrimination |
|---|---|---|---|---|
| Non-expert human assessment | 53% | Minutes to hours | Limited | Moderate |
| Expert developmental biologist | 79% | Minutes to hours | Moderate | Good |
| EmbryoNet deep learning | 91% | Seconds | Excellent | Excellent |
| Integrated biophysical-biochemical analysis | Not quantitatively assessed | Hours to days | Good | Excellent |
Mathematical Modeling of Developmental Dynamics
The creation of "digital twin" embryo models provides a computational framework for simulating how biochemical signals and mechanical forces interact to shape developmental outcomes [2]. These mathematical models, built using actual measurements of mechanical tension and signaling pathway activity, can predict how signaling patterns and tissue organization lead to specific cell layer formation during gastrulation [2]. The close alignment between these simulations and experimental observations provides strong validation for the integrated role of mechanical and biochemical signaling in development.
These models typically incorporate reaction-diffusion schemes similar to Turing patterns, which simulate how self-organized patterns emerge from initially homogeneous systems [16]. By quantitatively formalizing developmental circuitry, these models can predict experimental outcomes following specific perturbations, such as knockout or overexpression of signaling inhibitors like NOGGIN in the BMP4 pathway [16]. The predictive power of these models makes them invaluable for generating testable hypotheses about phenotypic outcomes under different genetic and environmental conditions.
Experimental Protocols and Methodologies
Optogenetic Control of Signaling Pathways
The precise dissection of signaling dynamics requires tools that can manipulate pathway activity with high spatiotemporal resolution. Optogenetic systems provide this capability by enabling light-activated control of developmental genes. The following protocol describes the establishment of a light-inducible BMP4 system in human embryonic stem cells (hESCs) [16]:
- Genetic Engineering: Insert the human BMP4 coding sequence downstream of a loxP-flanked stop cassette in a piggyBac vector, enabling controlled gene expression via light-induced loxP recombination.
- Light Sensitivity Induction: Treat cells with doxycycline (DOX) to confer light sensitivity by inducing the expression of a light-activated Cre recombinase.
- Spatial Patterning: Expose cells to specific wavelengths of light using digital micromirror devices to create precise patterns of BMP4 activation within cell colonies.
- Mechanical Context Manipulation: Combine light activation with varied mechanical environments, including unconfined low-tension conditions versus confined high-tension environments created using micropatterned substrates or tension-inducing hydrogels.
- Outcome Assessment: Fix cells at specific timepoints and perform immunofluorescence staining for downstream pathway components (phospho-SMAD1/5, phospho-SMAD4) and lineage markers (BRA, GATA6, SOX17) to quantify response to spatially controlled BMP4 activation.
This approach enables researchers to test how tissue geometry and mechanical stress at specific physical locations influence developmental outcomes, revealing the interdependence of biochemical and mechanical signals [16] [2].
High-Content Screening with Automated Phenotype Classification
Large-scale phenotypic analysis requires standardized protocols for image acquisition and processing. The following methodology supports high-throughput screening of signaling defects:
- Sample Preparation: Generate signaling-defective embryos through chemical genetics approaches using specific pathway modulators or via genetic manipulation. For zebrafish studies, treatments include SB-505124 for Nodal inhibition, DMH1 for BMP inhibition, and XAV939 for Wnt inhibition [51].
- Image Acquisition: Capture bright-field movies of embryos in random orientations using high-throughput imaging systems, collecting hundreds of thousands to millions of images across developmental timecourses (e.g., 2-26 hours post-fertilization for zebrafish) [51].
- Data Annotation: Manually annotate subsets of images to create training datasets, assigning phenotype classes based on treatment conditions and developmental timepoints when phenotypes first become apparent.
- Network Training: Train deep convolutional neural networks (CNNs) using annotated datasets, incorporating timestamp information to account for developmental trajectories. Implement transition logic that reflects biological constraints (e.g., early embryos cannot display specific signaling phenotypes, dead embryos cannot revert to alive).
- Phenotype Classification: Apply trained networks to new datasets, using classification algorithms that consider both morphological features and developmental timing to assign phenotype classes with associated confidence scores.
This protocol enabled the creation of EmbryoNet, which was trained on more than 2 million images comprising thousands of trajectories of normally developing and signaling-defective zebrafish embryos [51].
Data Presentation and Visualization Framework
Quantitative Analysis of Signaling Pathway Interactions
Effective resolution of phenotypic complexity requires systematic quantification of how different signaling pathways interact to produce specific morphological outcomes. The table below summarizes characteristic phenotypes associated with disruption of major signaling pathways during early development:
Table 2: Characteristic Phenotypes of Signaling Pathway Disruption in Vertebrate Development
| Signaling Pathway | Primary Phenotypic Defects | Overlapping Phenotypes | Distinguishing Features | Onset Timing |
|---|---|---|---|---|
| BMP | Loss of ventral tissues, expanded dorsal structures | Shortened tail, axis patterning defects | Radialization of embryo, specific changes in dorsal-ventral marker expression | Early gastrulation |
| Nodal | Loss of mesendoderm, absence of anterior structures | Cyclopia, trunk elongation defects | Complete absence of mesoderm derivatives rather than patterning defects | Early gastrulation |
| Wnt | Anteriorization, loss of posterior structures | Shortened tail, axis patterning defects | Specific expansion of anterior neural tissues, distinct gene expression patterns | Mid-gastrulation |
| FGF | Impaired migration, convergence-extension defects | Shortened tail, mesoderm patterning defects | Specific disruption of cell movements without initial patterning defects | Mid-gastrulation |
| Shh | Loss of ventral neural tube, midline defects | Cyclopia, neural patterning defects | Late midline defects after initial mesoderm formation, distinct from Nodal | Late gastrulation |
| PCP | Impaired convergence-extension movements | Shortened tail, neural tube closure defects | Specific disruption of polarized cell behaviors without altered tissue specification | Late gastrulation |
Visualization of Signaling and Mechanical Integration
The integration of mechanical and biochemical signaling can be visualized through pathway diagrams that highlight key interaction points. The following Graphviz diagram illustrates the core signaling cascade involved in symmetry breaking during gastrulation:
Diagram 1: Signaling integration in gastrulation.
The experimental workflow for optogenetic analysis of signaling mechanics can be visualized as follows:
Diagram 2: Experimental workflow for signaling analysis.
Critical Reagents and Experimental Systems
Table 3: Essential Research Reagents for Signaling Phenotype Resolution
| Resource | Type | Function/Application | Key Features |
|---|---|---|---|
| Light-inducible BMP4 hESCs | Engineered cell line | Optogenetic control of BMP4 signaling | Enables spatiotemporal precision in pathway activation; reveals mechanical context dependence [16] |
| EmbryoNet | Deep learning classifier | Automated phenotyping of signaling defects | 91% classification accuracy; early detection capability; processes millions of images [51] |
| Micropatterned substrates | Engineering platform | Control of geometrical confinement | Standardized mechanical environments; reveals role of tissue architecture in signaling [16] |
| YAP/TAZ inhibitors | Chemical probes | Mechanosensing pathway inhibition | Dissects mechanical contribution to signaling; identifies mechanical competence requirements [2] |
| Digital embryo models | Mathematical framework | Simulation of signaling mechanics integration | Predictive power for phenotypic outcomes; "digital twin" experimentation [2] |
| Signaling pathway modulators | Chemical genetics toolkit | Specific pathway activation/inhibition | BMP4 (activation); CHIR99021 (Wnt activation); SB-505124 (Nodal inhibition) [51] [52] |
The resolution of phenotypic complexity in signaling mutants requires a multidimensional approach that integrates biochemical, mechanical, and computational methodologies. The traditional focus on individual signaling pathways in isolation has given way to a more sophisticated understanding of how mechanical forces shape cellular responses to morphogens, creating interdependent signaling circuits that ensure robust developmental outcomes [16] [2]. This integrated perspective not only explains previous challenges in distinguishing overlapping phenotypes but also provides a roadmap for future research aimed at decoding the complex language of embryonic development.
The methodological advances described in this work—particularly deep learning-based phenotyping and optogenetic pathway control—represent transformative tools for developmental biology and drug discovery. By enabling researchers to move beyond subjective phenotypic classification to quantitative, high-resolution analysis of developmental trajectories, these approaches accelerate both basic research into signaling mechanisms and applied screening for teratogenic compounds [51]. As these methodologies continue to evolve, they promise to further illuminate the exquisite precision with which signaling dynamics orchestrate the emergence of form and function in the developing embryo.
Gastrulation is a fundamental process in early embryonic development where a pluripotent cell mass transforms into the three primary germ layers—ectoderm, mesoderm, and endoderm—that give rise to all fetal tissues. This process is governed by precise spatiotemporal signaling dynamics involving a complex interplay of biochemical and mechanical cues. In recent years, stem cell researchers have worked to recapitulate these developmental events in vitro to generate specific cell types for regenerative medicine, disease modeling, and drug development. The efficiency and specificity of germ layer induction remain significant challenges, requiring optimized protocols that mimic embryonic signaling environments. This technical guide synthesizes current advances in germ layer induction protocols, focusing on strategies to enhance efficiency, speed, and specificity through controlled manipulation of key developmental pathways including BMP, WNT, Nodal, and FGF/ERK signaling.
The following table summarizes key quantitative data from recent studies demonstrating efficient germ layer induction from human pluripotent stem cells (hPSCs). These protocols achieve high efficiency through precise modulation of developmental signaling pathways.
Table 1: Quantitative Efficiency of Recent Germ Layer Induction Protocols
| Germ Layer | Induction Strategy | Time Frame | Efficiency | Key Signaling Pathways Modulated | Reference |
|---|---|---|---|---|---|
| Definitive Endoderm (DE) | Chemically defined, small-molecule system | Multiple days | High (Protocol optimized for cost-effectiveness) | WNT, TGF-β [53] | |
| Extraembryonic Mesoderm (ExM) | BMP4 + CHIR99021 (CB) on naive hESCs | 4-5 days | ~90% (GATA6+/SNAIL+ cells) | BMP, WNT, Nodal [52] | |
| ExM from Primed hESCs | WNT activation | 4-5 days | High efficiency | WNT [52] | |
| Mesoderm & Endoderm | BMP4 + Mechanical confinement | Variable | Required for specification | BMP, WNT, Nodal + Mechanical tension [2] |
Experimental Protocols for Germ Layer Induction
Definitive Endoderm Differentiation from hPSCs
This protocol describes a recombinant protein-free, chemically defined system for generating definitive endoderm from human pluripotent stem cells [53].
Materials and Equipment
- Cell Lines: H1 or H9 hESCs (WiCell); human induced pluripotent stem cells (hiPSCs)
- Basal Medium: DMEM/F12
- Maintenance Medium: TeSR-E8 kit (STEMCELL Technologies #05990)
- Small Molecules: CHIR99021 (10 mM, Selleck #S2924), Y-27632 2HCI (Selleck #S1049)
- Passaging Reagent: Accutase (STEMCELL Technologies #07920)
- Substrate Options: Matrigel (BD Biosciences #354277), Vitronectin (Gibco #CTS279S3), or Synthemax II-SC (Corning #3535)
- Supplements: Vitamin C (Sigma #A8960)
- Equipment: Humidified incubator (37°C, 5% CO2), Zeiss LSM780 confocal microscope, CytoFLEX-S flow cytometer (Beckman Coulter)
Step-by-Step Protocol
hPSC Culture and Passaging:
- Maintain hPSCs in TeSR-E8 medium on Matrigel-coated plates
- For passaging, use Accutase dissociation
- Include 10 μM Y-27632 (ROCK inhibitor) for the first 24 hours after passaging to enhance survival
Matrigel Coating Preparation:
- Thaw Matrigel overnight on ice at 2°C to 8°C
- Dilute one 200 μL aliquot with 20 mL ice-cold DMEM/F12 medium
- Coat culture vessels with diluted Matrigel (1-2 hours at room temperature or overnight at 4°C)
- Pre-chill all pipette tips and labware that will contact Matrigel
Definitive Endoderm Induction:
- Prepare DE induction medium: DMEM/F12 supplemented with 3 μM CHIR99021 and 71 μg/mL Vitamin C
- Filter-sterilize through 0.22 μm filter before use
- Plate hPSCs at appropriate density and culture in DE induction medium
- Change medium daily for the duration of differentiation
Validation of DE Differentiation:
- Immunofluorescence Staining: Fix cells with 4% PFA and stain for DE markers including FOXA2 (1:200), SOX17 (1:200), and GATA4 (1:400)
- Flow Cytometry: Analyze using antibodies against CD184 (CXCR4)-APC (1:100 dilution)
- Imaging and Analysis: Use confocal microscopy (e.g., Zeiss LSM780) and analyze with ImageJ or ZEN2.3 software
Rapid Extraembryonic Mesoderm Specification
This protocol enables highly efficient ExM differentiation from both naive and primed hPSCs within 4-5 days [52].
Materials
- Cell Lines: Naive hESCs (AIC-N lines) or primed hPSCs
- Medium: FH-N2B27 (modified N2B27 with FGF4 and heparin)
- Induction Factors: CHIR99021 (GSK3 inhibitor), BMP4
- Coating: Matrigel-coated dishes
Step-by-Step Protocol
Cell Preparation:
- Dissociate naive or primed hPSCs using appropriate enzyme
- Inoculate onto Matrigel-coated dishes in FH-N2B27 medium
ExM Induction:
- Supplement FH-N2B27 medium with CHIR99021 and BMP4 (CB treatment)
- Culture for 4-5 days with daily medium changes
- Monitor morphological change from colony to mesenchymal phenotype
Validation and Expansion:
- Immunofluorescence: Stain for ExM markers (GATA6, SNAIL, VIM, KDR, FLT1)
- Flow Cytometry: Quantify GATA6+/SNAIL+ population (typically >90%)
- scRNA-seq: For comprehensive characterization, perform single-cell RNA sequencing using 10X Genomics platform
Signaling Pathways in Germ Layer Specification
Biochemical Signaling Networks
Germ layer specification is controlled by conserved developmental signaling pathways that act in concert to direct cell fate decisions. The following diagram illustrates the key pathways and their interactions in germ layer patterning:
Diagram 1: Signaling pathways in germ layer specification.
Integration of Mechanical and Biochemical Signaling
Recent research has revealed that mechanical forces play an essential role in conjunction with biochemical signals during germ layer specification. Studies using optogenetic tools to activate BMP4 signaling demonstrated that chemical cues alone are insufficient to initiate proper gastrulation; correct mechanical conditions are equally critical [2]. The mechanosensory protein YAP1 acts as a molecular brake on gastrulation, preventing these transformations from occurring prematurely. Gastrulation proceeds only when molecular signals and mechanical tension align, indicating cells must be both chemically prepared and physically primed for differentiation.
The Scientist's Toolkit: Essential Research Reagents
The following table compiles key reagents and their applications in germ layer induction protocols, serving as a quick reference for researchers designing differentiation experiments.
Table 2: Essential Research Reagents for Germ Layer Induction Studies
| Reagent Category | Specific Examples | Function in Differentiation | Application Notes |
|---|---|---|---|
| Small Molecule Inhibitors/Activators | CHIR99021 (GSK3 inhibitor) | Activates WNT signaling, promotes mesendodermal fates | Use at 3 μM for definitive endoderm induction [53] |
| LDN193189 (BMP inhibitor) | Inhibits BMP signaling, promotes neural ectoderm | Useful for neuroectoderm specification | |
| Y-27632 (ROCK inhibitor) | Enhances cell survival after passaging | Include first 24h after cell dissociation [53] | |
| Recombinant Proteins | BMP4 | Induces mesodermal and extraembryonic lineages | Combined with CHIR for ExM specification [52] |
| Extracellular Matrix Substrates | Matrigel | Provides basement membrane for cell attachment | Handle on ice to prevent premature gelling [53] |
| Vitronectin | Defined substrate for pluripotent stem cell culture | Alternative to Matrigel for xeno-free conditions [53] | |
| Synthemax | Synthetic peptide substrate for defined culture | Reconstitute at 1 mg/mL, dilute to 0.025 mg/mL for use [53] | |
| Cell Surface Markers for Validation | CD184 (CXCR4)-APC | Definitive endoderm marker | Use flow cytometry for quantification [53] |
| GATA6, SNAIL | Extraembryonic mesoderm markers | >90% positive cells indicates efficient ExM induction [52] | |
| FOXA2, SOX17 | Definitive endoderm markers | Use immunofluorescence for spatial distribution [53] |
The pursuit of efficient germ layer induction protocols requires careful consideration of both biochemical and biophysical cues that guide developmental processes. The protocols outlined in this guide demonstrate that rapid and specific differentiation can be achieved through precise manipulation of signaling pathways, with efficiency metrics exceeding 90% in optimized systems. As research continues to unravel the complexities of gastrulation, particularly the emerging role of mechanical forces and epigenetic pre-patterning, differentiation protocols will become increasingly refined. These advances will enhance our ability to generate specific cell types for therapeutic applications and improve the physiological relevance of stem cell-based disease models. The integration of engineering approaches with developmental biology principles will continue to drive progress in controlling cell fate decisions with spatiotemporal precision.
A fundamental question in developmental biology is how a robust and invariable developmental phenotype emerges despite fluctuations in environmental conditions such as nutrient availability. Central to this process is the phenomenon of epigenetic priming, wherein conserved transcription factors initiate chromatin remodeling events hours or even days before the major wave of transcriptional activation of their target genes. This preparatory mechanism establishes a permissive chromatin landscape that precisely regulates both the timing and amplitude of subsequent gene expression, thereby canalizing cell-fate specification and ensuring developmental precision [54].
Within the context of gastrulation research—a period of extensive cellular differentiation and morphogenesis—understanding these anticipatory epigenetic mechanisms is particularly crucial. The integration of multi-omics datasets provides the necessary resolution to capture the dynamic interplay between early chromatin reorganization and later gene expression, effectively bridging the temporal gap, or 'time lag', between priming and activation. This technical guide explores the mechanistic basis of this lag and outlines the computational and experimental frameworks required to study it, with a specific focus on applications in gastrulation and early embryonic development.
The Core Mechanism: BLMP-1 as a Paradigm for Transcriptional Priming
Pioneer Factor Activity and Chromatin Decompaction
The conserved transcription factor BLMP-1 (Blimp1) offers a canonical model for understanding epigenetic priming. In C. elegans, BLMP-1 functions as a pioneer factor that initiates chromatin decompaction at its target loci during embryogenesis—hours before major transcriptional activation occurs in post-embryonic development [54]. This primary, mechanistic step involves:
- Anticipatory Chromatin Remodeling: BLMP-1 binds its target loci and facilitates a more open chromatin configuration, characterized by reduced compaction.
- Regulation of Transcriptional Output: This initial decompaction does not immediately trigger transcription but sets the precise level for both the duration and amplitude of target gene expression that occurs later during larval stages.
- Genetic Separability: The priming function of BLMP-1 is genetically distinct from the mechanisms that determine the precise timing of gene expression induction, indicating a dedicated pathway for setting transcriptional potential.
Integration with Nutrient-Sensing Pathways
A critical feature of the BLMP-1 dependent priming mechanism is its modulation by the environment. The open chromatin state established during embryogenesis is maintained throughout larval development via nutrient-sensing pathways [54]. This integration of nutritional status ensures that the transcriptional output of key developmental genes is calibrated to environmental conditions, providing a mechanistic basis for developmental robustness. This is particularly important for resuming normal temporal patterning after animals exit nutrient-mediated developmental arrests, a process relevant to the metabolic regulation of gastrulation.
Table 1: Key Characteristics of the BLMP-1 Mediated Priming Mechanism
| Feature | Description | Functional Consequence |
|---|---|---|
| Developmental Timing | Chromatin decompaction begins in embryogenesis | Prepares for gene expression in post-embryonic larval stages [54] |
| Molecular Function | Acts as a pioneer factor to decompact chromatin | Establishes a permissive chromatin landscape for activation [54] |
| Environmental Input | Maintained by nutrient-sensing signals | Integrates transcriptional output with environmental conditions [54] |
| Phenotypic Role | Canalizes cell-fate specification, animal size, and molting | Ensures developmental robustness and precision [54] |
Computational Frameworks for Multi-Omics Integration
To capture the relationship between early chromatin changes and later gene expression, researchers must integrate data from multiple omics layers, such as genome conformation (Hi-C), chromatin accessibility (ATAC-seq), and gene expression (RNA-seq). The choice of integration method depends on the data structure and the biological question.
Categorizing Data Integration Strategies
Single-cell multimodal omics data can be integrated through several prototypical strategies, each suited to different experimental designs [55]:
- Vertical Integration: Combines different molecular modalities (e.g., RNA, ATAC, ADT) profiled from the same single cell.
- Diagonal Integration: Aims to integrate datasets across different modalities and different batches or conditions.
- Mosaic Integration: Integrates data from different molecular assays applied to different samples from the same biological system.
- Cross Integration: Involves transferring information or labels across different modalities measured in different cells.
For studying temporal dynamics like epigenetic priming, vertical integration is often the starting point, as it directly links the chromatin state of a cell with its transcriptional output.
Benchmarking Integration Methods for Key Tasks
A comprehensive benchmark of 40 integration methods across 64 real and 22 simulated datasets provides guidance for selecting the right tool [55]. Performance is highly dependent on the data modalities and the specific analytical task.
Table 2: Top-Performing Multi-Omics Integration Methods for Key Tasks
| Integration Category | Key Task | High-Performing Methods | Notable Strengths |
|---|---|---|---|
| Vertical (RNA+ADT) | Dimension Reduction & Clustering | Seurat WNN, sciPENN, Multigrate | Effectively preserves biological variation of cell types [55] |
| Vertical (RNA+ATAC) | Dimension Reduction & Clustering | Seurat WNN, Multigrate, UnitedNet | Robust performance across diverse datasets [55] |
| Vertical | Feature Selection | Matilda, scMoMaT, MOFA+ | Identifies cell-type-specific molecular markers (Matilda, scMoMaT) or a reproducible set of markers (MOFA+) [55] |
| Hi-C & RNA-seq | Network Analysis | HiEdge, HiC-Pro, hicCorrelate | Identifies statistically significant chromatin interactions; enables integration with transcriptional networks [56] |
A Network-Based Approach to Hi-C and RNA-seq Integration
A powerful method for unraveling the functional consequences of 3D genome architecture involves representing Hi-C data as a network. This approach was successfully applied to study chromatin interactions in Triple-Negative Breast Cancer (TNBC), revealing distinct interaction patterns compared to healthy tissue [56]. The workflow can be adapted for developmental systems like gastrulation:
- Process Hi-C Data: Use
HiC-Proto align raw sequencing reads, filter for valid interactions, and construct raw contact matrices [56]. - Normalize Matrices: Apply Iterative Correction and Eigenvector decomposition (ICE) to correct for technical biases (e.g., GC content, mappability) [56].
- Identify Significant Interactions: Use a statistical model like HiEdge to distinguish biologically meaningful chromatin interactions from random background contacts. HiEdge uses a non-central hypergeometric distribution to account for the distance-dependent decay of interaction frequency [56].
- Construct Chromatin Networks: Represent significant interactions as a graph where nodes are genomic bins and edges are significant chromatin interactions [56].
- Integrate with Transcriptional Data: Overlay gene expression data from RNA-seq onto the chromatin network to identify regulatory relationships, such as enhancer-promoter contacts that may be established during priming and activated later.
Network Integration of Hi-C and RNA-seq Data
Experimental Models & Signaling Pathways in Gastrulation
hESC Models for Studying Extraembryonic Mesoderm Specification
Human embryonic stem cell (hESC)-based models are powerful tools for investigating signaling dynamics during early developmental events analogous to gastrulation. Recent research has established that modulating BMP, WNT, and Nodal signaling pathways can rapidly (within 4-5 days) and efficiently (∼90%) induce both naive and primed hESCs into extraembryonic mesoderm (ExM)-like cells [3]. This specification process predominantly transits through a primitive streak-like intermediate (PSLI), a key developmental stage during gastrulation [3].
A critical finding is that the initial pluripotent state of the hESCs (naive vs. primed) influences the subsequent differentiation by affecting signal response, cellular composition, and developmental progression of the resulting ExM-like cells [3]. This underscores the importance of the pre-existing epigenetic and transcriptional landscape—a form of priming—in shaping developmental outcomes.
Visualizing the Core Signaling Pathway
The following diagram illustrates the core signaling pathway that guides the differentiation of hESCs through a primitive streak-like intermediate toward extraembryonic mesoderm, as informed by current research [3].
Signaling in ExM Specification
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagent Solutions for Multi-Omics Gastrulation Research
| Reagent / Tool | Function | Application in Priming Studies |
|---|---|---|
| CHIR99021 | GSK-3 inhibitor, activates WNT signaling | Directs hESC differentiation through primitive streak-like intermediate toward ExM [3] |
| BMP4 | Recombinant bone morphogenetic protein 4 | Works with CHIR99021 to induce ExM specification from hESCs [3] |
| AIC-N hESCs | Naive human embryonic stem cells | Model for studying pre-gastrulation ExM specification and the influence of pluripotency state [3] |
| HiC-Pro | End-to-end Hi-C data processing pipeline | Processes raw sequencing reads into normalized contact matrices for chromatin network analysis [56] |
| HiEdge | Statistical tool for Hi-C analysis | Identifies significant chromatin interactions while modeling distance-dependent decay, revealing priming-associated loops [56] |
| Seurat WNN | Weighted Nearest Neighbors integration | A top-performing method for vertical integration of RNA and ATAC modalities from single cells [55] |
| Matilda | Multi-omics feature selection method | Identifies cell-type-specific molecular markers from integrated single-cell data [55] |
The 'time lag' between epigenetic priming and gene expression is not a passive interval but an active period of chromatin reorganization that is critical for setting the parameters of subsequent transcriptional activation. As demonstrated in models from C. elegans to human embryonic stem cells, pioneer factors like BLMP-1 and external signals from pathways like WNT and BMP initiate and maintain this primed state. Effectively capturing this dynamic process requires the sophisticated integration of multi-omics datasets using robust computational methods, such as those benchmarked for single-cell data and network-based Hi-C analysis. By applying these technical guidelines, researchers can systematically dissect the spatiotemporal control of gene expression during gastrulation, with profound implications for understanding developmental biology and regenerative medicine.
Validation and Cross-Species Insights: Conserved Mechanisms and Species-Specific Adaptations
The precise orchestration of signaling dynamics during gastrulation is fundamental to establishing the vertebrate body plan. Disruptions in key evolutionarily conserved pathways—such as Nodal, BMP, and Wnt—result in characteristic morphological defects, but their classification by human experts remains challenging, subjective, and time-consuming [51] [57]. EmbryoNet addresses this bottleneck by leveraging a deep convolutional neural network (CNN) to provide automated, unbiased, and quantitative phenotypic classification [51]. This technical guide details the architecture, performance, and application of EmbryoNet, framing it as an essential tool for validating signaling defects within gastrulation research.
EmbryoNet Architecture & Workflow
Core Deep Learning Model
EmbryoNet's core is a modified ResNet18 CNN architecture, specifically adapted to process time-series image data of developing embryos [51]. A key innovation is the incorporation of a timestamp for each input image, enabling the model to learn and account for developmental trajectories and the precise timing of phenotypic emergence [51] [57]. The network was trained on a massive dataset of over 2 million images, comprising thousands of trajectories of both normally developing and signaling-defective zebrafish embryos [51] [57].
Phenotype Classification Logic
To enhance classification robustness, a model transition logic was implemented based on developmental biological principles. This logic assigns costs to state transitions in an individual embryo's track over time. For instance, early embryos are typically classified as "Unknown" before phenotypes manifest, and transitions from "Dead" to "Normal" are considered impossible. The sequence of transitions with the lowest cumulative cost is selected for the final classification, contributing to the model's high accuracy [51].
Table: EmbryoNet Classification Categories
| Category | Description |
|---|---|
| Normal | Wild-type, unperturbed embryonic development |
| -BMP | Bone Morphogenetic Protein pathway loss-of-function |
| +RA | Retinoic Acid pathway gain-of-function |
| -Wnt | Wnt pathway loss-of-function |
| -FGF | Fibroblast Growth Factor pathway loss-of-function |
| -Nodal | Nodal pathway loss-of-function |
| -Shh | Sonic Hedgehog pathway loss-of-function |
| -PCP | Planar Cell Polarity pathway loss-of-function |
| Unknown | Insufficient information for classification (typically early stages) |
| Dead | Embryo has disintegrated or died |
Quantitative Performance & Validation
Benchmarking Against Human Assessors
EmbryoNet's performance was rigorously benchmarked in a blinded classification task against human evaluators with varying levels of expertise [51]. The model significantly outperformed all human groups.
Table: EmbryoNet vs. Human Classification Performance
| Classifier Type | Accuracy | F-score | Classification Speed |
|---|---|---|---|
| EmbryoNet (AI) | 91% | 0.90 | Several hundred images in milliseconds [57] |
| Expert Developmental Biologist | 79% | 0.78 | Several days for hundreds of images [57] |
| Trained Non-Experts (with temporal info) | 54% | 0.52 | Not Specified |
| Trained Non-Experts (without temporal info) | 53% | 0.52 | Not Specified |
| Random Guessing | 9% | 0.09 | Instantaneous |
Experimental Validation of Signaling Defects
Beyond inhibitor studies, EmbryoNet validated its classifications using orthogonal genetic approaches. The network robustly identified phenotypes caused by the injection of mRNAs encoding pathway inhibitors, such as:
- Lefty1 mRNA: A Nodal pathway inhibitor, recapitulating the
-Nodalphenotype [51]. - Chordin mRNA: A BMP pathway inhibitor, recapitulating the
-BMPphenotype [51]. This demonstrated EmbryoNet's ability to correctly link morphological defects to specific signaling pathways, irrespective of how the pathway disruption was induced.
Key Experimental Protocols
Dataset Curation and Preparation
Purpose: To create a high-confidence, annotated dataset for training and validating the EmbryoNet model. Methodology:
- Perturbation: Zebrafish embryos were treated with highly specific small-molecule modulators (e.g., SB-505124 for Nodal inhibition) or mRNA injections to disrupt one of seven major signaling pathways (BMP, RA, Wnt, FGF, Nodal, Shh, PCP) [51].
- Imaging: Bright-field time-lapse movies of developing embryos were acquired in random orientations from 2 to 26 hours post-fertilization (h.p.f.) [51].
- Annotation: Expert curators, aware of the treatments, manually assigned phenotype classes (
-BMP,+RA, etc.,Unknown,Dead) at the developmental timepoint when the phenotype first became apparent for each embryo [51].
High-Throughput Drug Screening Protocol
Purpose: To identify novel teratogenic effects and mechanisms of action of pharmaceutical substances. Methodology:
- Exposure: Embryos are exposed to a library of compounds, such as FDA-approved drugs, at various concentrations.
- Automated Analysis: EmbryoNet automatically analyzes and classifies the resulting phenotypes from time-lapse imaging data.
- Mechanism Elucidation: The specific phenotypic class assigned by EmbryoNet (e.g., a shortened body axis characteristic of
-FGF) is used to infer the signaling pathway affected by the compound. For example, the tool identified that statins impact the FGF signaling pathway [57].
The Scientist's Toolkit: Research Reagent Solutions
Table: Essential Resources for EmbryoNet-Based Research
| Research Reagent / Solution | Function in Experimental Protocol |
|---|---|
| Specific Pathway Modulators (e.g., SB-505124) [51] | Induces precise loss-of-function or gain-of-function phenotypes for specific signaling pathways (BMP, RA, Wnt, etc.) in zebrafish embryos. |
| mRNA for Pathway Inhibitors (e.g., Lefty1, Chordin) [51] | Provides an orthogonal, genetic method to perturb specific pathways (Nodal, BMP) for experimental validation. |
| Zebrafish Embryos | A vertebrate model organism with transparent embryos, ideal for high-throughput imaging and phenotypic analysis [58] [57]. |
| High-Throughput Microscope | Enables automated, large-scale acquisition of bright-field time-lapse movies for hundreds of embryos simultaneously [57]. |
| EmbryoNet Software & GUI | The open-source software provides detection, tracking, and automated classification of embryonic phenotypes [51]. |
Signaling Pathways in Gastrulation & Phenotypic Outcomes
EmbryoNet was specifically designed to classify defects in seven major signaling pathways that orchestrate early vertebrate development, many of which are critically active during gastrulation [51] [57].
Applications in Drug Discovery & Cross-Species Analysis
De-risking Drug Development
EmbryoNet excels in high-throughput drug screening, providing detailed toxicological profiles and identifying teratogenic risks early in the product lifecycle. It can resolve a compound's mechanism of action by linking the induced phenotype to a specific pathway disruption. For instance, it was used to discover that the cholesterol-lowering drug Lovastatin affects embryonic development by disrupting the FGF signaling pathway, a finding with potential relevance beyond development to diseases like cancer [58] [57].
Generalizability Across Biological Systems
A key strength of EmbryoNet is its ability to generalize beyond the zebrafish model on which it was trained. The classification algorithms have been shown to robustly identify signaling defects in evolutionarily distant species, making it a versatile tool for comparative evolutionary developmental biology [51]. Furthermore, the platform is also applicable to the analysis of organoids (miniature, in vitro-grown organ models), supporting the advancement of animal-free research [58] [57].
The transformation of a simple embryonic disc into a complex, multi-layered body plan during gastrulation represents one of the most crucial morphogenetic events in vertebrate development. This process requires the precise coordination of tissue patterning and cell movements, directed by a handful of evolutionarily conserved signaling pathways. Among these, Nodal and Planar Cell Polarity (PCP) signaling have emerged as critical regulators that interface to coordinate gastrulation dynamics across vertebrate species. This technical review examines the conserved and divergent functions of Nodal and PCP signaling in zebrafish and Xenopus, two principal model organisms for vertebrate developmental biology research. We synthesize current understanding of how these pathways interact to control convergent extension (C&E) movements, analyze quantitative cellular behaviors, and provide detailed methodological frameworks for investigating these processes. The insights gained from these comparative analyses have significant implications for understanding congenital birth defects and designing targeted therapeutic interventions in human development and disease.
Gastrulation constitutes a fundamental process in embryonic development where the three primary germ layers—ectoderm, mesoderm, and endoderm—are established and shaped through highly coordinated cellular movements including epiboly, internalization, and C&E [59]. In vertebrate embryos, C&E movements simultaneously narrow tissues mediolaterally (convergence) while elongating them anteroposteriorly (extension), driven primarily by polarized mediolateral intercalation behavior (MIB) where cells insert between their anterior and posterior neighbors [60]. The molecular regulation of these processes involves an intricate interplay between patterning signals that establish embryonic axes and polarity signals that orient cellular behaviors.
The PCP pathway, first discovered in Drosophila, provides a molecular compass that polarizes cells within the tissue plane, enabling directional cell intercalations and migrations [61]. Core PCP components including Van Gogh (Vangl), Frizzled (Fz), Dishevelled (Dvl), and Prickle (Pk) acquire asymmetric subcellular distributions that orient cell polarity [60] [61]. Meanwhile, Nodal, a TGF-β superfamily morphogen, plays indispensable roles in mesendoderm induction and patterning along the anteroposterior axis [62] [63]. Emerging evidence demonstrates that these pathways cooperate extensively during gastrulation, though their functional relationships and conserved mechanisms across species remain active areas of investigation.
This review systematically compares the roles and interactions of Nodal and PCP signaling in zebrafish and Xenopus gastrulation, providing a comprehensive resource for researchers studying vertebrate morphogenesis. We integrate quantitative cellular data, detailed experimental methodologies, and visual signaling pathway maps to facilitate cross-species comparative analyses and experimental design.
Core Signaling Pathways: Molecular Architecture and Conservation
Planar Cell Polarity (PCP) Signaling Framework
The core PCP signaling pathway constitutes an evolutionarily conserved system that coordinates polarized cellular behaviors across tissue planes. Molecular genetic analyses, primarily in Drosophila, have identified six core proteins that form two distinct complexes at opposing cell membranes:
Table 1: Core PCP Pathway Components
| Drosophila | Vertebrates | Protein Type | Function |
|---|---|---|---|
| Fz (Frizzled) | Fz3, Fz2, Fz7, Fz6 | VII-pass transmembrane receptor | Distal complex component; Wnt receptor |
| Stan/Fmi (Starry night/Flamingo) | Celsr1, Celsr2, Celsr3 | VII-pass transmembrane cadherin | Cell adhesion; bridges Fz-Vang complexes |
| Dsh (Dishevelled) | Dvl1, Dvl2, Dvl3 | Cytoplasmic (PDZ, DIX, DEP domains) | Downstream signal transduction |
| Pk (Prickle) | Pk1, Pk2 | Cytoplasmic (PET, LIM domains) | Proximal complex component |
| Vang Gogh/Strabismus | Vangl1, Vangl2 | IV-pass transmembrane receptor | Proximal complex component |
| Dgo (Diego) | Inv (Inversin) | Cytoplasmic (ankyrin repeats) | Stabilizes distal complex |
These components form intercellular complexes where Fz-Dsh-Dgo localize to the distal side of cells while Vang-Pk localize proximally, with Fmi homodimers bridging adjacent cells [61]. This asymmetric distribution creates a molecular polarity that directs cytoskeletal reorganization and polarized protrusive activity.
Figure 1: Core PCP signaling pathway. PCP signals establish asymmetric localization of Fz/Dvl (green) and Vangl/Prickle (red) complexes across cell membranes, with Celsr (blue) facilitating intercellular communication. This molecular asymmetry directs cytoskeletal reorganization and cellular polarization.
Nodal Signaling Cascade
Nodal signaling initiates through ligand-receptor interactions that activate downstream transcriptional effectors. Nodal ligands form heterodimers with Gdf3 (Vg1) and bind a receptor complex comprising Type I (Alk4/Alk7) and Type II (ActRIIA/ActRIIB) serine-threonine kinase receptors with the Tdgf (Oep) co-receptor [60] [62]. This activates Smad2/Smad3 phosphorylation, complex formation with Smad4, and nuclear translocation where they partner with transcription factors like FoxH1 to regulate target gene expression including ndr1, ndr2, sox17, eomes, foxa2, and gata4-6 [62] [63].
Nodal signaling is spatially and temporally regulated by multiple mechanisms. Maternal factors including Hwa (Huluwa) activate β-catenin signaling to initiate dorsal ndr1 expression, while maternal Eomesodermin A (Eomesa) promotes lateroventral ndr1 and ndr2 expression [63]. Nodal also establishes positive feedback loops through autoregulation of its own expression and is antagonized by extracellular inhibitors like Lefty [62] [63].
Figure 2: Nodal signaling pathway. Nodal ligands bind receptor complexes leading to Smad2/3 phosphorylation and complex formation with Smad4. The complex translocates to the nucleus to activate target genes, including those involved in mesendoderm specification and Nodal itself, creating a positive feedback loop.
Comparative Analysis of Nodal and PCP Functions
Gastrulation Movements: Conserved Cellular Behaviors
Zebrafish and Xenopus utilize remarkably similar gastrulation movements despite differences in embryonic architecture. Both require C&E driven by MIB, wherein cells become mediolaterally elongated, form bipolar protrusions, and intercalate between anterior and posterior neighbors [60] [59]. These behaviors narrow tissues mediolaterally while extending them anteroposteriorly, and are similarly disrupted by PCP pathway impairment in both species [60] [64].
Table 2: Quantitative Analysis of Cellular Behaviors During Gastrulation
| Cellular Behavior | Zebrafish Measurement | Xenopus Measurement | PCP Dependency | Nodal Dependency |
|---|---|---|---|---|
| ML Cell Intercalation | Severely reduced in MZoep mutants [60] | Blocked by ErbB inhibition [65] | Essential in both [60] [64] | Required in both [60] [65] |
| Protrusion Orientation | Partially disrupted in MZoep mutants [60] | Not explicitly quantified | Core pathway function [60] | Partial regulation [60] |
| Cell Elongation (ML) | Impaired in Nodal-deficient embryos [60] | Bipolar shape impaired by ErbB inhibition [65] | Core pathway function [61] | Modulated in both [60] [65] |
| Radial Intercalation | Not prominently reported | Required for ectoderm epiboly; disrupted by Vangl2/Pk3 deficiency [64] | Essential [64] | Not explicitly demonstrated |
Beyond C&E, PCP signaling regulates radial intercalation behaviors in Xenopus ectoderm, where Vangl2/Prickle3 complexes concentrate apically in multiciliated cell progenitors and deep layer neural plate cells, enabling their intercalation toward the surface [64]. This expanded role for PCP signaling in radial intercalation may represent a species-specific adaptation or could reflect tissue-specific functions conserved across vertebrates.
Genetic Interactions and Pathway Hierarchy
The functional relationship between Nodal and PCP signaling demonstrates both conserved and species-specific characteristics. In zebrafish, Nodal and PCP signaling cooperate during gastrulation, with Nodal functioning both upstream of and in parallel to PCP [60]. Several lines of evidence support this model:
- PCP signaling remains partially active in Nodal-deficient zebrafish mutants, as demonstrated by anteriorly biased Prickle-GFP localization, albeit with increased non-polarized puncta [60]
- Genetic inhibition of Vangl2 in MZoep (Nodal co-receptor mutant) embryos exacerbates C&E defects [60]
- Nodal can induce PCP-dependent C&E in naïve zebrafish blastoderm explants [60]
In Xenopus, Activin (which signals through Nodal receptors) is sufficient to induce C&E and planar polarity in animal cap explants [60]. Furthermore, Nodal/Activin signaling promotes membrane translocation of Dvl, a core PCP component, suggesting a conserved upstream positioning of Nodal relative to PCP in certain contexts [60].
However, important differences emerge in the specific regulatory mechanisms. In zebrafish, Nodal regulation of C&E occurs through both cell-autonomous and non-autonomous mechanisms, with MZoep-/- cells only partially rescued when transplanted into wild-type hosts [60]. Additionally, Nodal-deficient zebrafish exhibit partially reduced expression of the core PCP component prickle1b, suggesting transcriptional regulation of PCP elements [66].
Experimental Frameworks: Methodological Comparisons
Zebrafish Gastrulation Studies
Embryo Manipulation and Phenotypic Analysis:
- Genetic models: Maternal-zygotic oep (MZoep) mutants lacking Nodal signaling exhibit severe C&E defects with reduced ML cell alignment and protrusive activity [60]. Vangl2 mutants (e.g., trilobite) display characteristic PCP-related phenotypes including shortened axes and disrupted protrusion polarity [60].
- Morpholino knockdown: Antisense morpholino oligonucleotides provide transient gene suppression, such as ErbB4-MO (5'-TTCCCTCCAAAAACTCTGGATCTCC-3') targeting the translational start site of XErbB4 [65].
- Cell transplantation: MZoep-/- cells transplanted into WT hosts to assess autonomy of Nodal signaling; WT donor cells can be labeled with membrane-tethered EGFP or tdTomato using CAAX fusions [60] [65].
- Explant assays: Animal pole blastoderm explants from shield-stage embryos cultured in fibronectin-coated dishes to assess autonomous extension capability; Nodal mRNA injection induces robust, PCP-dependent extension [60].
Quantitative Imaging and Analysis:
- 4D time-lapse microscopy: Tracking of individual cell behaviors using transgenic lines like Tg(myl7:EGFP-CAAX) for myocardial cells or ubiquitous membrane markers [67].
- Protrusion analysis: Quantification of protrusion orientation, persistence, and polarity relative to embryonic axes [60].
- Cell shape quantification: Measurement of mediolateral elongation ratios and orientation angles using segmented cell boundaries [60] [67].
- PCP protein localization: Asymmetric distribution analysis of fluorescently tagged core components (e.g., Prickle-GFP) at anterior vs. posterior cell membranes [60].
Xenopus Gastrulation Studies
Tissue Explant and Manipulation Approaches:
- Animal cap assays: Dissection of animal pole tissue at blastula stages; treatment with Activin or Nodal induces C&E movements and ML cell polarization [60].
- DMZ (dorsal marginal zone) explants: Open-face explants prepared by removing ectoderm to observe mesoderm cell behaviors during C&E [65].
- Head mesoderm migration assays: Anterior DMZ explants cultured on fibronectin-coated dishes to assess directional migration toward animal pole [65].
- Pharmacological inhibition: Small molecule inhibitors including SB431542 for Nodal/Activin/TGF-β receptor inhibition [63].
Cell Behavior Quantification:
- Cell intercalation assays: Dual labeling of adjacent cells with membrane-tethered EGFP and tdTomato to track neighbor exchanges [65].
- Cell adhesion assays: Dissociation of DMZ explants in calcium/magnesium-free buffer followed by reaggregation under shaking conditions [65].
- Membrane protrusion analysis: Time-lapse imaging of cells plated on fibronectin, classifying protrusion types (lamellipodia, filopodia) and dynamics [65].
- Convergent extension measurement: Quantifying length-width ratios of explants over time as readout of C&E efficiency [65].
Figure 3: Experimental workflow for comparative analysis of Nodal and PCP signaling. The schematic outlines key methodological approaches in zebrafish and Xenopus models, from initial experimental design through quantitative analysis of cellular, tissue, and molecular metrics.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Nodal and PCP Signaling Studies
| Reagent | Type | Application | Function/Mechanism |
|---|---|---|---|
| SB431542 | Small molecule inhibitor | Nodal signaling inhibition | TGF-β type I receptor inhibitor; blocks Smad2/3 phosphorylation [63] |
| MZoep mutants | Genetic model | Nodal loss-of-function | Lack Tdgf/Oep co-receptor; defective mesendoderm specification and C&E [60] |
| Vangl2 mutants | Genetic model | PCP loss-of-function | Disrupted core PCP function; defective cell polarity and intercalation [60] [64] |
| Membrane-tethered fluorescent proteins | Reporter constructs | Cell labeling and tracking | EGFP/tdTomato fused to CAAX motif for membrane localization; cell behavior analysis [65] [67] |
| Prickle-GFP | Localization biosensor | PCP signaling readout | Visualizes asymmetric protein distribution at anterior cell membranes [60] |
| ErbB4-MO | Morpholino oligonucleotide | Gene knockdown in Xenopus | Targets ErbB4 translation start site; inhibits ErbB signaling in gastrulation [65] |
| DN-ErbBs | Dominant negative receptors | ErbB pathway inhibition | Truncated receptors lacking intracellular domains; block endogenous signaling [65] |
Discussion: Integrated Signaling Models and Future Directions
The comparative analysis of Nodal and PCP signaling in zebrafish and Xenopus reveals a conserved functional architecture with context-dependent modifications. Both systems demonstrate that Nodal and PCP signaling cooperate to regulate gastrulation movements, with Nodal positioned both upstream of and in parallel to PCP. The conserved role of Nodal in promoting Dvl membrane localization and PCP component asymmetry suggests an ancient mechanistic relationship that predates the divergence of these species [60].
Important differences emerge in the specific implementation of these pathways. Zebrafish utilize distinct Nodal ligands (Ndr1/Squint and Ndr2/Cyclops) with partially overlapping functions, while Xenopus employs multiple Nodal-related genes (Xnr1-6) with potentially greater functional specialization [60] [62]. The maternal regulation of Nodal expression also differs, with zebrafish relying on maternal Hwa and Eomesa [63], while Xenopus utilizes VegT and β-catenin [62]. These variations likely reflect species-specific adaptations in embryonic development while maintaining core functional relationships.
Future research should address several unresolved questions. First, the complete molecular machinery linking Nodal signaling to PCP component localization remains incompletely characterized. Second, the potential conservation of the newly identified role for PCP signaling in radial intercalation [64] across vertebrate species warrants investigation. Third, the interplay between Nodal/PCP and other pathways regulating gastrulation, such as ErbB [65], Wnt, and FGF signaling, requires further elucidation. Finally, quantitative computational models integrating these signaling networks with biomechanical properties of embryonic tissues would significantly advance predictive understanding of gastrulation dynamics.
From a technical perspective, emerging approaches including CRISPR-based genetic engineering, improved biosensors for signaling activity, and advanced live imaging with light-sheet microscopy will enable more precise manipulation and observation of these processes. Application of these technologies in both zebrafish and Xenopus will continue to provide complementary insights into the conserved principles of vertebrate morphogenesis.
The comparative analysis of Nodal and PCP signaling in zebrafish and Xenopus reveals a deeply conserved functional relationship that orchestrates gastrulation movements through integrated control of tissue patterning and cellular polarity. While species-specific adaptations exist in the precise implementation of these pathways, the core architecture of Nodal and PCP cooperation in regulating convergent extension appears fundamentally conserved across vertebrates. The experimental frameworks and reagent tools described here provide a foundation for continued investigation into these crucial developmental processes, with significant implications for understanding human congenital disorders and evolutionary developmental biology. Continued comparative analysis across model systems will remain essential for distinguishing fundamental principles of vertebrate development from species-specific adaptations.
The morphogenetic processes that shape an embryo represent a fundamental marvel of biological systems, integrating biochemical signaling with physical forces to orchestrate the emergence of form and function. Among these signaling systems, Bone Morphogenetic Protein (BMP) pathways and their dynamic interplay with tissue mechanics have emerged as a deeply conserved regulatory module across bilaterian organisms. This conservation spans from Drosophila melanogaster to Homo sapiens, suggesting an ancient, fundamental mechanism for patterning and morphogenesis. Within the context of gastrulation research—a pivotal developmental window when the homogeneous embryonic plate self-organizes into the foundational germ layers—understanding this mechanochemical interdependence provides critical insights into the fundamental principles of embryonic patterning. The emerging paradigm reveals that BMP signaling operates not merely as a biochemical cue but as an integrated mechanochemical processor that translates physical constraints and forces into precise transcriptional responses and cellular behaviors, ultimately guiding the emergence of the body plan across diverse species.
Fundamental Mechanisms: BMP Signaling and Mechanical Transduction
The Molecular Architecture of BMP Signaling
BMPs belong to the Transforming Growth Factor-β (TGF-β) superfamily and are synthesized as large precursor molecules that undergo proteolytic cleavage to form mature, biologically active dimers [68]. These dimers can be either homodimers (e.g., BMP4/BMP4) or heterodimers (e.g., BMP4/BMP7), a flexibility that expands their functional repertoire and receptor binding specificity [68]. The canonical signaling cascade initiates when these ligand dimers bind to cell surface receptors, forming a heterotetrameric complex composed of two type I and two type II serine/threonine kinase receptors [69] [70]. This assembly triggers the phosphorylation of the type I receptor by the constitutively active type II receptor, subsequently activating receptor-regulated SMADs (R-SMADs: Smad1, Smad5, Smad8) [69] [70]. These phosphorylated R-SMADs then complex with the common-mediator Smad4 and translocate to the nucleus to regulate target gene expression [68] [69]. This pathway is extensively regulated at multiple levels by extracellular antagonists (e.g., NOGGIN, Chordin), intracellular inhibitors (e.g., Smad6, Smad7), and cross-talk with other signaling pathways [69] [70].
Mechanical Sensing and Transduction Pathways
Parallel to the biochemical signaling machinery, cells possess sophisticated mechanisms to sense and respond to mechanical cues from their microenvironment. The mechanosensitive transcription factor YAP1 (Yes-associated protein 1) serves as a key regulator in this process, shuttling between the cytoplasm and nucleus in response to mechanical stimuli and cellular tension [16] [71]. During gastrulation, tensile forces along the embryonic margin drive large-scale tissue movements that are essential for proper embryonic organization [16]. These mechanical signals regulate downstream pathways controlling cellular differentiation and structure formation, often through the modulation of cytoskeletal components, cell adhesion molecules, and force-generating proteins such as non-muscle myosin II [16] [72].
Integrated Mechanochemical Signaling Nodes
The integration of BMP signaling and mechanical transduction occurs at multiple nodal points, creating a coupled regulatory network. Research using human gastrula models has demonstrated that BMP4 signaling directly influences the subcellular localization of YAP1, promoting its nuclear accumulation where it can repress WNT3 mRNA expression, thereby shaping the emergent germ layer patterning [16] [71]. Conversely, tissue mechanics can modulate the BMP signaling cascade; geometrical confinement and localized tension regions establish specific mechanical states that regulate cellular responsiveness to BMP signals and subsequent fate transitions [16]. This reciprocal regulation creates a dynamic feedback loop wherein biochemical signals influence mechanical properties and vice versa, enabling the self-organizing capabilities observed in developing tissues.
Table 1: Core Components of the BMP-Mechanics Signaling Network
| Component Type | Key Elements | Primary Functions |
|---|---|---|
| BMP Ligands | BMP2, BMP4, BMP7 | Morphogen signaling, receptor activation, fate specification |
| Receptors | BMPR-IA/ALK3, BMPR-IB/ALK6, BMPR-II | Signal transduction, kinase activity, SMAD phosphorylation |
| Intracellular Transducers | Smad1/5/8, Smad4 | Nuclear translocation, gene regulation, transcriptional control |
| Mechanosensors | YAP1, TAZ | Force sensing, nuclear shuttling, mechanical feedback |
| Effector Proteins | Non-muscle myosin II, E-cadherin | Force generation, cell adhesion, cytoskeletal organization |
Conserved Dynamics in Gastrulation: From Drosophila to Human
Drosophila Germ-Band Extension
The conservation of BMP-mechanics interdependence is strikingly evident during Drosophila germ-band extension (GBE), a paradigmatic convergent-extension process that elongates the embryonic body plan. Through quantitative analysis of morphodynamic atlases and machine learning approaches, researchers have delineated a precise signaling cascade wherein BMP establishes dorsoventral (DV) pair-rule-gene patterns that subsequently establish an E-cadherin gradient [72]. This adhesion molecule gradient then creates an opposite-direction myosin gradient through mechanochemical feedback loops, ultimately driving the polarized cell intercalations that underlie tissue elongation [72]. This coordinated process demonstrates how BMP signaling provides the initial positional information that is subsequently translated into mechanical forces through the modulation of adhesion and contractility.
Mammalian Gastrulation Models
In mammalian systems, including human gastruloids, a remarkably similar interdependence governs symmetry breaking and germ layer specification. When human embryonic stem cells (hESCs) are grown in geometrical confinement and exposed to BMP4, they self-organize into radially symmetric patterns reminiscent of the human gastrula, with distinct concentric rings representing embryonic and extra-embryonic lineages [16] [73]. In these models, mechanical signals are crucial regulators of the BMP4 signaling cascade, which in turn regulates WNT and YAP activity, ultimately shaping the fate patterns of gastrulation [16] [71]. The confinement-dependent architecture generates localized high-tension regions and stress fibers that establish mechanical states permissive for specific fate transitions, demonstrating how physical constraints actively participate in developmental patterning rather than merely constraining it.
Signaling Dynamics and Fate Patterning
Quantitative studies of BMP, WNT, and NODAL signaling dynamics in human gastruloids have revealed an intricate temporal sequence wherein BMP signaling initiates waves of WNT and NODAL signaling activity that move toward the colony center at a constant rate [73] [74]. These signaling dynamics, rather than stable spatial gradients, control cell fate patterning—with longer durations of WNT and NODAL signaling promoting mesoderm differentiation, while sustained BMP signaling directs differentiation toward CDX2-positive extra-embryonic cells [73] [74]. This dynamic interpretation mechanism represents a sophisticated strategy for encoding positional information, with tissue mechanics serving as an essential modulator of the signaling kinetics and thresholds.
Table 2: Quantitative Parameters of Signaling Dynamics in Human Gastruloids
| Signaling Pathway | Activation Sequence | Spatiotemporal Pattern | Primary Fate Outcomes |
|---|---|---|---|
| BMP | First responder (0-6h) | Restricted to colony edges by 12h | Extra-embryonic differentiation (CDX2+) |
| WNT | Secondary wave (6-12h) | Wavefront moving toward center | Mesendoderm competence |
| NODAL | Tertiary wave (12-18h) | Wavefront moving toward center | Mesoderm specification |
| YAP | Mechanically regulated | Nuclear localization in high-tension regions | WNT repression, germ layer balancing |
Experimental Approaches and Methodologies
Micropatterned Gastruloid Systems
The development of micropatterned human gastruloid platforms has provided a standardized, quantitative system for investigating BMP-mechanics interactions with high reproducibility. In this approach, hESCs are confined to precisely defined geometric patterns (typically circular substrates ranging from 200-800 μm in diameter) using microfabrication techniques [16] [73]. These confined colonies are then stimulated with recombinant BMP4 ligand (at concentrations typically ranging from 1-10 ng/ml), triggering self-organization that recapitulates aspects of embryonic patterning [16] [73]. The geometric confinement generates a specific tissue architecture with localized high-tension regions at the periphery, establishing a mechanical context that modulates cellular responses to morphogen signals [16]. This system enables high-resolution live imaging, fixed-timepoint immunostaining, and molecular profiling to decipher the coupled dynamics of signaling and mechanics.
Optogenetic BMP4 Control
To achieve spatiotemporal precision in BMP4 activation, researchers have developed light-inducible BMP4 expression systems in hESCs [16] [71]. This optogenetic approach involves inserting the human BMP4 coding sequence downstream of a loxP-flanked stop cassette in a piggyBac vector, enabling controlled gene expression via light-induced loxP recombination [16]. Upon doxycycline treatment to confer light sensitivity, brief pulses of blue light (typically 1-5 minutes) trigger BMP4 expression with spatial resolution determined by the illumination pattern [16] [71]. This technology enables unprecedented dissection of BMP signaling requirements, demonstrating that localized BMP4 activation is sufficient to induce SMAD1/5 phosphorylation and subsequent amnion differentiation, while also revealing the tension-dependent induction of WNT and NODAL for mesoderm specification [16] [71].
Physical Perturbation and Mechanical Measurements
Complementary physical perturbation approaches are essential for establishing causal relationships between mechanics and signaling. These include pharmacological modulation of cytoskeletal tension (e.g., ROCK inhibitors to reduce actomyosin contractility), substrate stiffness manipulation, and direct mechanical compression or stretching of tissues [16]. Concurrently, traction force microscopy, fluorescence recovery after photobleaching (FRAP) of adhesion proteins, and laser ablation techniques enable quantification of mechanical properties including internal stresses, tissue tension, and junctional dynamics [16] [72]. These biophysical measurements have revealed that tensile forces along the embryonic margin drive large-scale tissue movements during gastrulation, with the mechanosensory gene YAP1 differentially regulated in the primitive streak region compared with surrounding epiblast cells [16].
Computational Modeling and Theoretical Frameworks
Mathematical Models of Mechanochemical Patterning
The integration of experimental data with quantitative mathematical models has been instrumental in formalizing our understanding of BMP-mechanics interdependence. These models typically incorporate reaction-diffusion dynamics for morphogen signaling coupled with mechanical force balance equations and feedback regulations [16] [73]. For instance, Laurent et al. developed a model that integrates tissue mechanics into BMP4 morphogen dynamics, successfully predicting tissue-scale responses to BMP4 signaling and explaining how mechanical signals regulate the BMP4 signaling cascade, which in turn controls WNT and YAP activity [16]. These models have demonstrated that the observed wave-like propagation of WNT and NODAL signaling is inconsistent with a pure reaction-diffusion Turing system, indicating instead that the final signaling state is homogeneous with spatial differences arising primarily from boundary effects and mechanical constraints [73] [74].
Machine Learning Approaches
Recent advances have incorporated machine learning methodologies to mine complex morphodynamic datasets and identify predictive relationships between biochemical and mechanical factors. For example, neural networks trained on Drosophila morphogenetic atlases have revealed that knowledge of the initial myosin distribution is sufficient to forecast subsequent tissue flow patterns during germ-band extension [72]. Principal component analysis further demonstrated that the coarse-grained dynamics of gastrulation are low-dimensional and reproducible across embryos, suggesting that simple rules can describe the complex interplay between BMP patterning and tissue mechanics [72]. These data-driven approaches complement mechanistic modeling by identifying minimal predictive sets of variables and revealing conserved dynamical features across species.
The Scientist's Toolkit: Essential Research Reagents and Methodologies
Table 3: Key Research Reagent Solutions for BMP-Mechanics Investigations
| Reagent/Method | Specific Example | Primary Application | Technical Notes |
|---|---|---|---|
| Micropatterned Substrates | CYTOOchips, custom PDMS stamps | Geometric confinement studies | Standardized sizes (200-800μm) enable reproducible patterning |
| Optogenetic BMP Systems | iBMP4-hESCs (piggyBac vector) | Spatiotemporal BMP control | Blue light activation (1-5min pulses) after doxycycline sensitization |
| Mechanical Reporters | FRET-based tension sensors, YAP/TAZ localization | Live imaging of mechanical states | Antibodies for phospho-YAP/TAZ available for fixed samples |
| Signaling Reporters | SMAD1/5/8 phosphorylation, BRE-Luc reporter | BMP pathway activity quantification | Commercial phospho-SMAD1/5 antibodies validated for imaging/Western |
| Cytoskeletal Modulators | ROCK inhibitor (Y-27632), Blebbistatin | Actomyosin perturbation | Dose titration essential to avoid complete cytoskeletal disruption |
| Mathematical Modeling | Custom Python/Matlab scripts (GitHub available) | Quantitative model simulation | Combines reaction-diffusion with mechanical force balance |
The conserved interdependence between BMP signaling and tissue mechanics represents a fundamental principle of embryonic development, with profound implications for both basic biology and regenerative medicine applications. From Drosophila germ-band extension to human gastruloid patterning, this mechanochemical integration enables the robust self-organization of complex structures from initially homogeneous cell populations. The mechanistic insights gleaned from these studies not only advance our understanding of embryogenesis but also inform engineering approaches for tissue fabrication and organoid development. As research progresses, the continued integration of advanced molecular tools, biophysical measurements, and computational modeling will further elucidate how these conserved pathways interact across scales to transform biochemical information and physical forces into the exquisite patterns of life.
Diagram 1: Core BMP-Mechanics Signaling Circuit. This integrated pathway illustrates the reciprocal regulation between BMP signaling (yellow nodes) and mechanical components (green nodes), with key inhibitory interaction shown in red.
Diagram 2: Gastruloid Pattering Experimental Workflow. This diagram outlines the key experimental steps and interactions in micropatterned gastruloid systems, showing how geometric confinement (yellow) initiates mechanical signaling (green) that patterns biochemical waves (red) to specify cell fates (blue).
The use of model organisms, particularly the mouse (Mus musculus), has been a cornerstone of developmental biology, operating on the critical underlying assumption that gene functions and developmental systems are conserved between these models and humans [75]. This paradigm has enabled tremendous advances in our understanding of the molecular basis of development, physiology, and disease pathogenesis. The biological justification for this approach is strong, rooted in evolutionary homology—the shared ancestry of tissue and organ systems across diverse animals [75]. Remarkable examples of functional conservation, such as the Pax6/eyeless gene master-regulating eye development across phyla as divergent as vertebrates and Drosophila, appear to validate this approach [75]. However, a growing body of evidence challenges the universality of this assumption, revealing that protein functions and gene regulatory networks can diverge significantly through evolutionary time [75]. This review examines the mechanisms of such divergence, with a specific focus on insights from gastrulation research, and explores the implications for interpreting model organism data in the context of human developmental biology.
Theoretical Framework: Mechanisms of Developmental Systems Divergence
Molecular Mechanisms of Evolutionary Divergence
Evolutionary developmental biology (evo-devo) investigates how animal morphology evolves by determining how cell types and their spatio-temporal placement change over evolutionary time [76]. This divergence occurs through several key mechanisms:
Positive Selection and Functional Divergence: Adaptive molecular evolution can modify ancestral gene functions or generate novel ones. Statistical methods comparing synonymous and non-synonymous nucleotide substitution rates have identified numerous cases of molecular adaptation, often associated with novel functions or evolutionary "arms races" [75]. For example, the progesterone receptor (PGR) evolved extremely rapidly in humans and chimpanzees, with amino acid substitutions occurring in regions critical for transcriptional activity, coincident with changes in the mechanism of parturition in higher apes [75]. This suggests that rodent models may not faithfully replicate PGR actions in primates.
Divergence in Gene Regulatory Networks (GRNs): Evolutionary modifications are often mediated through changes in how genes are regulated and how their products function within networks [75]. The field is moving toward system-level descriptions of cell specification, which involves determining the networks of gene regulatory interactions that direct the formation of cell types [76]. Comparisons of these GRNs are revealing new insights into the mechanisms of evolutionary change in development [76].
Table 1: Documented Cases of Functional Divergence in Key Developmental Genes
| Gene | Function in Mouse | Divergent Function in Other Species | Functional Consequence |
|---|---|---|---|
| HoxA-11 | Activates prolactin (PRL) expression in endometrial stromal cells [75] | Only represses PRL expression in opossum, platypus, and chicken [75] | Novel transcriptional activation function in placental mammals |
| Phospholipase C zeta 1 (PLCZ1) | Contains nuclear localization signal (NLS); enters nucleus after fertilization [75] | Lacks NLS in other mammals; does not enter nucleus [75] | Divergent subcellular localization and potential signaling |
| Progesterone Receptor (PGR) | Standard transcriptional regulation | Rapid evolution in humans/chimpanzees with altered transcriptional activity [75] | Associated with changes in primate parturition mechanisms |
The Plasticity of Gastrulation as a Model for Understanding Divergence
Gastrulation represents a critical juncture in embryonic development where the pluripotent epiblast self-organizes into the three primary germ layers—ectoderm, mesoderm, and endoderm [73]. The morphology of gastrulation is highly variable across the animal kingdom, suggesting this fundamental process is not rigidly constrained by evolutionary pressures [77]. This diversity provides a powerful framework for understanding developmental system divergence.
The mode of mesendoderm internalization is a major determinant of gastrulation morphology [77]. In most invertebrates and anamniote vertebrates, mesoderm internalizes as a continuous epithelial layer through invagination (e.g., sea urchins, Drosophila) or involution (e.g., Xenopus) [77]. In contrast, amniotes (reptiles, birds, mammals) employ ingression, where mesoderm precursors undergo an epithelial-to-mesenchymal transition (EMT) and enter the embryo as individual cells [77]. The extent of EMT exists on a spectrum, with cells exhibiting varying degrees of epithelial or mesenchymal characteristics that profoundly influence gastrulation mechanics [77].
Signaling Dynamics in Gastrulation: Insights from Human Models
Conserved Signaling Cascades and Their Divergent Implementation
Decades of mouse research have established that a signaling cascade involving Bone Morphogenetic Protein (BMP), WNT, and NODAL pathways is necessary for gastrulation [73]. These ligands are typically expressed near the site of gastrulation, leading to the prevailing view that signaling gradients pattern the embryo [73]. However, recent research using human gastruloids—in vitro models of human gastrulation—has revealed both conserved and potentially divergent features of this process.
Quantitative studies in human gastruloids demonstrate that BMP signaling initiates waves of WNT and NODAL signaling activity that move toward the colony center at a constant rate [73]. Mathematical modeling indicates this wave-like behavior is inconsistent with a reaction-diffusion-based Turing system, suggesting no stable WNT/NODAL gradient forms [73]. Instead, the final signaling state appears homogeneous, with spatial differences arising primarily from boundary effects [73].
Dynamics Over Gradients: A New Paradigm for Fate Patterning
In human gastruloids, the dynamics of signaling events, rather than stable spatial gradients, appear to control cell fate patterning [73]:
- The duration of WNT and NODAL signaling controls mesoderm differentiation
- The duration of BMP signaling controls differentiation of CDX2-positive extra-embryonic cells
- The identity of these BMP-induced extra-embryonic cells closely resembles human trophoblast cells in vivo
- Mesoderm differentiation is controlled dynamically by the combinatorial effect of multiple signals rather than mapping directly to a pre-patterned spatial domain
This represents a potential divergence from traditional models based on mouse studies and highlights the importance of direct analysis of human developmental systems.
Table 2: Quantitative Signaling Dynamics in Human Gastruloid Fate Patterning
| Signaling Pathway | Dynamic Pattern | Role in Fate Patterning | Experimental Manipulation |
|---|---|---|---|
| BMP | Initiates signaling wave; defines domain of extra-embryonic differentiation [73] | Duration controls differentiation of CDX2-positive trophoblast-like cells [73] | Inhibition prevents extra-embryonic differentiation; constitutive signaling expands domain |
| WNT | Wave propagates toward colony center following BMP initiation [73] | Duration controls mesoderm differentiation [73] | Pathway agonists/antagonists modulate mesoderm specification |
| NODAL | Wave propagates toward colony center following BMP initiation [73] | Duration works combinatorially with WNT to control mesoderm differentiation [73] | Receptor inhibitors block mesoderm formation; altered dynamics shift fate boundaries |
Experimental Approaches: Synthesizing Gastrulation Modes and Technical Protocols
Experimental Evidence for Gastrulation Plasticity
Recent experiments demonstrate that gastrulation modes can be transitioned by perturbing a small number of components, supporting the evolutionary plasticity of this process [77]:
- In the sea anemone Nematostella vectensis, which normally internalizes endoderm by invagination, disruption of the PAR polarity complex causes disassembly of adherens junctions. Some cells then acquire a mesenchymal phenotype and internalize via ingression rather than invagination [77].
- When N. vectensis embryonic cells are dissociated and reaggregated, altering the embryonic geometry from a hollow sphere to a compact ball, the embryo abandons coherent invagination in favor of multipolar ingression and cavitation [77].
- These findings suggest that transitions between gastrulation modes do not present a strong evolutionary constraint and can be achieved through relatively modest changes in cell adhesion and polarity.
Detailed Experimental Protocol: Human Gastruloid Generation and Signaling Analysis
Objective: To generate human pluripotent stem cell-derived gastruloids for quantifying BMP, WNT, and NODAL signaling dynamics during self-organized fate patterning [73].
Methodology:
Gastruloid Generation:
- Culture human pluripotent stem cells in essential 8 medium until 70-80% confluent.
- Dissociate to single cells using EDTA or enzymatic dissociation.
- Seed 300-500 cells per well in ultra-low attachment 96-well plates in gastruloid differentiation medium.
- Centrifuge plates at 300-400g for 3-5 minutes to promote aggregate formation.
- Culture for up to 96 hours, monitoring aggregate formation daily.
Live Signaling Monitoring:
- Generate reporter cell lines expressing fluorescent proteins under the control of BMP (BRE), WNT (TCF/LEF), and NODAL (ARE) responsive elements.
- Alternatively, use immunofluorescence staining for phosphorylated SMAD1/5/8 (BMP), β-catenin (WNT), and SMAD2/3 (NODAL) on fixed samples at multiple time points.
- Image gastruloids using confocal microscopy every 2-4 hours for 72-96 hours to capture signaling dynamics.
Data Quantification and Modeling:
- Quantify fluorescence intensity in concentric regions from the gastruloid periphery to the center.
- Calculate signaling wave velocity and duration for each pathway.
- Apply mathematical modeling to determine if dynamics conform to reaction-diffusion systems or boundary-effect dominated systems.
- Correlate signaling dynamics with spatial differentiation markers (e.g., BRACHYURY for mesoderm, CDX2 for trophoblast-like cells) via immunostaining or single-cell RNA sequencing.
The Scientist's Toolkit: Essential Research Reagents and Solutions
Table 3: Essential Research Reagents for Gastrulation and Developmental Divergence Studies
| Reagent/Category | Specific Examples | Function/Application | Considerations for Divergence Studies |
|---|---|---|---|
| Cell Lines | Human Pluripotent Stem Cells (hPSCs), Mouse Embryonic Stem Cells (mESCs) [73] | In vitro model generation (gastruloids); comparative functional studies | Species-specific differences in signaling requirements, pluripotency states, and differentiation efficiency |
| Signaling Reporters | BRE-Luc (BMP), TCF/LEF-GFP (WNT), ARE-mCherry (NODAL) [73] | Live monitoring of pathway activation dynamics; quantitative comparison | Potential differences in response element conservation between species |
| Pathway Modulators | Recombinant BMP4/WNT3A/NODAL proteins; Small molecule inhibitors (LDN193189, IWP2, SB431542) [73] | Experimental perturbation of signaling; testing functional requirements | Species-specific differences in inhibitor sensitivity and ligand-receptor interactions |
| Cell Fate Markers | Antibodies against BRACHYURY (mesoderm), SOX17 (endoderm), CDX2 (trophoblast) [73] | Characterization of differentiation outcomes; correlation with signaling | Timing and combinatorial expression may differ between models |
| Gene Editing Tools | CRISPR/Cas9 systems, siRNA/shRNA [75] | Functional validation of gene-specific roles; testing functional equivalence | Off-target effects; species-specific efficiency; potential compensatory mechanisms |
The evidence for evolutionary divergence in developmental systems, particularly in dynamically regulated processes like gastrulation, has profound implications for biomedical research and therapeutic development. While model organisms remain indispensable tools, the assumption of functional conservation must be critically evaluated on a case-by-case basis [75]. This is especially crucial for:
- Drug Target Validation: Targets identified in model organisms must be carefully validated in human systems, particularly for rapidly evolving genes involved in reproduction, immunity, and brain function [75].
- Developmental Toxicity Testing: Current animal models may not fully recapitulate human-specific developmental vulnerabilities, particularly for drugs targeting signaling pathways with divergent dynamics.
- Stem Cell-Based Therapies: Understanding human-specific aspects of developmental signaling is essential for directing cell fate therapeutically.
- Disease Modeling: The translational relevance of disease models depends on the conservation of underlying gene networks and their interactions.
The emerging paradigm suggests that while core developmental pathways are often conserved, their regulation, dynamics, and functional outputs can diverge significantly. Embracing human-based models like gastruloids, while maintaining a comparative evolutionary perspective, will be essential for advancing our understanding of human development and its implications for medicine.
The process of gastrulation, during which the three primary germ layers—ectoderm, mesoderm, and endoderm—are established, represents a pivotal milestone in early embryonic development. Studying these events in humans presents significant ethical and technical challenges due to limited tissue accessibility and ethical oversight [44]. Within this context, gastruloids—three-dimensional in vitro structures derived from pluripotent stem cells that recapitulate aspects of gastrulation—have emerged as powerful model systems [44] [6]. This technical guide frames the benchmarking of these models within a broader thesis on the fundamental role of signaling dynamics in governing gastrulation, providing researchers with methodologies to rigorously validate gastruloid patterning against in vivo developmental standards.
The morphogenetic events of gastrulation are orchestrated by complex signaling centers that pattern the embryo through precise spatiotemporal gradients [44]. Engineered gastruloid models, including both two-dimensional (2D) micropatterned systems and three-dimensional (3D) gastruloid constructs, provide valuable insights into cell differentiation, signaling pathways, and tissue organization during germ layer formation [44] [6]. The accurate recapitulation of these signaling dynamics serves as the primary benchmark for evaluating the physiological relevance of in vitro models.
Key Benchmarking Criteria for Gastruloid Models
Morphological and Molecular Benchmarks
Validated gastruloid models must demonstrate faithful recapitulation of key morphological and molecular features observed in vivo. Benchmarking requires a multi-parameter approach that assesses both structural organization and molecular patterning.
Table 1: Essential Benchmarking Criteria for Gastruloid Validation
| Benchmark Category | Specific Parameters | In Vivo Reference (Carnegie Stage) | Validation Methods |
|---|---|---|---|
| Spatial Organization | Germ layer arrangement, Axial organization | CS7-9 (Trilaminar disc formation) | Immunofluorescence, Spatial transcriptomics [44] [3] |
| Gene Expression | Pluripotency exit markers (OCT4, NANOG downregulation), Germ layer-specific markers (SOX17, GATA6) | CS5-7 (Pre- to post-implantation transition) | scRNA-seq, Bulk RNA-seq, qPCR [44] [3] |
| Signaling Pathway Activity | BMP, WNT, Nodal pathway activation | CS6 (Primitive streak formation) | Phospho-protein staining, Reporter cell lines, Pathway inhibitors [3] |
| Developmental Timeline | Pace of germ layer specification, Somitogenesis onset | Species-specific developmental milestones | Live imaging, Time-course transcriptomics [44] |
Quantitative Assessment of Patterning Efficiency
Robust benchmarking requires quantitative metrics to evaluate the efficiency and reproducibility of germ layer patterning. Recent advances have established protocols that achieve high differentiation efficiency.
Table 2: Quantitative Metrics for Gastruloid Patterning Efficiency
| Cell Type | Key Markers | Reporting Studies | Efficiency Range | Timeframe |
|---|---|---|---|---|
| Extraembryonic Mesoderm (ExM) | GATA6, SNAIL, VIM, KDR, FLT1, HAND1 [3] | Kime et al., 2025; Pham et al., 2024 | ~90% [3] | 4-5 days [3] |
| Primitive Streak-like Intermediates | T (Brachyury), MIXL1, E-CADHERIN (downregulation) [3] | Multiple gastruloid protocols | Variable (Protocol-dependent) | 24-72 hours |
| Amnion-like Cells | IGFBP3, GATA3 [3] | Kime et al., 2025 | ~6.4% of total population [3] | 4-5 days [3] |
Experimental Protocols for Gastruloid Benchmarking
Protocol 1: Rapid Induction of Extraembryonic Mesoderm from hESCs
This protocol enables the efficient derivation of extraembryonic mesoderm (ExM) from human embryonic stem cells (hESCs) within 4-5 days, achieving approximately 90% efficiency [3].
Key Steps:
- Cell Preparation: Dissociate naive hESCs (e.g., AIC-N hESCs) and inoculate onto Matrigel-coated dishes.
- Culture Medium: Use modified N2B27 medium supplemented with FGF4 and heparin (FH-N2B27).
- Signaling Modulation: Add CHIR99021 (CHIR, a GSK3 inhibitor) and BMP4 (CB treatment) to induce differentiation.
- Monitoring: Culture for 4 days, during which cells rapidly lose colony morphology and convert to mesenchymal phenotype.
- Validation: Assess via immunofluorescence for ExM markers (GATA6, SNAIL, VIM, KDR, FLT1) and flow cytometry to quantify efficiency.
Technical Notes: This system induces ExM specification through primitive streak-like intermediates (PSLI), delineating the regulatory roles of WNT and Nodal signaling in this process [3]. The initial pluripotent state of the hESCs influences signal response, cellular composition, and developmental progression of the resulting ExM cells.
Protocol 2: Generation of 3D Gastruloids with Axial Organization
This protocol describes the generation of 3D gastruloids that mimic posterior embryonic development, including somitogenesis and axial elongation.
Key Steps:
- Aggregation: Use forced aggregation techniques in U-bottom or AggreWell plates to generate uniform embryonic organoids [44].
- Size Control: Standardize aggregate size to ensure reproducible differentiation trajectories and morphogenic behavior.
- Signaling Patterning: Apply precise temporal sequences of WNT, BMP, and Nodal pathway activation to pattern the aggregates.
- Extended Culture: Transfer aggregates to rotary or static culture conditions, optionally with ECM hydrogel embedding, to support complex morphogenesis [44].
- Axial elongation: Culture gastruloids for up to 21 days to observe advanced developmental events including somite formation and neural tube patterning [44].
Technical Notes: These 3D constructs can express an outer layer of trophoblast (KRT7+) cells, an inner layer of epiblast cells (OCT4+), and a mesendoderm population (GATA6+), similar to Carnegie stage 7 embryos, as evidenced by RNA-sequencing [44].
Signaling Pathways in Gastruloid Patterning
The following diagram illustrates the core signaling pathways and their interactions in gastruloid patterning, highlighting the dynamic interplay between key developmental signaling centers.
Signaling Pathway Dynamics: The diagram illustrates how coordinated BMP, WNT, and Nodal signaling directs the differentiation of pluripotent stem cells through a primitive streak-like intermediate toward various cell fates, predominantly extraembryonic mesoderm [3]. Studies reveal that ExM specification from hESCs predominantly proceeds through these primitive streak-like intermediates, with WNT and Nodal signaling playing distinct regulatory roles in this process [3].
Research Reagent Solutions for Gastruloid Studies
The following table details essential research reagents and their applications in gastruloid differentiation and patterning studies.
Table 3: Essential Research Reagents for Gastruloid Differentiation
| Reagent Category | Specific Examples | Function in Gastruloid Patterning | Application Notes |
|---|---|---|---|
| Small Molecule Inhibitors/Activators | CHIR99021 (GSK3 inhibitor), BMP4, A83-01 (TGF-β inhibitor) | Modulate key signaling pathways (WNT, BMP, Nodal) to direct differentiation [3] | Concentration and timing critically influence cell fate; CHIR+BMP4 combination rapidly induces ExM [3] |
| Cell Culture Matrices | Matrigel, Synthetic PEG hydrogels, Laminin-521 | Provide structural support and biochemical cues for morphogenesis [44] | Matrigel supports attachment and growth of naive hESCs during ExM differentiation [3] |
| Cytokines and Growth Factors | FGF4, Heparin, VEGF | Support mesoderm specification and vascular differentiation [3] | FGF4 and heparin (FH-N2B27 medium) used in combination with CHIR and BMP4 for ExM induction [3] |
| Cell Line Reporters | Fluorescent reporters for Brachyury (T), SOX17, GATA6 | Enable real-time monitoring of lineage specification and patterning dynamics [44] | Critical for quantifying patterning efficiency and isolating specific populations for analysis |
| Aggregation Platforms | U-bottom wells, AggreWell plates, Pyramidal microwells | Standardize gastruloid size and uniformity through forced aggregation [44] | Control over spheroid size dictates differentiation trajectory and morphogenic behavior [44] |
The rigorous benchmarking of gastruloid models against in vivo developmental standards is paramount for establishing their physiological relevance and utility in developmental biology and drug development. By focusing on the recapitulation of signaling dynamics—through morphological assessment, molecular characterization, and functional validation—researchers can leverage these powerful models to dissect human gastrulation mechanisms. The protocols and benchmarks outlined in this guide provide a framework for standardized validation across research platforms, enabling more reproducible and physiologically relevant studies of human development and disease modeling. As the field advances, continued refinement of these benchmarking standards will be essential for realizing the full potential of gastruloid technologies in both basic research and translational applications.
Conclusion
The study of signaling dynamics in gastrulation has evolved from a purely biochemical perspective to an integrated paradigm where mechanical forces, temporal signaling patterns, and epigenetic regulation are inextricably linked. The synergy between pathways like Nodal and PCP, and the critical role of tissue mechanics, as revealed by advanced tools like Brillouin microscopy and optogenetics, underscore that successful morphogenesis requires both chemical and physical priming. The rise of sophisticated in vitro models, such as gastruloids, combined with AI-driven phenotyping and single-cell multi-omics, provides an unprecedented, ethically feasible window into human development. Future research must focus on building more complex integrated models, further elucidating the 'mechanical organizer,' and translating these fundamental insights into clinical applications for regenerative medicine, understanding congenital disorders, and improving reproductive health outcomes.
References
- 1. Embryology, Gastrulation - StatPearls - NCBI Bookshelf - NIH [https://www.ncbi.nlm.nih.gov/books/NBK554394/]
- 2. Study uncovers role of mechanical forces in human ... [https://www.news-medical.net/news/20251122/Study-uncovers-role-of-mechanical-forces-in-human-gastrulation.aspx]
- 3. Deciphering signaling mechanisms and developmental ... [https://www.nature.com/articles/s41467-025-59491-x]
- 4. Zebrafish Gastrulation: Cell Movements, Signals, and ... [https://www.sciencedirect.com/science/article/abs/pii/S0074769607610043]
- 5. The Physical Mechanisms of Drosophila Gastrulation [https://pmc.ncbi.nlm.nih.gov/articles/PMC7054018/]
- 6. Advances in engineered models of peri-gastrulation [https://www.sciencedirect.com/science/article/pii/S2589004225009204]
- 7. Dissecting the dynamics of signaling events in the BMP ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6814242/]
- 8. Nodal Signaling and Congenital Heart Defects - NCBI - NIH [https://www.ncbi.nlm.nih.gov/books/NBK500272/]
- 9. Optimizing Nodal, Wnt and BMP signaling pathways for robust ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11182123/]
- 10. Wnt signaling pathways in biology and disease [https://www.nature.com/articles/s41392-025-02142-w]
- 11. Planar Cell Polarity Signaling: The Developing Cell's Compass [https://pmc.ncbi.nlm.nih.gov/articles/PMC2773631/]
- 12. Cell autonomous polarization by the planar cell polarity ... [https://www.nature.com/articles/s41467-025-64563-z]
- 13. Wnt, Activin, and BMP Signaling Regulate Distinct Stages ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2533280/]
- 14. Combinatorial interpretation of BMP and WNT controls the ... [https://www.sciencedirect.com/science/article/pii/S2405471224001169]
- 15. Advances in mechanochemical modelling of vertebrate ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12409983/]
- 16. Crosstalk between tissue mechanics and BMP4 signaling ... [https://www.sciencedirect.com/science/article/abs/pii/S1934590925003376]
- 17. Highly dynamic mechanical transitions in embryonic cell ... [https://www.nature.com/articles/s41467-025-61702-4]
- 18. Linking signaling dynamics and cell fate decisions through ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12588912/]
- 19. Revealing Dynamic Mechanisms of Cell Fate Decisions ... [https://www.frontiersin.org/journals/genetics/articles/10.3389/fgene.2019.01280/full]
- 20. Wires in the soup: quantitative models of cell signaling - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3895447/]
- 21. Comprehensive review of signaling pathways and ... [https://pubmed.ncbi.nlm.nih.gov/39653094/]
- 22. Epigenetic regulation of cell fate transition: learning from early ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9310515/]
- 23. Epigenetic control of cell identities from epiblast to gastrulation [https://pmc.ncbi.nlm.nih.gov/articles/PMC12525014/]
- 24. Coordinated action of multiple active histone modifications ... [https://www.nature.com/articles/s41467-025-60246-x]
- 25. Epigenetic and transcriptional regulations prime cell fate ... [https://www.nature.com/articles/s41467-023-36116-9]
- 26. A Comparative Overview of Epigenomic Profiling Methods [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2021.714687/full]
- 27. : an emerging tool for... | Nature Methods Brillouin microscopy [https://www.nature.com/articles/s41592-019-0543-3?error=cookies_not_supported&code=fa5e4dd9-fbdd-4c5d-b6c9-97937432a95d]
- 28. - PMC Brillouin microscopy [https://pmc.ncbi.nlm.nih.gov/articles/PMC11465583/]
- 29. High-resolution line-scan Brillouin microscopy for live ... [https://www.nature.com/articles/s41592-023-01822-1]
- 30. Full-field Brillouin microscopy based on an imaging Fourier ... [https://www.nature.com/articles/s41566-025-01619-y]
- 31. Imaging the mechanical properties of a developing ... [https://www.sciencedirect.com/science/article/pii/S1742706125006543]
- 32. Optogenetic tools for dissecting complex intracellular ... [https://www.sciencedirect.com/science/article/abs/pii/S0006291X20301005]
- 33. Optogenetic Application to Investigating Cell Behavior and ... [https://www.frontiersin.org/journals/cellular-neuroscience/articles/10.3389/fncel.2022.811493/full]
- 34. The Art of Illuminating Complex Signaling Pathways - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8425415/]
- 35. Optogenetic control of Nodal signaling patterns [https://pmc.ncbi.nlm.nih.gov/articles/PMC11030342/]
- 36. Chemogenetic and optogenetic strategies for ... [https://pubmed.ncbi.nlm.nih.gov/40777439/]
- 37. Color-tuned Channelrhodopsins for Multiwavelength ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3442514/]
- 38. Dual-color optical activation and suppression of neurons ... [https://elifesciences.org/reviewed-preprints/90327]
- 39. Stem cell-based human embryo models: current knowledge ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04581-2]
- 40. Pluripotent stem cell-derived model of the post- ... [https://www.nature.com/articles/s41586-023-06368-y]
- 41. An explainable map of human gastruloid morphospace ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11463602/]
- 42. Embryo model completes gastrulation to neurulation and ... [https://www.nature.com/articles/s41586-022-05246-3]
- 43. Signaling Pathways in Human Blastocyst Development [https://pmc.ncbi.nlm.nih.gov/articles/PMC12452331/]
- 44. Advances in engineered models of peri-gastrulation - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12159512/]
- 45. Single-cell multi-omics delineates the dynamics of distinct ... [https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-025-11619-5]
- 46. Advances in single-cell omics and multiomics for high- ... [https://www.nature.com/articles/s12276-024-01186-2]
- 47. The technological landscape and applications of single-cell ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10242609/]
- 48. Emerging technologies of single-cell multi-omics [https://haematologica.org/article/view/11928]
- 49. A multi-omic single-cell landscape reveals transcription and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12288317/]
- 50. Quantitative Comparison of the Anterior-Posterior Patterning ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6643877/]
- 51. EmbryoNet: using deep learning to link embryonic ... [https://www.nature.com/articles/s41592-023-01873-4]
- 52. Deciphering signaling mechanisms and developmental ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12095623/]
- 53. Protocol for differentiation of human pluripotent stem cells into ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12205342/]
- 54. An Epigenetic Priming Mechanism Mediated by Nutrient ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7904604/]
- 55. Multitask benchmarking of single-cell multimodal omics ... [https://www.nature.com/articles/s41592-025-02856-3]
- 56. Integration of chromosome conformation and gene ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1597245/full]
- 57. EmbryoNet AI automatically identifies developmental ... [https://www.gesundheitsindustrie-bw.de/en/article/news/embryonet-ai-automatically-identifies-developmental-disorders]
- 58. EmbryoNet AI Technologies [http://embryonet.de/]
- 59. Zebrafish Gastrulation: Cell Movements, Signals, and ... [https://www.sciencedirect.com/science/chapter/bookseries/abs/pii/S0074769607610043]
- 60. Nodal and planar cell polarity signaling cooperate to ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7250581/]
- 61. Planar Cell Polarity Signaling: Coordinated Crosstalk for Cell ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11130840/]
- 62. Nodal Signaling - an overview [https://www.sciencedirect.com/topics/medicine-and-dentistry/nodal-signaling]
- 63. Maternal Factors and Nodal Autoregulation Orchestrate ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2022.887987/full]
- 64. The involvement of PCP proteins in radial cell ... [https://www.sciencedirect.com/science/article/pii/S001216061530018X]
- 65. Regulation of Xenopus gastrulation by ErbB signaling - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4939279/]
- 66. Peer review in Nodal and planar cell polarity signaling ... [https://elifesciences.org/articles/54445/peer-reviews]
- 67. Nodal signaling regulates asymmetric cellular behaviors ... [https://www.nature.com/articles/s42003-022-03826-7]
- 68. Insights into Bone Morphogenetic Protein—(BMP-) Signaling ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8533954/]
- 69. Bone Morphogenetic Protein (BMP) signaling in development ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4232216/]
- 70. The roles and regulatory mechanisms of TGF-β and BMP ... [https://www.nature.com/articles/s41422-023-00918-9]
- 71. Crosstalk between tissue mechanics and BMP4 signaling ... [https://pubmed.ncbi.nlm.nih.gov/41086807]
- 72. Learning a conserved mechanism for early neuroectoderm ... [https://arxiv.org/html/2405.18382v1]
- 73. Dissecting the dynamics of signaling events in the BMP, WNT ... [https://pubmed.ncbi.nlm.nih.gov/31613879/]
- 74. Dissecting the dynamics of signaling events in the BMP, WNT ... [https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3000498]
- 75. Use with caution: Developmental systems divergence and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2701150/]
- 76. Evolutionary Developmental Biology - an overview [https://www.sciencedirect.com/topics/immunology-and-microbiology/evolutionary-developmental-biology]
- 77. The evolution of gastrulation morphologies - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC10216749/]