The Blueprint of Life
How Homology Reveals Evolution's Master Code
From butterfly wings to whale flippers, a single revolutionary concept unlocks nature's deepest design secrets.
When 16th-century anatomist Pierre Belon sketched the skeletons of humans and birds side by side, he saw an astonishing pattern: despite vast differences in size and function, their bones aligned like architectural blueprints of a master builder. This discovery ignited a scientific quest that would unravel evolution's greatest secret—homology, the biological principle that similar structures in different species reveal shared ancestry 2 4 .
For centuries, biologists have debated how a bat's wing, a mole's digging paw, and a human hand could share the same bone structure. This article traces the explosive development of homology theory—from its pre-Darwinian origins to cutting-edge genetics—and reveals why it remains biology's most powerful tool for decoding life's interconnected history.
The Birth of a Biological Concept
Ideal Archetypes to Evolutionary Maps
The term "homology" was coined in 1843 by Richard Owen, a brilliant anatomist who opposed Darwin's evolutionary ideas. For Owen, homologous structures—like mammalian forelimbs—reflected an abstract "archetype" in the mind of a Creator 2 7 . His definition was purely structural:
"The same organ in different animals under every variety of form and function." 3

Richard Owen (1804-1892)
British anatomist who coined the term "homology" and developed the concept of archetypes in comparative anatomy.

Charles Darwin (1809-1882)
Revolutionized homology by interpreting it as evidence for common descent through natural selection.
Three criteria emerged to identify homologs:
- Position: Bones or organs occupy equivalent spatial relationships (e.g., humerus always proximal to radius/ulna).
- Development: Structures arise from similar embryonic tissues.
- Composition: Shared detailed organization (e.g., five-digit limb structure) 2 4 .
Darwin's 1859 Origin of Species revolutionized this concept. Homology became evidence for common descent—the bat's wing and human hand were similar not by divine template, but because both descended from a prehistoric mammal's limb 4 .
Key Milestones in Homology Theory
1555
Pierre Belon publishes comparative anatomy of human and bird skeletons
1843
Richard Owen coins term "homology" and develops archetype theory
1859
Darwin reinterprets homology as evidence for common descent
1995
Pax6 gene experiment demonstrates deep genetic homology
Homology's Evolutionary Toolkit
From Bones to Genes
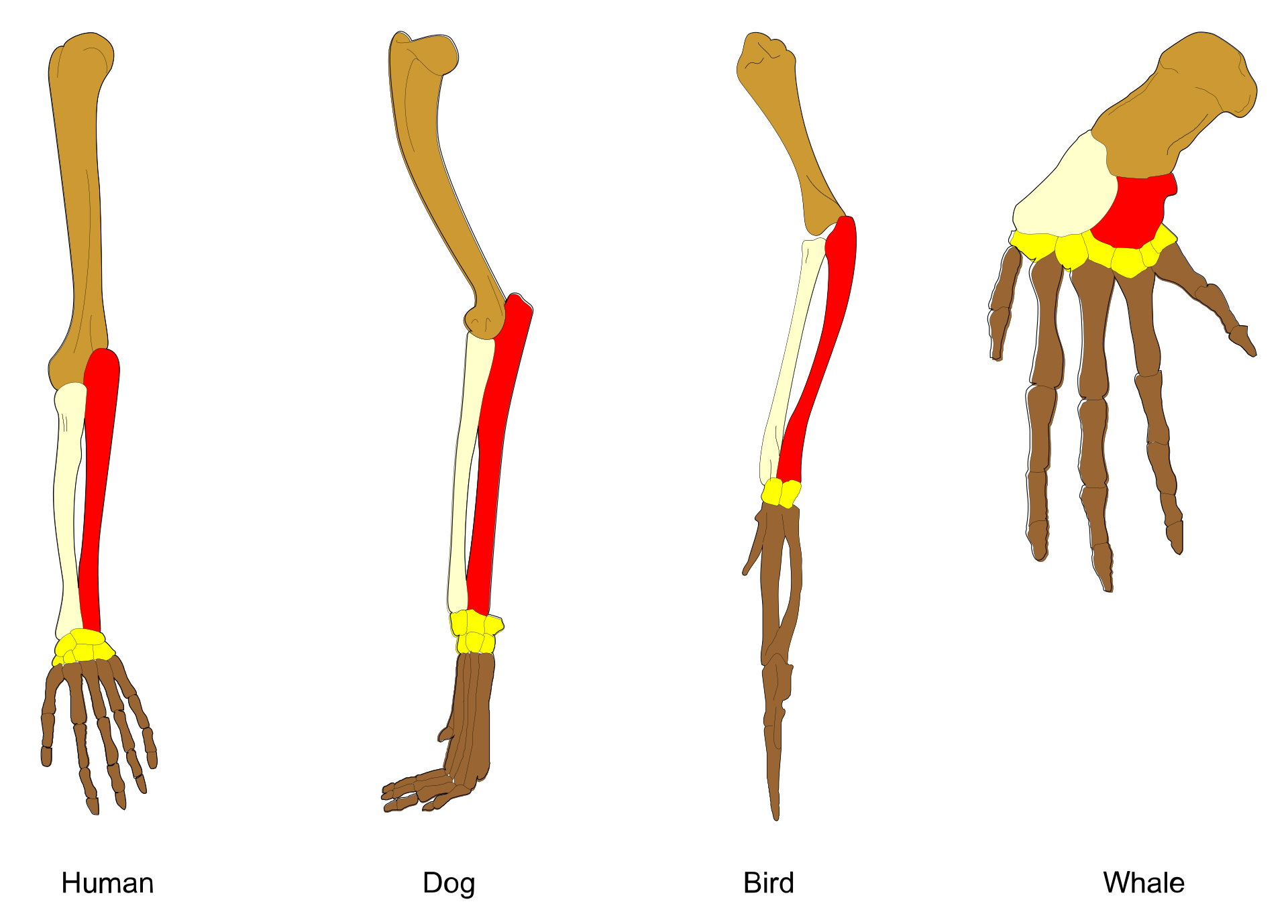
1. Structural Homology: The Vertebrate Triumph
The pentadactyl limb (five-digit appendage) became homology's poster child. Despite radical functional shifts, the same bones persist:
- Horses: Stand on a single elongated digit (middle finger/toe)
- Whales: Fingers encased in flipper blubber
- Bats: Digits stretched to support wing membranes 4 7
| Species | Forelimb Function | Homologous Bones | Modifications |
|---|---|---|---|
| Human | Grasping | Humerus, radius, ulna, carpals, phalanges | Thumb opposability |
| Horse | Running | Humerus, radius, ulna, metacarpals | Single weight-bearing digit |
| Bat | Flight | Humerus, radius, digits 2-5 | Digit elongation for wing membrane |
| Whale | Swimming | Humerus, radius, ulna, phalanges | Shortened bones, hyper-phalanges |
2. Serial Homology: Nature's Repeating Patterns
Within individual organisms, repeated structures like insect segments or vertebrae proved homologous through developmental pathways. Centipede legs, insect mouthparts, and crustacean appendages all derive from ancestral body segments 2 .
3. Molecular Homology: The DNA Revolution
The discovery of pax6 genes controlling eye development in vertebrates and insects revealed "deep homology"—shared genetic machinery despite 500 million years of divergence 2 5 .
| Term | Definition | Example |
|---|---|---|
| Orthology | Genes diverged after speciation | Human & mouse hemoglobin genes |
| Paralogy | Genes diverged after duplication within a genome | Hemoglobin vs. myoglobin in humans |
| Xenology | Genes transferred horizontally between species | Antibiotic resistance genes in bacteria |
The Crucial Experiment: How a Fruit Fly Gene Built a Mouse Eye
Methodology: Genetic Cross-Testing
In a landmark 1995 experiment, researchers tested whether the pax6 gene—essential for eye development in fruit flies—could trigger eye formation in distantly related vertebrates 5 6 :
- Gene Isolation: Pax6 extracted from mice.
- Embryonic Injection: Mouse pax6 inserted into Drosophila embryos.
- Phenotype Analysis: Examined developing fly tissues for eye structures.

Drosophila melanogaster
The common fruit fly, a model organism in genetic research where the pax6 experiment was conducted.

Mus musculus
The house mouse, whose pax6 gene was used in the cross-species experiment.
Results and Analysis
Shockingly, mouse pax6 induced ectopic eyes in flies—including compound lenses and photoreceptors—despite 550 million years of evolutionary separation 6 . This proved:
- Eye development relies on conserved genetic pathways.
- Homology operates at molecular levels unseen by anatomists.
- Evolution repurposes deep ancestral "toolkits" for new functions.
| Parameter | Fruit Fly (Control) | Fly + Mouse pax6 | Significance |
|---|---|---|---|
| Eye Formation | Normal compound eyes | Extra functional eyes | Mouse gene works in arthropods |
| Tissue Specificity | Eye-antennal disc only | Legs, wings, antennae | pax6 is a master eye regulator |
| Structural Accuracy | Native fly eye structure | Correct lens cells | Genetic instructions conserved |
The Scientist's Homology Toolkit
| Tool | Function | Example Use |
|---|---|---|
| CRISPR-Cas9 | Gene editing | Disrupt pax6 to test eye homology |
| GFP Tagging | Fluorescent protein tracking | Visualize homologous cell lineages |
| Phylogenetic Software | DNA sequence alignment & tree-building | Distinguish orthologs from paralogs |
| Micro-CT Scanning | 3D anatomical reconstruction | Compare bone homologies across fossils |
| RNA Interference | Gene silencing | Test developmental homology in embryos |
Challenges and Controversies
Homology faces ongoing debates:
- Genetic Dissociations: Barnacle shells and mollusk shells look homologous but form via different genes—convergent evolution masquerading as true homology .
- Hierarchy Conflicts: A structure may be homologous at one level (e.g., genetic) but not another (e.g., anatomical) 5 6 .
- Circular Reasoning Critics: Creationists argue homology assumes evolution to prove it—though molecular phylogenetics now provides independent validation .

Convergent vs. Homologous Traits
The wings of bats (mammals) and birds evolved independently, showing similar function but different evolutionary origins—an example of convergent evolution.

Molecular Phylogenetics
Modern DNA sequencing provides independent validation of homology relationships inferred from anatomy.
Homology Today: Beyond Bones
Modern biology integrates homology across scales:
- Evo-Devo: How homologous genes orchestrate embryonic development.
- Bioinformatics: Algorithms detecting DNA homologies predict protein functions.
- Hierarchical Framework:
- Morphological Homology (organism level)
- Genealogical Homology (population genetics)
- Phylogenetic Homology (species divergence) 5
As Nobel laureate François Jacob noted:
"Evolution works like a tinkerer recycling old parts into new inventions."
Homology remains that unifying thread—connecting Aristotle's comparative sketches to CRISPR-edited genomes—revealing life's breathtaking unity amid its staggering diversity.

Image: Pierre Belon's 1555 comparative skeleton diagram of a human and bird, one of the earliest homology illustrations.