Research Articles
Optogenetic Control of Embryonic Signaling: Precision Tools for Developmental Biology and Disease Modeling
This article explores the transformative application of optogenetics for manipulating signaling pathways in embryonic development.
miRNAscope for microRNA Detection: A Comprehensive Guide to Spatial miRNA Analysis in Research and Diagnostics
This article provides a comprehensive analysis of the miRNAscope assay, an advanced in situ hybridization technology for detecting microRNAs with single-cell resolution in FFPE and frozen tissues.
A Comprehensive Guide to Interpreting RNAscope Control Probe Results for Accurate Gene Expression Analysis
This article provides researchers, scientists, and drug development professionals with a complete framework for interpreting RNAscope control probe results, a critical step for ensuring data integrity in spatial gene expression...
RNAscope vs. SABER FISH: A Detailed Comparison of Sensitivity and Application
This article provides a comprehensive comparison for researchers and drug development professionals evaluating high-sensitivity RNA in situ hybridization (ISH) platforms.
Mastering RNAscope for FFPE Samples: A Comprehensive Guide from Basics to Advanced Applications
This article provides a comprehensive guide for researchers and drug development professionals utilizing RNAscope in situ hybridization technology on formalin-fixed paraffin-embedded (FFPE) samples.
RNAscope Hybridization Temperature: The Critical Factor for Optimal Signal and Specificity
This article provides a comprehensive guide for researchers and drug development professionals on the pivotal role of hybridization temperature in the RNAscope in situ hybridization assay.
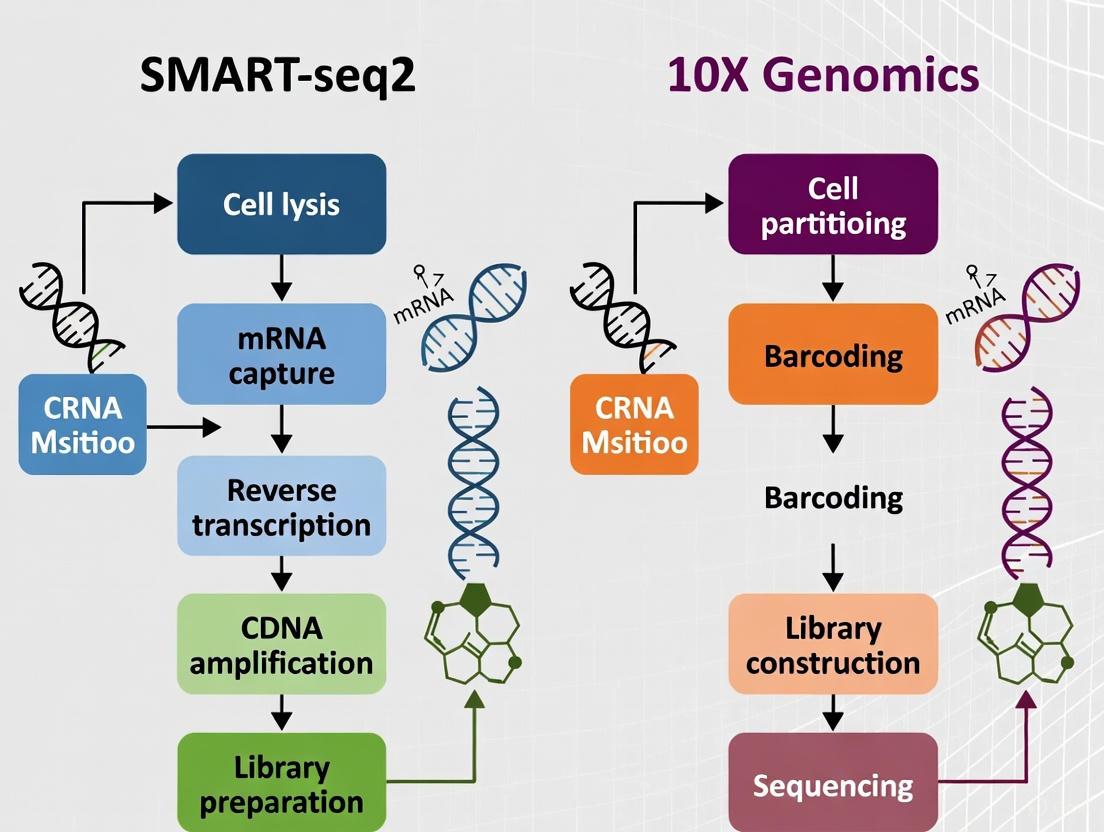
SMART-seq2 vs. 10X Genomics for Embryo Cells: A Definitive Guide for Developmental Biologists
Selecting the optimal single-cell RNA sequencing platform is crucial for advancing research in human embryonic development and stem cell-based embryo models.
In Situ Hybridization Sensitivity Comparison: A Guide for Researchers and Developers
This article provides a comprehensive comparison of in situ hybridization (ISH) sensitivity for researchers, scientists, and drug development professionals.
Batch Effect Correction in Multi-Site Embryo Studies: Strategies for Robust Data Integration and Biological Discovery
Integrating single-cell and spatial transcriptomic data from multiple sites and studies is essential for building comprehensive models of embryonic development but is severely challenged by technical batch effects.
Preserving Integrity: A Comprehensive Guide to Preventing RNA Degradation in Embryo Samples
This article provides a systematic guide for researchers and drug development professionals on preventing RNA degradation during embryo sample preparation.